Strong inferences about trait state evolution can be initiated by tracing the number and direction of current trait state evolution from the most likely ancestral trait state (i.e., homology), if a highly supported species tree among different species is provided. Furthermore, such phylogeny-driven trait evolution can also provide insight into convergence or reversionary evolution (i.e., homoplasy) (Wake et al., 2011; Guerreroa and Hahna, 2018). However, phylogeny-based trait inference is often limited by the widespread occurrence of gene-tree discordance (GTD). GTD has been widely found in organismal phylogenies, especially in phylogenomics, where thousands to millions of genes are typically used to reconstruct phylogenies. The rampant GTD, especially when the incongruent traits occur in these discordant gene regions, known as hemiplasy (Guerrero and Hahn, 2018; Wu et al., 2018), can amplify false trait state inference even in highly supported species trees. However, although such effects may be intrinsic and profound (Wu et al., 2018), few studies have documented the effects of gene tree discordance on phylogeny-based trait state inference.
The most common explanations for GTD are incomplete lineage sorting (ILS) and introgressive hybridization (IH) (Degnan and Rosenberg, 2009), aside from gene tree estimation error which likely was caused by a paucity of informative sites at selected gene loci, misidentification of orthologs due to paralogy, and poor fit of the substitution model to the sequence data. ILS is strongly influenced by effective population size and evolutionary time (Pamilo and Nei, 1988), as it occurs through the random transmission of segregating ancestral variation between successive nodes in a species tree (Maddison, 1997). As a consequence, ILS would arguably be exaggerated in groups that underwent recent fast radiations (e.g., Pease et al., 2016; Wang et al., 2018; Wu et al., 2018; Li et al., 2021). However, IH can also lead to GTD when ancestral or recent loci are shared between diverging lineages and there is a conflict between evolutionary history of introduced genes and those from native genomes (e.g., Wu et al., 2018; Feng et al., 2019). If ILS is a major source of GTD across the phylogeny, the probability of hemiplasy for incongruent trait evolution along a targeted phylogeny should be considered (Robinson and Anne, 2011; Mendes et al., 2016; Hahn and Nakhleh, 2016; Copetti et al., 2017; Guerrero and Hahn, 2018; Wu et al., 2018), rather than simply mapping the states of traits on a single species tree to deduce their evolutionary trajectory (Wake et al., 2011; Guerrero and Hahn, 2018). In particular, it is important to note that if the genes underlying trait variation occur in these discordant genomic regions, unrelated species are likely to coalesce, potentially leading to incorrect inferences about ancestral trait states. When successive speciation events and associated lineage sorting of genetic polymorphisms leads to a discordance between species and gene trees, this is known as hemiplasy (Avise and Robinson, 2008). In this sense, hemiplasy is a genuine form of trait homology (due to common ancestry) in a species tree, but it is still not homoplasy at the level of the gene tree (Avise and Robinson, 2008; Wu et al., 2018). To better explain the evolution of incongruent traits when GTD occurs, Guerrero and Hahn (2018) developed a method, the hemiplasy risk factor (HRF), to highlight individual branches of the phylogenetic tree along which hemiplasy is likely to occur and contrast them with the probability of convergence of the observed incongruent trait. Thus, quantifying the HRFs in a coalescent unit tree can help us explore the evolution of many incongruent states of traits of interest.
The phylogenetic relationship between the monotypic Himalayan genus Milula Prain and the globally distributed genus Allium L. has been widely debated. The taxonomic notation of Milula has always been problematic due to the conspicuously elongated spicate compared to the mostly umbel or capitate inflorescences in Allium (Krause, 1930; Hutchinson, 1959; Stearn, 1960; Traub, 1972; Takhtajan, 1987; Friesen et al., 2000). The growing body of integrated evidence strongly supports the revision of the monotypic species M. spicata Prain to A. spicatum (Prain) N. Friesen (Friesen et al., 2000, 2006). Molecular evidence suggests that A. spicatum belongs to subg. Cyathophora R.M. Fritsch, which consists of five diploid species, including A. mairei Léveillé, A. rhynchogynum Diels, A. spicatum, A. cyathophorum Bureau & Franch., and A. farreri Stearn, and one tetraploid species of hybrid origin (A. tetraploideum M.J. Li & X.J. He) (Huang et al., 2014; Li et al., 2019a). Despite the abundance of evidence supporting the genetic affinities between A. spicatum and its relatives, the occurrence of GTD confounds evolutionary trajectories of traits between them. For example, A. mairei and A. spicatum are successively sister to the most recently diverged sister species A. cyathophorum and A. farreri in the nuclear ITS tree; however, A. spicatum and A. farreri constitute the most recently diverged sister species, and A. mairei and A. cyathophorum are successively sister to them in the chloroplast DNA (cpDNA) tree and the low copy nuclear gene (At103) tree (Li et al., 2016). In many studies, such GTDs have been treated as phylogenetic noise (Knowles and Kubatko, 2010), and only a few studies have explained the underlying mechanisms of such GTDs and evaluated the impact of such GTDs on the phylogeny-based inference of trait states.
Here, using 1262 single-copy genes (SCGs), chloroplast genomes and population genetic data, we further investigated GTD across the phylogeny of Allium subg. Cyathophora. Furthermore, we used coalescent simulations to explore the underlying mechanisms of such GTD within subg. Cyathophora and turned the clearly present GTD into an information source of ILS to elucidate the controversial character evolutionary history of subg. Cyathophora.
2. Materials and methodsIn order to clearly illustrate the operating procedures in this study, a simple analytical flowchart has been generated in Fig. S1. The detailed information is described as follows.
2.1. Transcriptome and chloroplast genome collectionIn this study, a total of five species were selected, including four diploid species (Allium mairei, A. spicatum, A. cyathophorum, and A. farreri) of Allium subg. Cyathophora, and one diploid individual of A. przewalskianum Regel, which was used as an outgroup. Sampling information is provided in Table S1. The transcriptomes and chloroplast genomes of these five species were obtained from NCBI under accession numbers SAMN13698330–SAMN13698335 and MN882559–MN882564, respectively (all from Li et al., 2021).
2.2. Chloroplast assemblyFast-Plast v.1.2.6 (https://doi.org/10.5281/zenodo.973887) was used to assemble the whole chloroplast genomes (cpDNA). Geneious v.10.2.6 (Kearse et al., 2012) was then used for manual validation and consensus sequence generation. The assembled chloroplast genomes were aligned using MAFFT (Katoh and Standley, 2013).
2.3. Transcriptome assembly and SCGs identificationThe paired-end clean reads of each transcriptome were de novo assembled using Trinity v.2.1.5 (Grabherr et al., 2011) with default parameters. TRANSDECODER-v.5.0.2 was used to predict the longest putative CDS and protein sequences in the open read frame (ORF). CD-HIT (Huang et al., 2010) was used to generate the non-redundant CDS and protein sequences with a threshold of 0.95. OrthoMCL (Li et al., 2003) was used to identify orthologous sequence groups for the five species, and then SCGs were extracted for downstream analyses using a custom Perl script. Each orthologous sequence group was aligned using PRANK (Löytynoja and Goldman, 2008) with the '-codon' parameter. The orthologous clusters were filtered by removing those with length < 300 bp and '-' character in each sequence > 50%.
2.4. Phylogenetic analysisThe concatenation and coalescence methods were used to infer the species tree. For the concatenation approach, all SCGs were concatenated into a supermatrix with the alignment length in 950, 648 bp, and then a maximum likelihood (ML) tree was inferred in RAxML v.8 (Stamatakis, 2014) using the GTR-GAMMA model and '-f a' for rapid 100 bootstrapping to search for the ML tree with the highest score (hereafter referred to as the nuclear ML tree). For the coalescent approach, we first reconstructed individual ML trees for each of the SCGs in RAxML with the same parameters as described above. After filtering the SCG trees with bootstrap support (BS) less than 70% at each node using a custom Perl script, we used the remaining SCG trees to infer a coalesced tree in MP-EST 2.0 (Liu et al., 2010a) with a maximized pseudo-likelihood function (henceforth referred to as the nuclear MP-EST tree). Similarly, a ML tree based on the aligned chloroplast genomes was also inferred in RAxML v.8 (hereafter called cpDNA tree). We then inputted the SCG trees with BS ≥ 70% and the topology of the cpDNA tree into MP-EST 2.0 to generate a cpDNA MP-EST tree with the same branch length in coalescent units and topology as the cpDNA tree. The "tree.noclock2clock" function in the R package phybase (Liu et al., 2010b) was used to convert a non-clocklike tree into a clocklike (ultra-metric) tree using an ad hoc approach. Densitree (Bouckaert, 2010) was used to describe all ultra-metric trees together to illustrate phylogenetic heterogeneity.
2.5. Synonymous substitutions per synonymous site (KS)To further validate the phylogenetic relationships among the target species, we calculated the percentage of synonymous substitutions per synonymous site (KS) between given pairwise orthologs (e.g., Guan et al., 2019). We used INPARANOID v.4.1.1 (Sonnhammer and Östlund, 2015) to infer interspecific orthologous sequences, and single-copy orthologs were extracted using a custom Perl script. All pairwise orthologous sequences were aligned using PRANK with the '-codon' parameter. Then, a ML method was implemented in the YN00 program of the PAML package v.4.1 (Yang, 2007) to compute KS between orthologous sequence pairs. The resulting KS were plotted as a histogram (KS plot) with KS ≥ 1.0 removed due to sparse data. We implemented Gaussian mixture distributions using an expectation-maximization (EM) algorithm to infer significant peaks in each observed KS distribution in the R package mixtools (Benaglia et al., 2009). A parametric bootstrap (B = 100) of the likelihood ratio statistic was performed using the "boot.comp" function in the R package mixtools to examine the most likely number of Gaussian components that fit each KS distribution. Sizer tests (Chaudhuri and Marron, 1999) were used to further confirm the Gaussian mixture model components across a distribution at various (log transformed) bandwidths to distinguish true data features from noise by visually testing between increases, decreases, or values not significantly different from zero. True peaks were identified as those Gaussian plots that shifted significantly from increase to decrease in the sizer. The significant (alpha = 0.05) features in each observed KS distribution were tested when KS values were ≤ 1.0 and KS bandwidths were 0.01–1.00. The corresponding age of each KS plot was calculated using the formula: KS/generation time/mutation rate, with a generation time of two years and an average mutation rate of 1.25 × 10−8 (which varies from 1 × 10−8 to 1.5 × 10−8) for plants (Ossowski et al., 2010).
2.6. Population phylogenies based on ten nuclear SCGsTo further confirm the widespread occurrence of GTD within Allium subg. Cyathophora, ten SCGs (9266-2, 9201-5, 10076-6, 9341-10, 10096-12, 9360-14, 10110-18, 9259-23, 7189-26, and 7214-27) were downloaded from NCBI under accession numbers MT479228–MT481754 (all from Li et al., 2021) for population phylogenetic analyses. A total of 46 individuals were sampled, including 18 A. cyathophorum, 22 A. farreri, five A. spicatum, and one A. mairei (Table S1). Alignment of each gene was performed in MEGA 6.0 (https://www.megasoftware.net/) and checked by eye. The haplotype file for each gene was generated in DNAsp v.6.0 (http://www.ub.edu/dnasp). The ML tree for each haplotype file was reconstructed in PhyML (Guindon, 2010) using a likelihood-based method.
2.7. Coalescence simulationsTo test whether ILS was a major factor causing GTD across the Allium subg. Cyathophora phylogeny, we performed coalescent simulations following the method developed by Wang et al. (2018; https://github.com/wk8910/ILS_simulation). In brief, we started by counting the topology of the SCG trees using the R package ape (Paradis and Schliep, 2019). Then, we simulated 500, 000 gene trees for both the nuclear and cpDNA MP-EST trees under the multispecies coalescent model using the "sim.coaltree.sp" function in R package phybase. Several approaches were used to test the topological consistency or difference between the observed SCG trees and the simulated gene trees. First, we compared the distributions of the topological frequencies of the observed SCG trees and the simulated gene trees. Second, we fitted a linear regression equation in R for the frequencies of the observed SCG trees and the probabilities of the simulated gene trees. Third, we first calculated the Robinson-Foulds (RF) distances from each of the observed SCG trees respectively to the nuclear (RF1) and cpDNA (RF2) MP-EST trees using the "tree.distance" function in the R package phybase, and then calculated ΔRF = RF1 − RF2 to measure whether each SCG topology was more similar to the nuclear MP-EST tree or the cpDNA MP-EST tree. Finally, a likelihood ratio test (LRT) was performed to assess the goodness of fit of the two simulated gene tree distributions to the observed gene tree distribution by testing the statistic t = −2(L(τn) − L(τm)) with 1000 parametric bootstrap replicates, where τm was a null hypothesis about the goodness of fit of the simulated cpDNA trees to the observed SCG trees, τn was an alternative hypothesis about the goodness of fit of the simulated nuclear gene trees to the observed SCG trees, and L(τm) and L(τn) were their respective log-likelihood tests.
For our coalescent simulations, we expected a higher frequency of the topology same with the cpDNA MP-EST tree if ILS alone could explain the topological incongruence between the cpDNA and nuclear trees. In contrast, an absence or lower frequency of the topology same from the cpDNA MP-EST tree would be expected if IH was present in the evolutionary history of subg. Cyathophora. We therefore counted the number of additional lineages for both the observed SCG trees and the simulated nuclear gene trees using the "deep_coal_count" function in phyloNet v.2.4 (Than et al., 2008). If IH had occurred in the evolutionary history of subg. Cyathophora, we would expect to see more extra lineages in the observed SCG trees than in the simulated nuclear gene trees. To further test whether IH contributed to GTD within subg. Cyathophora, we estimated interspecific relationships by analyzing all observed SCG trees using parsimonious inference of hybridization in the presence of ILS (Yu et al., 2013) in phyloNet, assuming one versus two past hybridization events.
2.8. Reconstructing ancestral trait statePhylogeographic investigation showed that spicate and umbel inflorescences are clearly geographically segregated across the 500 mm Qinghai-Tibet Plateau isohyet (QTP, Li et al., 2019b). Accordingly, their ecological niche (here represented by altitude), seed size, flowering phase, and ovary size also differ (Li et al., 2016). Among these traits, geographic distribution (western QTP/eastern QTP) and inflorescence (spicate/umbel) are categorical; seed size (thousand seed weight: 1.317, 0.948, 0.878, 1.691, 2.009 g), flowering phase (month: 6–9, 8–10, 8–10, 6–8, 6–8), ovary size (3.3, 2.9, 2.1, 4.0, 4.2 mm) and altitude (mean values: 3400, 2287, 4219, 3899, 2703 m) for A. przewalskianum, A. mairei, A. spicatum, A. cyathophorum and A. farreri, respectively, are quantitative traits (Table S2). These traits are considered irreversible because they are stable in the natural populations. We then used the Bayesian Binary Method (BBM) in RASP (Yu et al., 2020) based on the nuclear ML tree to infer the ancestral state of the categorical traits. We ran a total of one million generations, nine hot Markov chains and one cold chain with temperature increments of 0.1 for each analysis. For quantitative traits, we first calculated their square root values and then used the "fastAnc" function in the R package phytools (Revell, 2012) to quickly estimate their ancient state based on the nuclear ML tree.
2.9. Identifying genes under natural selectionTo examine the effect of natural selection on spicate-specific lineages throughout the phylogeny of Allium subg. Cyathophora, we calculated ω (dN/dS ratios), a criterion of selection pressure, using the CODEML program in the PAML package on the observed SCG trees with BS ≥ 70%. In general, ω = 1 indicates neutral selection, ω > 1 positive selection, and ω < 1 negative selection (Yang, 2007). The ω values were calculated by implementing a null model (Ma0, no lineages with specifically high nonsynonymous substitutions due to natural selection) and an alternative model (Ma, spicate was counted as a lineage-specific trait and was likely under positive selection, which consequently could have led to GTD) on each of the SCG gene trees using a strict branch-site approach used to estimate and predict recent radical evolution (Zhang, 2000). The LRTs were used to estimate the significance of ω values (P < 0.05) by calculating the chi-squared (χ2) distribution between 2△L and df. The significance P value was adjusted using the false discovery rate (FDR). The positively selected sites were identified using Bayes Observed Bayes (BEB) with posterior probability less than 0.08 (Yang et al., 2005).
2.10. Quantifying and validating hemiplasyILS and neutral selection are two important assumptions for quantifying hemiplasy (Guerreroa and Hahna, 2018). Therefore, we just used the SCGs under neutral selection to infer a MP-EST tree (hereafter called neutral MP-EST tree). In the present of ILS, the Hemiplasy Risk Factors (HRFs) were calculated and visualized for the internal nodes of the neutral MP-EST tree using R package pepo (https://github.com/guerreror/pepo), where an average mutation rate of 1.25 × 10−8 (the substitution rate per nucleotide site per year, s/s/y) for plants (ranging from 1 × 10−8 to 1.5 × 10−8; Ossowski et al., 2010), and the effect population size (Ne) of 133, 870 (with 95% highest posterior density interval 6310–978, 952) for the most recently common ancestor of subg. Cyathophora (Wang et al., 2015) were used to calculate the 2Ne mutation rate of 3.21 × 10−3, which varied from 1.5 × 10−4 to 2.35 × 10−2.
To evaluate the role of hemiplasy on the incongruent characters along the phylogeny of Allium subg. Cyathophora, we selected the genes likely to be correlated with inflorescence development to infer tree topologies. We first ran the SCG functions with the default parameters using the TrEMBLEMBL database (http://kr.expasy.org/sprot/) as a background. To further detect the biological processes of the SCGs, Gene Ontology (GO) enrichment was performed. R package clusterProfiler (Yu et al., 2012) was used for GO pathway enrichment analysis. The GO background of the genome of the species was obtained using the eggNOG mapper (Huerta-Cepas et al., 2017) with the default parameters. To further find genes related to inflorescence development, we used Interproscan (https://www.ebi.ac.uk/interpro/about/interproscan/) to search all SCGs to map genes of the ARF (auxin response factor) family, the SST family (Li, 2012), the PLC (phosphoinositide phospholipase C) family (Li et al. 2018), the FT/TFL1 (flowering locus T/terminal flower 1) family, the SPB (squamosa promoter binding protein) family, the MADS box, and the PEBP (phosphatidy lethanolamine-binding protein) family, respectively.
3. Results 3.1. Transcriptome assemblyClean sequences comprising of 7.34, 7.67, 6.76, 6.67 and 13.1 Gb cDNA bases were obtained for Allium cyathophorum, A. farreri, A. spicatum, A. mairei and A. przewalskianum. The de novo assembly produced 469, 514, 368, 926, 267, 859, 300, 360 and 477, 740 contigs with N50 values of 833, 1157, 736, 957 and 1178 bp for these species, respectively (Table S3).
3.2. Phylogeny of Allium subg. CyathophoraA total of 1262 SCGs were identified using OrthoMCL for the four diploid species of Allium subg. Cyathophora, with A. przewalskianum used as the outgroup. Among these 1262 SCG trees, 840 were found to have BS ≥ 70%. The consistent topology between the concatenation and coalescence-based trees (Fig. S3) strongly supported that A. cyathophorum and A. farreri were the closest species, and A. mairei and A. spicatum were successively sister to them (Fig. 1B and C). However, the cpDNA tree revealed that A. spicatum and A. farreri were the closest relatives, and they were each sister to A. cyathophorum, and finally the three were sister to A. mairei (Fig. 1D). This was consistent with the topology based on the cpDNA fragments (Li et al., 2019a, b). Ultimately, the nuclear and cpDNA trees were clearly phylogenetically incongruent.

|
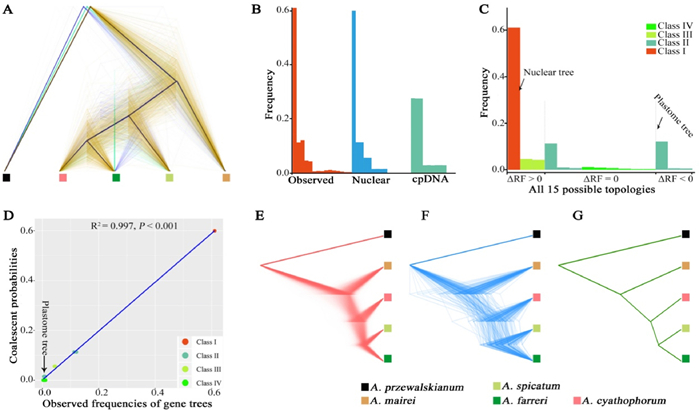
| Fig. 1 Phylogenetic relationships among the four diploid species of the genus Allium subg. Cyathophora based on transcriptome analyses. (A) The density plots of synonymous nucleotide substitutions (KS) of the pairwise orthologs. The divergence time was estimated using the formula: KS/generation time/mutation rate. (B) The ML tree based on the concatenation of 1262 SCGs. (C) The MP-EST tree based on 840 SCG trees with the bootstrap support (BS) ≥ 70% at each node. (D) The ML tree of the whole-chloroplast genome. BS at each node in these three trees are 100%. |
The species pairwise orthologs identified by INPARANOID were 13, 525, 13, 097 and 12, 761 in Allium mairei-A. spicatum, A. mairei-A. cyathophorum and A. mairei-A. farreri, respectively; the pairwise orthologs in A. spicatum-A. cyathophorum and A. spicatum-A. farreri were 14, 109 and 13, 711, respectively; and the pairwise orthologs of A. cyathophorum-A. farreri were 15, 204 (Table S4). Our Gaussian mixture modeling identified a unique peak for synonymous site divergence (KS) for each pairwise species, and each was significantly supported by the Sizer tests (Fig. S2 and Table S5). The interspecific divergence ages for in A. mairei-A. spicatum, A. mairei-A. cyathophorum and A. mairei-A. farreri were respectively estimated to be approximately 4.80 (4.00–6.00), 4.24 (3.53–5.30) and 4.48 (3.73–5.60) million years ago (Ma), respectively based on the corresponding KS peak values of 0.120, 0.106, and 0.112; the divergence ages for A. spicatum-A. cyathophorum and A. spicatum-A. farreri were at ~3.32 (2.77–4.15) and ~3.44 (2.87–4.30) Ma respectively, with the corresponding KS values of 0.083 and 0.086; the divergence time of A. cyathophorum and A. farreri around at 1.68 (1.40–2.10) Ma, with the KS peak of 0.042 (Fig. 1A and Tables S5–S6). Such KS-based interspecific divergence, such that the closer the phylogenetic relationships, the smaller the KS values corresponding to the latter divergence times, also provided support for the phylogeny of subg. Cyathophora revealed by the SCGs (Fig. 1).
3.4. Genome-wide GTDAmong the SCG trees, a high degree of phylogenetic discordance was found: 60.90% of all SCG trees were congruent with the nuclear MP-EST tree, while the remaining 30.91% of trees showed discordant topologies with the nuclear MP-EST tree (Fig. 2A and Table 1). A similar result was also observed among the SCG trees with BS ≥ 70% (Table 1). All SCG tree topologies were classified into 15 topological types (Table 1). According to the phylogenetic position of the spicate-specific Allium spicatum, these 15 types were further divided into four classes: Class Ⅰ, 1 out of 15 types, agreed with the nuclear MP-EST tree in which A. spicatum was directly sister to the A. cyathophorum-A. farreri clade; Class Ⅱ, 6 out of 15 types, showed that A. spicatum was sister to either A. cyathophorum or to A. farreri; Class Ⅲ, 2 out of 15 types, showed that A. spicatum was the most distant species in the phylogenies, or A. spicatum was clustered together with A. mairei, respectively; and Class Ⅳ, 6 out of 15 types, exhibited that A. mairei was sister to A. farreri, A. cyathophorum or A. spicatum, respectively, at the most recent node in the phylogenies (Table 1). The SCG trees with the second highest proportion (12.12% of all SCG trees; 9.76% of the SCG trees with BS ≥ 70%) had the same topology as the cpDNA tree (A. spicatum and A. farreri were the closest relatives at the most recent branch). Notably, the proportion of the gene trees resembling the cpDNA tree was slightly higher than the proportion of gene trees in which A. spicatum was directly sister to A. cyathophorum at the most recent node (11.25% of all SCG trees; 7.38% of the SCG trees with BS ≥ 70%) (Table 1). Furthermore, A. mairei was always positioned in the earliest diverged branch in the top three SCG tree topologies, accounting for the highest proportion (61.09% + 12.12% + 11.25% of all SCG trees; 72.98% + 9.76% + 7.38% of the SCG trees with BS ≥ 70%) (Table 1).
|
| Fig. 2 Incomplete lineage sorting (ILS) model for gene-tree discordance (GTD) across the phylogeny of Allium subg. Cyathophora, using A. przewalskianum as an outgroup. (A) The densitree of 1262 SCG trees. Black lines indicate the consensus tree. Yellow lines indicate the most frequent topology, followed by the second and the third ones indicted by blue and green lines (Table 1). (B) The distributions of the observed 1262 ML SCG trees, the simulated gene trees respectively from the nuclear and cpDNA MP-EST trees, respectively. (C) Ordering of the empirical frequencies of all 15 possible gene tree topologies according to the frequency of each topology in ΔRF > 0 (gene trees more similar to the nuclear MP-EST tree), ΔRF = 0 (trees equally similar to the nuclear and cpDNA MP-EST trees) and ΔRF < 0 (trees more similar to the cpDNA MP-EST tree). All 15 possible topologies are classified into four classes (detail in Table 1) and are indicated by different colors. (D) A fitted linear regression between the simulated frequencies of gene topologies of different classes based on the coalescence model and the corresponding observed frequencies. The arrow indicates the cpDNA topology. (E) Simulated gene trees with the same topology as the cpDNA tree. (F) Observed gene trees having the same topology as the cpDNA tree. (G) Observed cpDNA tree using the whole chloroplast sequence. All trees in (E, F, G) are scaled to the same total length. |
| Class | Topology | SCG trees | SCG Trees with BS ≥ 70% | Simulated nuclear trees | Simulated cpDNA trees |
| Class Ⅰ | ((((Cy, Fa), Sp), Ma), Pr)a | 61.09% | 72.98% | 59.98% | 27.60% |
| Class Ⅱ | ((((Fa, Sp), Cy), Ma), Pr)b | 12.12% | 9.76% | 11.33% | 27.52% |
| ((((Cy, Sp), Fa), Ma), Pr) | 11.25% | 7.38% | 11.34% | 2.80% | |
| (((Cy, Sp), (Fa, Ma)), Pr) | 0.95% | 0.36% | 1.51% | 2.88% | |
| (((Fa, Sp), (Cy, Ma)), Pr) | 0.63% | 0.48% | 1.49% | 2.89% | |
| ((((Cy, Sp), Ma), Fa), Pr) | 0.71% | 0.00% | 1.52% | 2.87% | |
| ((((Fa, Sp), Ma), Cy), Pr) | 0.55% | 0.36% | 1.49% | 2.86% | |
| Class Ⅲ | ((((Cy, Fa), Ma), Sp), Pr) | 4.60% | 3.57% | 5.60% | 2.86% |
| (((Cy, Fa), (Sp, Ma)), Pr) | 4.28% | 3.57% | 5.59% | 0.03% | |
| Class Ⅳ | ((((Ma, Sp), Fa), Cy), Pr) | 1.11% | 0.36% | 0.03% | 0.03% |
| ((((Ma, Cy), Fa), Sp), Pr) | 0.87% | 0.24% | 0.02% | 0.03% | |
| ((((Ma, Fa), Cy), Sp), Pr) | 0.71% | 0.24% | 0.02% | 0.03% | |
| ((((Ma, Cy), Sp), Fa), Pr) | 0.48% | 0.12% | 0.03% | 0.03% | |
| ((((Ma, Sp), Cy), Fa), Pr) | 0.32% | 0.00% | 0.02% | 0.03% | |
| ((((Ma, Fa), Sp), Cy), Pr) | 0.32% | 0.00% | 0.02% | 0.02% | |
| a Indicates the nuclear genome topology. b Indicates the chloroplast topology. |
|||||
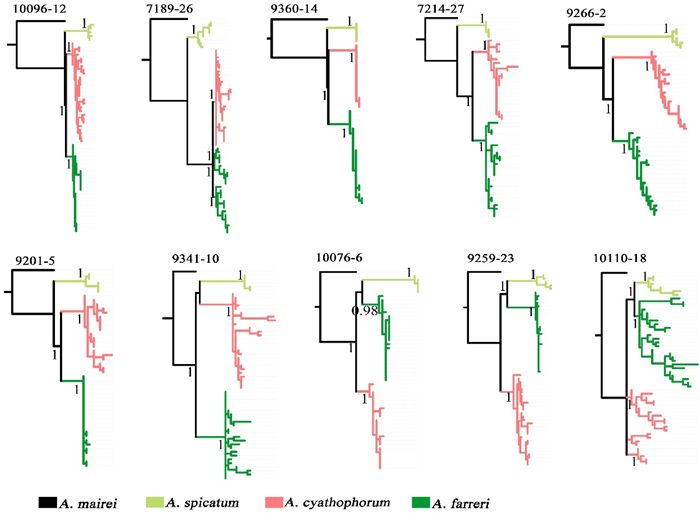
To further validate the phylogenetic discordance at the population level, we obtained sequence variations at ten SCG loci to reconstruct the phylogenies of multiple individuals for each of Allium spicatum, A. cyathophorum and A. farreri, all of which have high phylogenetic discordance, and an individual of A. mairei was used as an outgroup. The resulting phylogenies were highly consistent with the phylotranscriptomic analyses. No common haplotypes were found among these species (Figs. 2A and 3). We found that six (10096-12, 7189-26, 9360-14, 7214-27, 9266-2 and 9201-5) of the ten gene trees were topologically consistent with the nuclear MP-EST tree, in which A. mairei and A. spicatum were successively sister to the most recently diverged species A. cyathophorum and A. farreri (Fig. 3). Among the remaining four gene trees, two (9341-10 and 10076-6) showed that A. spicatum and A. cyathophorum were joined together at the youngest node, and both were sister to A. farreri. In the other two trees (9259-23 and 10110-18) A. spicatum and A. farreri were clustered together in the youngest node, and then both were sister to A. cyathophorum, and finally the three to A. mairei (Fig. 3).
|
| Fig. 3 Phylogenetic relationships for several individuals of each species of Allium subg. Cyathophora based on the ten SCGs using Sanger sequencing. Numbers on the branches indicate posterior probability (PP). |
Our statistical test showed that the distribution of the simulated gene trees based on the nuclear MP-EST tree had a much better fit to the distribution of the observed SCG trees than the simulated gene trees generated from the cpDNA tree (Fig. S4). In short, a striking agreement between the distributions of simulated nuclear trees and observed SCG trees was found when simulating 500, 000 gene trees each based on the nuclear MP-EST tree and the cpDNA tree just under ILS alone (Fig. 2B and Table 1). We found that ΔRF > 0 occurred in the majority of the observed SCG trees (~70.0%), suggesting that they had the same topology as the nuclear MP-EST tree. However, the proportion of SCG tree topologies closer to the cpDNA tree with ΔRF < 0 was small, about 13.3%, and the remaining 16.7% of all gene trees had the same RF distance (ΔRF = 0) from the nuclear MP-EST tree and the cpDNA tree (Fig. 2D).
The overall correlation coefficient for the coalescently simulated nuclear trees and the observed SCG trees was 0.997 (P < 0.001, Fig. 2D), which was very close to 1, indicating a good fit between the multispecies coalescent simulations and the observed SCG trees. The distributions of pairwise tree distances between the nuclear MP-EST tree and the gene tree for both the observed and simulated nuclear trees showed a clearly consistent pattern (Fig. S4). Furthermore, both the observed (30.71%, 258/840) and simulated (30.11%, 150, 553/500, 000) gene trees based on the nuclear MP-EST tree showed approximately the same number of extra lineages. All these results consistently suggested that ILS was a major factor of genome-wide GTD within subg. Cyathophora. Interestingly, among the simulated gene trees based on the cpDNA tree, we found that the proportion of gene trees like the cpDNA tree (27.52%) was approximately equal to that like the nuclear MP-EST tree (27.60%; Fig. 2B and Table 1). Our converted gene tree branches of the cpDNA-like tree were very similar to those of the cpDNA tree in both the observed and simulated gene trees (Fig. 2E, F, G). Both results supported that the anomalous cpDNA tree was a product of ILS.
We performed phylogenetic network analysis to further exclude the causal role of IH in genome-wide GTD. Our phyloNet-based phylogeny was consistent with the nuclear MP-EST tree if a past hybridization event occurred in the evolutionary history of subg. Cyathophora, in which no hybridization was found between the spicate-specific Allium spicatum and the other umbel species. However, an ancient genomic exchange from the common ancestor of the clade of A. spicatum–A. cyathophorum–A. farreri to A. mairei was detected with a small fraction of 17.27% (Table S7). When two past hybridization events occurred, a spurious phylogenetic network was generated that was highly discordant with the nuclear MP-EST tree, and thus we rejected the two hybridization event hypothesis (Table S7).
In conclusion, our multispecies coalescent simulations and phyloNet-based network analyses both support that ILS is the major cause of genome-wide GTD, as well as the phylogenetic incongruence between cpDNA and nuclear.
3.7. Genes under the positive selectionDepending upon the branch with spicate being tested across 840 SCG trees with BS ≥ 70%, some selection was identified on ~22.5% of all loci (189 out of 840); genes where ω > 1 and P < 0.05 were ~5.24% of all loci (44 out of 840), and genes with ω > 1 and P < 0.01 were ~2.98% (25 out of 840) in our dataset (Table S8). Five tree topological types were observed in these positively selected gene sites and they coincided with three of the four classes (Table S9). Interestingly, the proportion of each class in these 44 positively selected sites was highly consistent with their proportion in 840 SCG trees: class I accounted for 72.7%, class II for 18.2% (11.4% + 6.8%) and class III for 9.1% (6.8% + 2.3%) (Table S9); their corresponding abundances in 840 SCG trees were 72.98%, 18.34% (9.76% + 7.38% + 0.36% + 0.48% + 0.36%) and 7.14% (3.57% + 3.57%), respectively (Table 1). With respect to these results, we found no significant difference between the proportions of genes under positive selection within the nuclear MP-EST like trees (5.22%, 32/613) and within the cpDNA-like gene trees (5.19%, 8/154).
Gene Ontology (GO) enrichment showed that the gene functions of these positively selected genes were enriched in different molecular biological processes (Table S9). The positively selected genes that showed discordant topologies with the nuclear MP-EST tree, were enriched in detoxification proteins, oleosin, transmembrane proteins, chromosome metabolism, cell cycle control, binding and catalytic activity (Table S9).
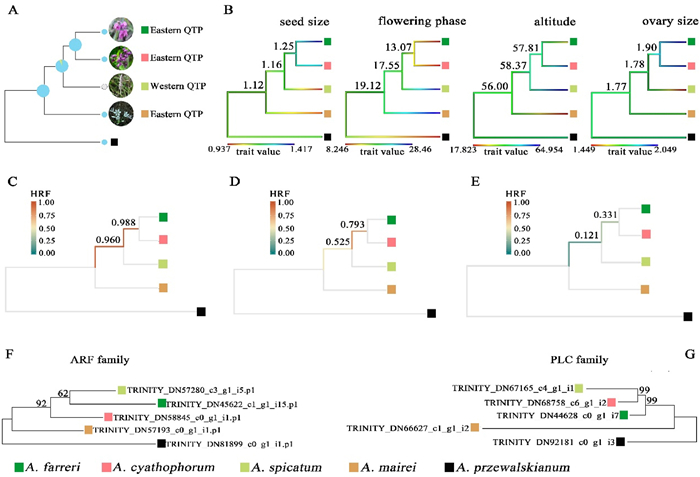
3.8. Ancestral trait states and hemiplasy for the incongruent traitsThe RASP-based ancestral trait inferences strongly indicated (BS = 100%) that the ancestral inflorescence in each internode was most likely in the umbel state and distributed in the eastern QTP (Fig. 4A). In short, we found that the most recent common ancestor (MRCA) of Allium cyathophorum and A. farreri was 99.53% umbel + 0.47% unknown; the MRCA of A. spicatum and A. cyathophorum-A. farreri was 86.98% umbel +13.02% spicate; and the MRCA of the subg. Cyathophora was 96.13% umbel +3.87% unknown. For the ancestral states of the quantitative traits, different values were quantified for each of the branch tips: 1) A. spicatum has the smallest seed size, which is similar to A. mairei, but significantly different with the larger seed size in A. cyathophorum, and the largest one in A. farreri; 2) A. spicatum shows a later flowering phase from August to October, which is similar to A. mairei, but different with the earlier flowering phase of A. cyathophorum and A. farreri, whose flowering period ranges from June to August; 3) A. spicatum is distributed in the regions with the highest altitudes, which is similar to A. cyathophorum, but different from A. farreri and A. mairei, which are chiefly distributed across lower altitudes; 4) A. spicatum has the smallest ovary size, which is different with the intermediate ovary size in A. mairei, and the largest ovary size in A. cyathophorum and A. farreri. We observed that trait values of the branch tips mostly were near end of observed value ranges, whereas the trait values of the internal nodes were nearer the mean of trait distributions (Fig. 4B). These results suggest that an ancestral trait may have evolved along two contrast directions to form derived traits.
|
| Fig. 4 Trait evolution and estimation of HRFs for hemiplasy. (A–B) The phylogeny-based ancestral trait inferences for the (A) categorical traits (inflorescence and geographical distribution) and (B) quantitative traits (seed size, flowering phase, altitude, and ovary size). Numbers on the branches indicate the ancestral trait values. (C–E) Quantification of hemiplasy for trait transition along the phylogeny with constant substitution rate (1.25 × 10−8) and different 2Ne (C) 1.5 × 10−4 (D) 3.21 × 10−3 and (E) 2.35 × 10−2. (F) A representative tree topology of the ARF gene family and (G) one of the PLC gene family, probably related with inflorescence development. |
We calculated the HRFs for each internal branch of the neutral MP-EST tree in the presence of ILS. For a given substitution rate per nucleotide site per year (s/s/y) of 1.25 × 10−8, we found HRF values decreased with increasing effective population size. In short, when parameterized with the lowest 2Ne mutation rate, we observed the highest values of HRF of 0.988 at the internal branch of Allium cyathophorum and A. farreri, and 0.960 at the internal branch of A. spicatum and A. cyathophorum-A. farreri (Fig. 4C). Similarly, when using the median 2Ne mutation rate, HRF at the internal branch of A. cyathophorum and A. farreri was 0.793, and the HRF for the internal branch of A. spicatum and A. cyathophorum-A. farreri was 0.525 (Fig. 4D). Perhaps unsurprisingly, parametrizing with the largest 2Ne mutation rate returned the lowest HRFs: 0.331 at the internal branch of A. cyathophorum and A. farreri, and 0.121 at the internal branch of A. spicatum and A. cyathophorum-A. farreri (Fig. 4E).
Gene Ontology (GO) enriched the neutral genes in biological process, cellular component and molecular function (Fig. S5). The ARF (auxin response factor, they bind to auxin response DNA elements in the promoters of auxin-regulated genes and either activate or repress transcription of these genes depending on a specific domain in the middle of the protein) and PLC (phospholipase C) families have been found to play key roles in regulating plant growth and development. We found that two genes each from the ARF and PLC families, respectively, showed the topologies discordant with the species tree (Fig. 4F and G). In these two trees, A. spicatum was either linked to A. farreri (Fig. 4F) or to A. cyathophorum (Fig. 4G).
4. Discussion 4.1. ILS for the genome-wide GTDPhylotranscriptomics has been widely used for phylogenetic analyses by extracting reliable orthologs from assembled transcripts (Cheon et al., 2020). This approach greatly improves our ability to infer species phylogenies when considering species with giant genomes such as the Allium species (Sun et al., 2020). Our phylotranscriptomics of the four diploid species of the genus Allium subg. Cyathophora yielded a strongly supported species relationship using both concatenation and coalescence methods. Thus, A. cyathophorum and A. farreri are the most closely related species, both are sister to the spicate-specific A. spicatum, and the three are sister to A. mairei (Fig. 1B and C). Our KS-based species divergence also demonstrated the reliability of this species phylogeny, indicating that the earliest diverged species was A. mairei around 4.8 Ma, the second species to diverge was A. spicatum around 3.44 Ma, and the most recently diverged species are A. cyathophorum and A. farerri around 1.68 Ma (Fig. 1A). These divergence times are similar to those inferred from the constant mutation rate (Li et al., 2019b). However, widespread gene tree disagreement (GTD, ~38.9% of 1262 SCG trees) was detected among orthologous gene trees, even those with high BS across all nodes (~27% of 840 SCG trees with BS ≥ 70, Fig. 2A). The GTD observed in the phylotranscriptomics was also detected in the population phylogeny (4 out of 10 SCGs, Fig. 3), similar to the discrepancies in the cpDNA and nuclear gene trees (Fig. 1; Li et al., 2016).
Our previous (Li et al., 2019b) and current (Fig. 3) population genetics both indicated that no shared haplotypes were found among the Allium subg. Cyathophora species, suggesting little gene flow among them. Our phyloNet-based phylogenetic network analysis also supported that (Table S7). We identified similar proportions of gene tree topology classes in positively selected and empirical genes, suggesting natural selection has little influence on the GTD associated with the subg. Cyathophora genomes (Table 1 and S9). The distribution of the observed gene trees showed that GTD occurred mainly among the recently diverged A. spicatum, A. cyathophorum and A. farreri probably due to ILS (Class Ⅱ, accounting for 82% of all the discordant gene trees), although individual gene trees presented highly variable bootstrap support for the specific position of individual species (Table 1). Indeed, our coalescent stimulations also strongly indicated that ILS could have linked A. spicatum together with different species (Fig. 2 and Table 1). The origin time of subg. Cyathophora coincided with the dramatic uplift of the QTP and associated ancient climate in the region (the eastern QTP; Fig. 4A; Li et al., 2019b). These geological events could have led a population bottleneck to the ancient population of A. spicatum (Wang et al., 2015), which could be in favor of the stochastic fixation of the ancient genetic polymorphisms across successive speciation events due to genetic drift. The discordance between the cpDNA tree and the nuclear MP-EST tree seems to be a good example of this due to the random geographic distribution of such differential fixation, such as the genetic affinity between the geographically disjunctive A. spicatum and A. farreri rather than the geographically adjacent A. spciatum and A. cyathophorum (Li et al., 2016). Such geographically disjunctive but genetic affinities in the cpDNA tree might suggest we should reject the chloroplast replacement hypothesis, in which the ancestor A. spicatum captured a chloroplast from the ancestor A. farreri, due to the lack of a second contact (Fig. S4).
4.2. Possibility of hemiplasy for the incongruent traitsPhylogeny-based ancestral trait state inference is an effective way to trace the trait evolution along the phylogeny, which provides a profound understanding to the phenotypic diversity in the nature. However, widespread GTD across the whole genome can recover different relationships among species, and even some unrelated species in a species tree are connected by some genes or regions. If trait changes occur in these discordant regions, a phylogeny-based inference of ancestral traits may produce a false pattern (Hahn and Nakhleh, 2016; Guerrero and Hahn, 2018; Wu et al., 2018). The evolution of the inflorescence has undoubtedly attracted our attention, especially in systematic and phylogenetic studies, because the enormous phenotypic diversity of the inflorescence is directly related to the reproductive success of plants. The conspicuously elongated spicate in Allium spicatum compared to the umbel in congeners has been considered as an alternative strategy to adapt to the extreme alpine habitat (here represented by altitude), accompanied by consequential changes in seed size, flowering phase, and ovary size (Li et al., 2016, 2019b). Our RASP-based ancestral state reconstruction showed the ancestral state of inflorescence at the internodes along the species tree of subg. Cyathophora was most likely at the umbel state and distributed across the eastern QTP with high confidence (BS = 100%) (Fig. 4A), which was consistent with the previous results using nuclear or cpDNA gene trees (Li et al., 2016). This result indicated that tree topology seemingly does not affect estimates of the ancestral state of the inflorescence, although the spicate inflorescence of A. spicatum has variable phylogenetic position, resulting in a paraphyletically distributed umbel inflorescence in the phylogeny of subg. Cyathophora. Similar to the categorical traits, the quantitative traits also showed paraphyletic distributions for the specific trait values of all branch tips, although there was consistency among ancestral trait values of internal nodes. All of these trait-state inferences seem to support a random fixation of derived trait values from ancient states (Fig. 4B). However, given the ~27%–38.9% GTD across the genomes of subg. Cyathophora, this however has inevitably led to hemiplasy (Degnan and Rosenberg, 2009; Guerrero and Hahn, 2018).
It has been documented that higher HRFs imply a higher probability of hemiplasy than homoplasy regarding the incongruent traits between closely related species. When the HRFs > 0.95, hemiplasy could be ten times more likely than homoplasy, and when HRFs < 0.50 homoplasy is up to ten times more likely than hemiplasy (Guerrero and Hahn, 2018). When given a constant substitution rate per nucleotide site per year, our HRFs were found to decrease as effective population size increased (Fig. 4). In our study, the Ne (133, 870, 95% HPDI [6310, 978, 952]) used to calculate spicate and umbel inflorescence internode was much larger than the current Ne of A. spicatum (9821, 95% HPDI [3162, 51, 591]), which has a population bottleneck (the ancient Ne is 334, 95% HPDI [32, 7986]) (Wang et al., 2015), and thus HRFs would reach approximately 1 in the context of a small Ne for the ancestor Allium spicatum, implying a higher probability of hemiplasy regarding the trait transition between A. spicatum and A. cyathophorum – A. farreri, and in contrast to a lower shared probability of derived alleles (Guerrero and Hahn, 2018). In addition, we found two neutral genes correlated with inflorescence development that show discordant tree topologies with the nuclear MP-EST tree (Fig. 4F and G). This may also give support for the occurrence of incongruent inflorescence in the small ancient population of A. spicatum due to the random fixation of the ancient polymorphisms. Of course, identifying the genes that directly determine the formation of spicate and umbel inflorescences would provide more robust evidence for this conclusion. In addition, we found that the HRF at the internode of A. cyathophorum and A. farreri, was higher than that at the internode of A. spicatum and A. cyathophorum-A. farreri (Fig. 4C, D, E). This is because hemiplasy most likely underlies the character state transition in the presence of ILS, suggesting the relevant variation occurred in an earlier clade and the following successive lineages are randomly sorted from these ancient polymorphisms (Guerrero and Hahn, 2018).
5. ConclusionsIn this study, phylotranscriptomic analyses of the four diploid species of Allium subg. Cyathophora revealed widespread genome-wide gene-tree discordance (GTD), as well as incongruence between plasmid and nuclear genomes. Our coalescent simulations supported that substantial intergenomic ILS could have caused such GTD. We found that in the presence of ILS, hemiplasy is more likely than homoplasy to explain the trait transition between the current consecutive lineages. This is due to differential fixation of ancestral segregating variants. Our study exemplifies how to convert such phylogenetic noise into evolutionary information. This will provide a deeper understanding of how character–state transitions drive speciation and diversification.
AcknowledgementsWe would like to thank Dr. Daniel Petticord at the University of Cornell for his assistance with English language and grammatical editing of the manuscript. This work was supported by the Key Science & Technology Project of Gansu Province (22ZD6NA007), the National Key Research and Development Program of China (2021YFD2200202). Computing support was provided by the Supercomputing Center of Lanzhou University.
Author contributions
MJL designed research. MJL collected samples. MJL, ZZZ and GL analyzed data. MJL and ZZZ writing original draft. All the authors revised and agreed to the published version of the manuscript.
Data availability statement
The transcriptomes under PRJNA598096 and complete chloroplast genomes have been deposited in the National Center for biotechnology information (NCBI), with public accession at https://www.ncbi.nlm.nih.gov/biosample.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.07.004.
Avise, J.C., Robinson, T.J., 2008. Hemiplasy: a new term in the lexicon of phylogenetics. Syst. Biol., 57: 503-507. DOI:10.1080/10635150802164587 |
Benaglia, T., Chauveau, D., Hunter, D., et al., 2009. mixtools: an R package for analyzing finite mixture models. J. Stat. Softw., 32: 1-29. |
Bouckaert, R.R., 2010. Densitree: making sense of sets of phylogenetic trees. Bioinformatics, 10: 10. |
Chaudhuri, P., Marron, J.S., 1999. SiZer for exploration of structures in curves. J. Am. Stat. Assoc., 94: 907. |
Cheon, S., Zhang, J.Z., Park, C., 2020. Is phylotranscriptomics as reliable as phylogenomics?. Mol. Biol. Evol., 37: 3672-3683. DOI:10.1093/molbev/msaa181 |
Copetti, D., Alberto, Búrquez, Bustamante, E., et al., 2017. Extensive gene-tree discordance and hemiplasy shaped the genomes of North American columnar cacti. Proc. Natl. Acad. Sci. U.S.A., 114: 12003-12008. DOI:10.1073/pnas.1706367114 |
Degnan, J.H., Rosenberg, N.A., 2009. Gene-tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol., 24: 332-340. DOI:10.1016/j.tree.2009.01.009 |
Feng, S., Ru, D.F., Sun, Y.S., et al., 2019. Trans-lineage polymorphism and nonbifurcating diversification of the genus Picea. New Phytol., 222: 576-587. DOI:10.1111/nph.15590 |
Friesen, N., Fritsch, R.M., Blattner, F.R., 2006. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso, 22: 372-395. DOI:10.5642/aliso.20062201.31 |
Friesen, N., Fritsch, R.M., Pollner, S., et al., 2000. Molecular and morphological evidence for an origin of the aberrant genus Milula within Himalayan species of Allium (Alliacae). Mol. Phylogenet. Evol., 17: 209-218. DOI:10.1006/mpev.2000.0844 |
Grabherr, M.G., Haas, B.J., Yassour, M., et al., 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol., 29: 644-652. DOI:10.1038/nbt.1883 |
Guan, C.F., Liu, S.Y., Wang, M.K., et al., 2019. Comparative transcriptomic analysis reveals genetic divergence and domestication genes in Diospyros. BMC Plant Biol., 19: 227. DOI:10.1186/s12870-019-1839-2 |
Guerreroa, R.F., Hahna, M.W., 2018. Quantifying the risk of hemiplasy in phylogenetic inference. Proc. Natl. Acad. Sci. U.S.A., 115: 12787-12792. DOI:10.1073/pnas.1811268115 |
Guindon, O., 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of phyml 3.0. Syst. Biol., 59: 307-321. DOI:10.1093/sysbio/syq010 |
Hahn, M.W., Nakhleh, L., 2016. Irrational exuberance for resolved species trees. Evolution, 70: 7-17. DOI:10.1111/evo.12832 |
Huang, Y., Niu, B.F., Gao, Y., et al., 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics, 26: 680-682. DOI:10.1093/bioinformatics/btq003 |
Huang, D.Q., Yang, J.T., Zhou, C.J., et al., 2014. Phylogenetic reappraisal of Allium subgenus Cyathophora (Amaryllidaceae) and related taxa, with a proposal of two new sections. J. Plant Res., 127: 275-286. DOI:10.1007/s10265-013-0617-8 |
Hutchinson, J., 1959. The Families of Flowering Plants. Clarendon Press, Oxford.
|
Huerta-Cepas, J., Forslund, K., Pedro, C.L., et al., 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol., 34: 2115-2122. DOI:10.1093/molbev/msx148 |
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Knowles, L.L., Kubatko, L.S. (Eds.), 2010. Estimating species trees: practical and theoretical aspects. Wiley-Blackwell, Hoboken, NJ, USA.
|
Krause, K., 1930. Liliaceae. In: Engler, A. (Ed.), Die Natürlichen Pflanzenfamilien.
Engelmann, Leipzig, pp. 227-386.
|
Li, L., , Roos, D.S., 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res., 13: 2178-2189. DOI:10.1101/gr.1224503 |
Li, L.L., Zhao, Y., Chang, R.H., et al., 2018. Expression and bioinformatics analysis of PLC gene family in Lm Type Ricinus communis inflorescence. Mol. Plant Breed., 16: 6604-6615. |
Li, M.J., Tan, J.B., Xie, D.F., et al., 2016. Revisiting the evolutionary events in Allium subgenus Cyathophora (Amaryllidaceae): insights into the effect of the Hengduan Mountains Region (HMR) uplift and Quaternary climatic fluctuations to the environmental changes in the Qinghai-Tibet Plateau. Mol. Phylogenet. Evol., 94: 802-813. DOI:10.1016/j.ympev.2015.10.002 |
Li, M.J., Liu, J.Q., Guo, X.L., et al., 2019a. Taxonomic revision of Allium cyathophorum (Amaryllidaceae). Phytotaxa, 415: 240-246. DOI:10.11646/phytotaxa.415.4.9 |
Li, M.J., Xie, D.F., Xie, C., et al., 2019b. A phytogeographic divide along the 500 mm isohyet in the Qinghai-Tibet Plateau: insights from the phylogeographic evidence of Chinese Alliums (Amaryllidaceae). Front. Plant Sci., 10: 149. |
Li, M.J., Yu, H.X., Guo, X.L., et al., 2021. Out of the Qinghai-Tibetan plateau and rapid radiation across Eurasia for Allium section Daghestanica (Amaryllidaceae). AoB Plants, 13: 3. DOI:10.1007/s00477-019-01672-4 |
Li, W.W., 2012. Functional analysis of SST family in sexual reproduction of Arabidopsis thaliana. Wuhan University Press, China.
|
Liu, L., Yu, L.L., Edwards, S.V., 2010a. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol. Biol., 10: 302. DOI:10.1186/1471-2148-10-302 |
Liu, L., Yu, L.L., Edwards, S.V., 2010b. PHYBASE: an R package for phylogenetic analysis. Bioinformatics, 26: 962-963. DOI:10.1093/bioinformatics/btq062 |
Löytynoja, A., Goldman, N., 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science, 320: 1632-1635. DOI:10.1126/science.1158395 |
Maddison, W.P., 1997. Gene trees in species trees. Syst. Biol., 46: 523-536. DOI:10.1093/sysbio/46.3.523 |
Mendes, F.K., Hahn, Y., Hahn, M.W., 2016. Gene-tree discordance can generate patterns of diminishing convergence over time. Mol. Biol. Evol., 33: 3299-3307. DOI:10.1093/molbev/msw197 |
Ossowski, S., Schneeberger, K., Lucas-Lledó, J.I., et al., 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science, 327: 92-94. DOI:10.1126/science.1180677 |
Pamilo, P., Nei, M., 1988. Relationships between gene trees and species trees. Mol. Biol. Evol., 5: 568-583. |
Paradis, E., Schliep, K., 2019. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics, 35: 526-528. DOI:10.1093/bioinformatics/bty633 |
Pease, J.B., Haak, D.C., Hahn, M.W., et al., 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biology, 14: e1002379. DOI:10.1371/journal.pbio.1002379 |
Revell, L.J., 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol., 3: 217-223. DOI:10.1111/j.2041-210X.2011.00169.x |
Robinson, T.J., Anne, R., 2011. Examination of hemiplasy, homoplasy and phylogenetic discordance in chromosomal evolution of the Bovidae. Syst. Biol., 60: 439-450. DOI:10.1093/sysbio/syr045 |
Sonnhammer, E.L., Östlund, G., 2015. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res., 43: D234-D239. DOI:10.1093/nar/gku1203 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Stearn, W.T., 1960. Allium and Milula in the Central and eastern Nepal Himalaya. Bull. Br. Museum, Nat. History, 2: 159-191. |
Sun, X.D., Zhu, S.Y., Li, N.Y., et al., 2020. A chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant, 13: 1328-1339. |
Takhtajan, A.L., 1987. Systema Magnoliophytorum. Nauka, Leningrad.
|
Than, C., Ruths, D., Nakhleh, L., 2008. PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinformatics, 9: 322. |
Traub, H.P., 1972. The order Alliales. Plant Life, 28: 129-132. |
Wake, D.B., Wake, M.H., Specht, C.D., 2011. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science, 331: 1032-1035. DOI:10.1126/science.1188545 |
Wang, K., Lenstra, J.A., Liu, L., et al., 2018. Incomplete lineage sorting rather than hybridization explains the inconsistent phylogeny of the wisent. Commun. Biol., 1: 169. DOI:10.2112/si84-023.1 |
Wang, X.Y., Li, Y.S., Liang, Q.L., et al., 2015. Contrasting responses to Pleistocene climate changes: a case study of two sister species Allium cyathophorum and A. spicata (Amaryllidaceae) distributed in the eastern and western Qinghai-Tibet Plateau. Ecol. Evol., 5: 1513-1524. DOI:10.1002/ece3.1449 |
Wu, M., Kostyun, J.L., Hahn, M.W., et al., 2018. Dissecting the basis of novel trait evolution in a radiation with widespread phylogenetic discordance. Mol. Ecol., 27: 3301-3316. DOI:10.1111/mec.14780 |
Yang, Z., Wong, W.S., Nielsen, R., 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol., 22: 1107-1118. DOI:10.1093/molbev/msi097 |
Yang, Z.H., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol., 24: 1586-1591. DOI:10.1093/molbev/msm088 |
Yu, G., Wang, L.G., Han, Y., et al., 2012. Clusterprofiler: an R package for comparing biological themes among gene clusters. Omics, 16: 284-287. DOI:10.1089/omi.2011.0118 |
Yu, Y., Barnett, R.M., Nakhleh, L., 2013. Parsimonious inference of hybridization in the presence of incomplete lineage sorting. Syst. Biol., 62: 738-751. DOI:10.1093/sysbio/syt037 |
Yu, Y., Blair, C., He, X.J., 2020. RASP (reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Biol. Evol., 37: 604-606. DOI:10.1093/molbev/msz257 |
Zhang, J.Z., 2000. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol., 50: 56-68. DOI:10.1007/s002399910007 |



