b. Department of Forest Biodiversity and Herbarium, Korea National Arboretum, Pocheon, 11186, Republic of Korea;
c. Department of Biology, Chungbuk National University, Cheongju, 28644, Republic of Korea;
d. International Joint Lab for Molecular Phylogeny and Biogeography, Institute of Botany, Academy of Sciences of Uzbekistan, Tashkent, 100125, Uzbekistan
The genus Allium L. is one of the largest genera in the Amaryllidaceae family, with approximately 1000 species that are naturally distributed in the Northern Hemisphere (Fritsch and Friesen, 2002; Friesen et al., 2020; Govaerts et al., 2021). Its main center of diversity is located between Southwest and Central Asia and the Mediterranean regions (Choi and Oh, 2011). Molecular phylogenies have demonstrated the monophyly of Allium (Friesen et al., 2006; Nguyen et al., 2008; Li et al., 2010). The most recent classification divided Allium into 15 subgenera and 72 sections based on molecular evidence (nrITS; Friesen et al., 2006). Subsequent studies have focused on the morphology, molecular-based phylogeny, evolutionary history, and biogeography of various ambiguous subgenera and sections of Allium (Seregin et al., 2015; Herden and Friesen, 2016; Li et al., 2016, 2021; Sinitsyna et al., 2016; Friesen et al., 2020; Xie et al., 2020a, 2020b; Yang et al., 2023), including several novel sections and subsections of Allium (Friesen et al., 2020, 2021; Khan et al., 2022; Yang et al., 2023). Several characteristics important for the taxonomy of Allium have been investigated in detail, including seed testa sculptures (Celep et al., 2012; Baasanmunkh et al., 2020, 2021; Yusupov et al., 2022), pollen morphology (Namin et al., 2009), bulb and scape anatomy (Khorasani et al., 2018), pistil morphology (Choi et al., 2012), septal nectary anatomy (Gurushidze et al., 2008), leaf anatomy (Mashayekhi and Columbus, 2014), and the chromosomes (Peruzi et al., 2016; Han et al., 2020). However, due to the enormous number of Allium species worldwide, studies that combine both morphological and molecular data of many species are still lacking.
Allium is characterized by the presence of bulbs enclosed in membranous tunics, flowers with six free, or almost free, tepals, and often a subgynobasic style (Fritsch and Friesen, 2002; Friesen et al., 2006; Choi et al., 2011; Choi and Oh, 2011). Recently, a number of Allium species have been described as new to science based on their morphology (e.g., Armağan, 2021; Khan et al., 2021), or mixed morphological, including molecular and karyological properties (e.g., Koçyiğit et al., 2016; Choi et al., 2019; Xie et al., 2020c; Friesen et al., 2020, 2022; Jang et al., 2021; Pandey et al., 2021; Bartolucci et al., 2022; Khan et al., 2022). In general, the major morphological traits used to assess Allium taxonomy include rhizome and bulb tunics, stamens, tepals, and filaments. Rhizome and bulb tunics are the most useful at the subgeneric level (Friesen et al., 1999; Friesen et al., 2006; Choi and Oh, 2011); pistil morphology is useful for clades within subgenera (Choi et al., 2012), and the floral characteristics of tepals and filaments are useful at the species level (Xu and Kamelin, 2000; Choi et al., 2011, 2012; Jang et al., 2021).
Floral structures of Allium are characterized by a fasciculate to umbel or head-like inflorescence, hypogynous flower, free or almost free tepals in two slightly differentiated whorls, and trilocular ovaries with single style (Fritsch and Friesen, 2002; Choi and Oh, 2011). However, there are very few studies that have focused on the flower morphology of the Allium genus (Zuraw, 2009; Choi et al., 2011, 2012; Aryakia et al., 2016). Fritsch (1992) reported various shapes and positions of the nectaries and their openings in more than 160 species. Gurushidze et al. (2008) investigated nectary anatomy in the subgenus Melanocrommyum with molecular analysis. In addition, several studies examined nectaries in various species (Zuraw et al., 2009, 2010). Choi et al. (2011) presented a micro- and macro-morphological comparative analysis of floral characteristics of four New World Allium species. Furthermore, Choi et al. (2012) studied pistil morphology of northeast Asian and northern North American Allium using molecular data. Recently, Jang et al. (2021) investigated floral parts specifically for species of section Rhizirideum in South Korea and northeast China. However, due to over-reliance on dried specimens, disagreements regarding the taxonomic importance of specific morphological traits, and the proliferation of synonyms (Hanelt et al., 1992; Khassanov and Fritsch, 1994; Mes et al., 1997; Gregory et al., 1998; Choi and Oh, 2011), the infrageneric taxonomy and evolution in the genus are still not entirely understood.
Here, we investigated the extensive floral morphology of Allium from the central and the east Asian countries, using quantitative and qualitative morphological characteristics. The aims of this study were to: (1) analyze floral morphology of previously unstudied species and different accessions of previously studied taxa, and (2) evaluate the taxonomic implications of the analyzed floral characters in Allium. This study, together with that of Choi et al. (2011, 2012), provides a strong basis for a future monograph and the systematic understanding of Allium.
2. Materials and methods 2.1. Plant materialsA total of 87 accessions of 74 taxa belonging to 30 sections and nine subgenera were evaluated in this study (Table S1). Field surveys were conducted between 2005 and 2021 in Asian countries, namely, Mongolia (26 accessions), South Korea (24), Kyrgyzstan (20), Uzbekistan (13), China (3), Russia (2), and Japan (1). In addition, we collected three cultivated species from South Korea (Table S1). All samples were stored in 70% ethanol during the field surveys and were subsequently deposited in the herbarium of Changwon National University (CWNU) (Table S1). The subgenera and sections were provided by Friesen et al. (2006). All taxa names were ordered alphabetically followed by subgenus and section (Table S1).
2.2. Morphological observation and measurementThe size and shape of flowers (at least 30–50 for each accession) were measured and photographed using an Olympus SZ61 stereoscope (Olympus Co., Tokyo, Japan) with a TrueChrome II camera (Tucsen Photonics Co., Ltd., Fuzhou, China). Quantitative characters of flowers, including length and width of perianth, tepal, stamen, and pistil, were measured (Fig. 1). Furthermore, we examined qualitative characteristics of flowers, including color of perianth, shape of tepals, filaments, ovaries, and anthers. The colors of perianth were determined using RHS color chart (Royal Horticultural Society, 2015).
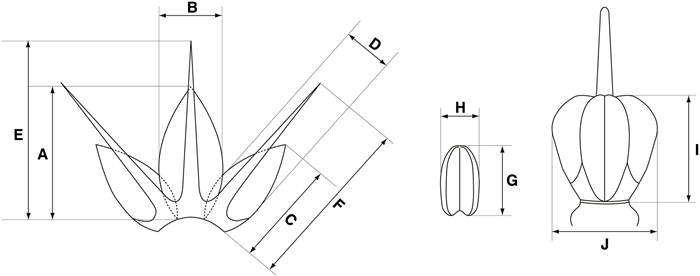
|
| Fig. 1 Illustration of flower structure in Allium with quantitative characteristics. (A) Inner tepal length (B) inner tepal width (C) outer tepal length (D) outer tepal width (E) inner filament length (F) outer filament length (G) anther length (H) anther width (I) ovary length (J) ovary width. |
All statistical analyses were conducted using FeatureImpCluster, flexclust, clustMixType, factoextra, cluster packages in R v.4.0.4 (R Core Team, 2020). FeatureImpCluster package offers a feature importance for partitional clustering. For clustering of mixed type data, the clustMixType package was used in R by using k-prototypes algorithm. The analysis was conducted with two raw data sets, one including only quantitative factor and another that included a combination of quantitative and qualitative factors. After calculating feature importance values, a second cluster plot was generated by adding certain qualitative traits with high values. We applied this analysis to all accessions and each subgenus. The cluster analysis for all accessions was performed with an average value of quantitative characteristics. All quantitative and qualitative characteristics are summarized in Table 1.
| No. | Abbreviations (Quantitative) | Morphological characteristics | No. | Abbreviations (Qualitative) | Morphological characteristics |
| 1 | ITL | inner tepal length | 1 | PSP | perianth shape |
| 2 | ITW | inner tepal width | 2 | PCO | perianth color |
| 3 | OTL | outer tepal length | 3 | ITS | inner tepal shape |
| 4 | OTW | outer tepal width | 4 | ITA | inner tepal apex |
| 5 | IFL | inner filament length | 5 | OTS | outer tepal shape |
| 6 | OFL | outer filament length | 6 | OTA | outer tepal apex |
| 7 | AL | anther length | 7 | RFP | reflexed point of tepal apex |
| 8 | AW | anther width | 8 | RL | filament relative length compared with perianth |
| 9 | OL | ovary length | 9 | IFS | inner filament shape |
| 10 | OW | ovary width | 10 | OFS | outer filament shape |
| 11 | OVU | number of ovules | 11 | MEN | filament margin entire |
| 12 | MTT | filament margin toothed | |||
| 13 | TP | filament toothed position | |||
| 14 | OVS | ovary shape | |||
| 15 | APH | appendages on ovary | |||
| 16 | STY | style type | |||
| 17 | STG | stigma type |
We obtained ITS sequences of 67 species belonging to 30 sections and nine subgenera from NCBI (Table S4). Two species including Nothoscordum bivalve and Tulbaghia violacea were selected as an outgroup according to Friesen et al. (2006) and Li et al. (2010). The sequences were aligned using ClustalW (Thompson et al., 1994) and the default settings and manual adjustments were made using BioEdit (Hall, 1999). Ambiguous nucleotide bases were corrected using the corresponding base of the sequence that was obtained using the reverse primer. The phylogenetic analyses were conducted using the Bayesian inference (BI), Maximum Likelihood (ML), and Maximum Parsimony (MP) methods. The BI analysis was conducted using MrBayes 3.2.2 (Ronquist et al., 2012), with two independent runs of four simultaneous chains, executed for 5, 000, 000 generations using the Markov chain Monte Carlo algorithm. ML was performed using RAxML v.8.2.11 (Stamatakis, 2006, 2014) as implemented in the Geneious v.10.2.2 (Kearse et al., 2012) search for the best-scoring ML tree algorithm, which included 1000 bootstrap replicates. MP analysis was conducted using PAUP v.4.0b10 (Swofford, 1989). The trees were determined from a 50% majority-rule consensus to estimate the posterior probabilities. The reconstructed trees were visualized using FigTree v.1.4.2 (Rambaut, 2012).
3. Results and discussionWe studied the floral morphology of 87 accessions of 74 Allium taxa belonging to 30 sections and nine subgenera from Central to East Asian countries. The shapes and sizes of floral morphology are provided in Tables S2 and S3. The detailed flower description of each taxon is provided based on our fresh samples (Supplementary data). In addition, we provide the global distribution and flowering season for each species based on extensive literatures and our own field information (Supplementary data).
3.1. Quantitative and qualitative characteristics 3.1.1. Inflorescence colorThe inflorescence colors varied among the investigated species. In general, we classified colors into five categories: blue, purple and pink, red, white, and yellow (Figs. 2-16; Table S2). For example, species of the subgenus Reticulatobulbosa have inflorescence of various colors, including reddish purple (Allium jodanthum, Fig. 11D), pale yellow (A. flavidum, Fig. 12B), and white (A. leucocephalum, Fig. 12D).

|
| Fig. 2 Floral morphology in Allium (A–E) Subg. Allium (A) A. atroviolaceum (B) A. filidens (C) A. filidentiforme (D) A. longicuspis (E) A. macrostemon (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
| Fig. 3 Floral morphology in Allium (A–E) Subg. Allium (A) A. griffithianum (B) A. caeruleum (C–D) A. caesium (E) A. glomeratum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
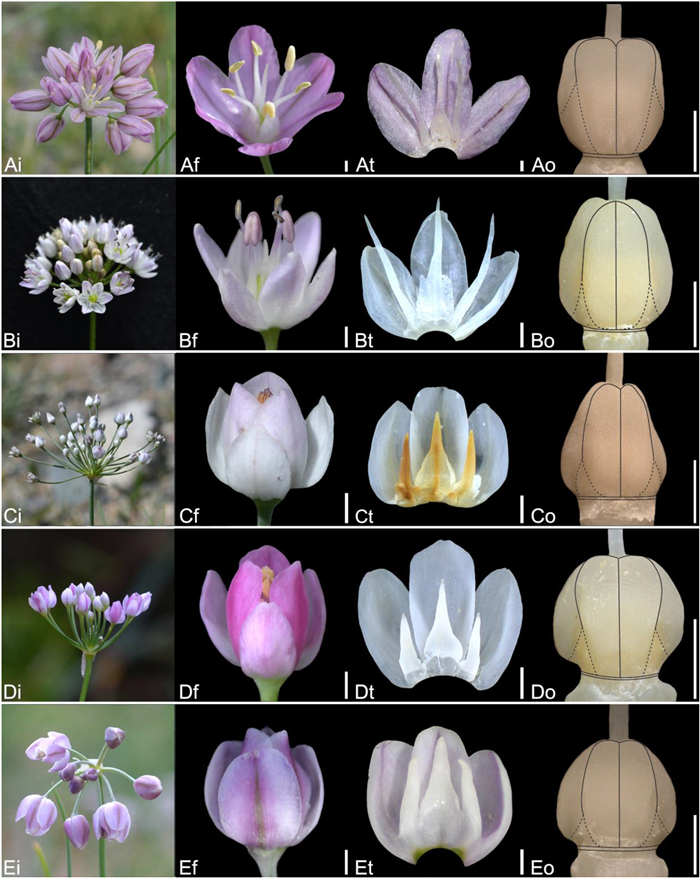
| Fig. 4 Floral morphology in Allium (A–D) Subg. Allium (A) A. sabulosum (B) A. haneltii (C) A. anisotepalum (D) A. caricifolium (E) Subg. Anguinum (E) A. microdictyon (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
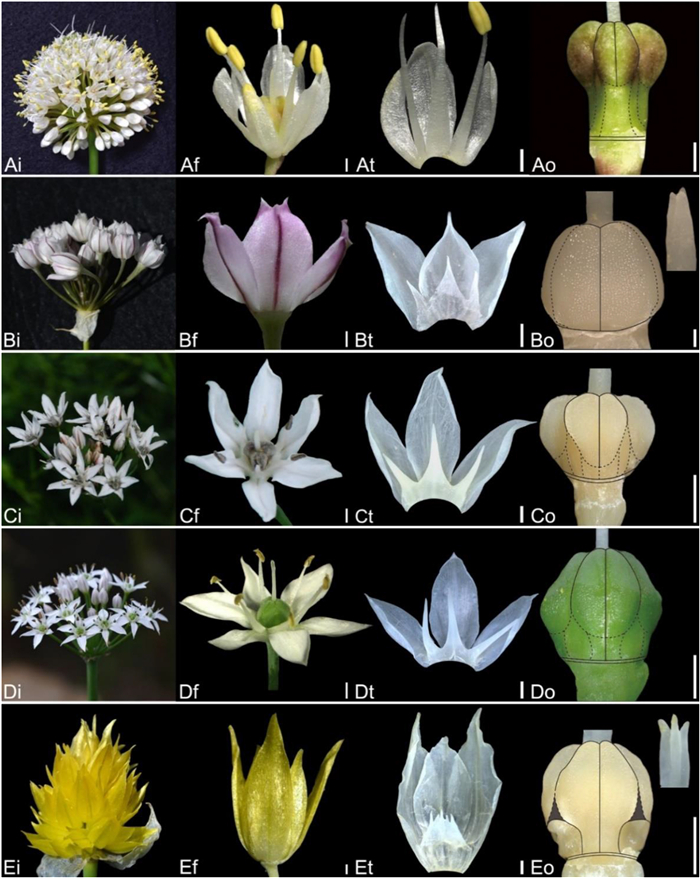
| Fig. 5 Floral morphology in Allium (A) Subg. Anguinum (A) A. ulleungense (B–D) Subg. Butomissa (B) A. oreoprasum (C) A. ramosum (D) A. tuberosum (E) Subg. Cepa (E) A. semenovii (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
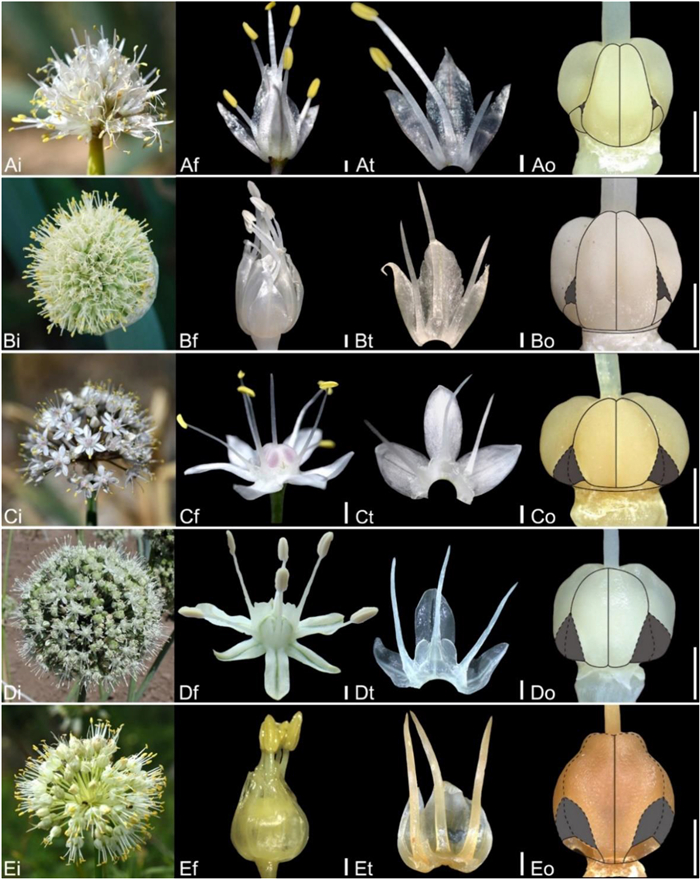
| Fig. 6 Floral morphology in Allium (A–E) Subg. Cepa (A) A. altaicum (B) A. fistulosum (C) A. galanthum (D) A. oschaninii (E) A. condensatum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
| Fig. 7 Floral morphology in Allium (A–E) Subg. Cepa (A) A. linearifolium (B) A. pseudojaponicum (C) A. taguetii (D) A. thunbergii var. teretifolium (E) A. thunbergii var. thunbergii (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
| Fig. 8 Floral morphology in Allium (A–D) Subg. Cepa (A) A. maximowiczii (B) A. schoenoprasum var. foliosum (C) A. schoenoprasum var. schoenoprasum (D) A. karelinii (E) Subg. Melanocrommyum (E) A. giganteum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
| Fig. 9 Floral morphology in Allium (A–C) Subg. Melanocrommyum (A) A. fetisowii (B) A. stipitatum (C) A. karataviense (D) Subg. Microscordum (D) A. monanthum (E) Subg. Polyprason (E) A. carolinianum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
| Fig. 10 Floral morphology in Allium (A–E) Subg. Polyprason (A) A. korolkowii (B) A. platyspathum subsp. amblyophyllum (C) A. platyspathum subsp. platyspathum (D) A. kirilovii (E) A. obliquum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
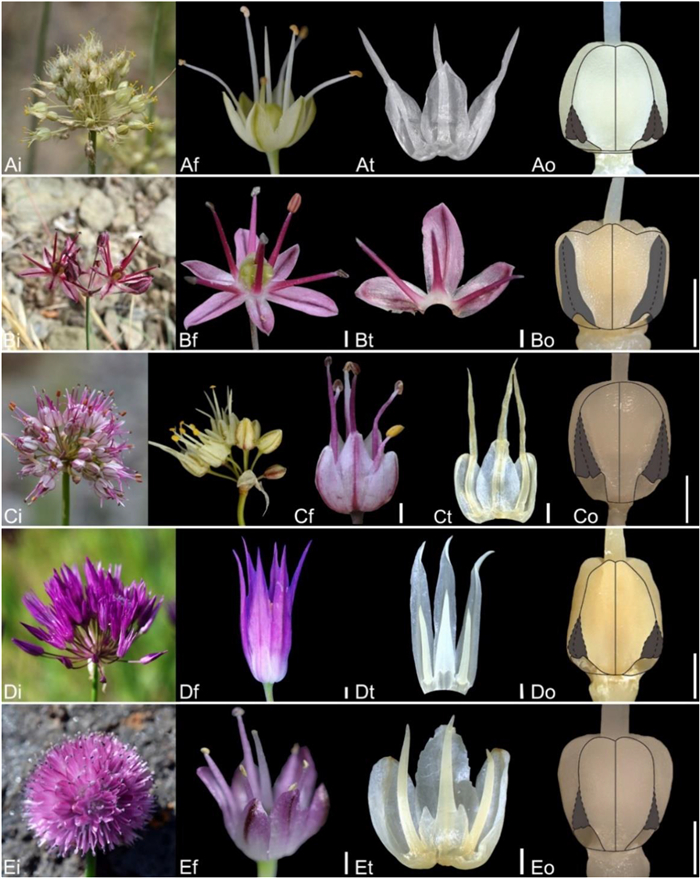
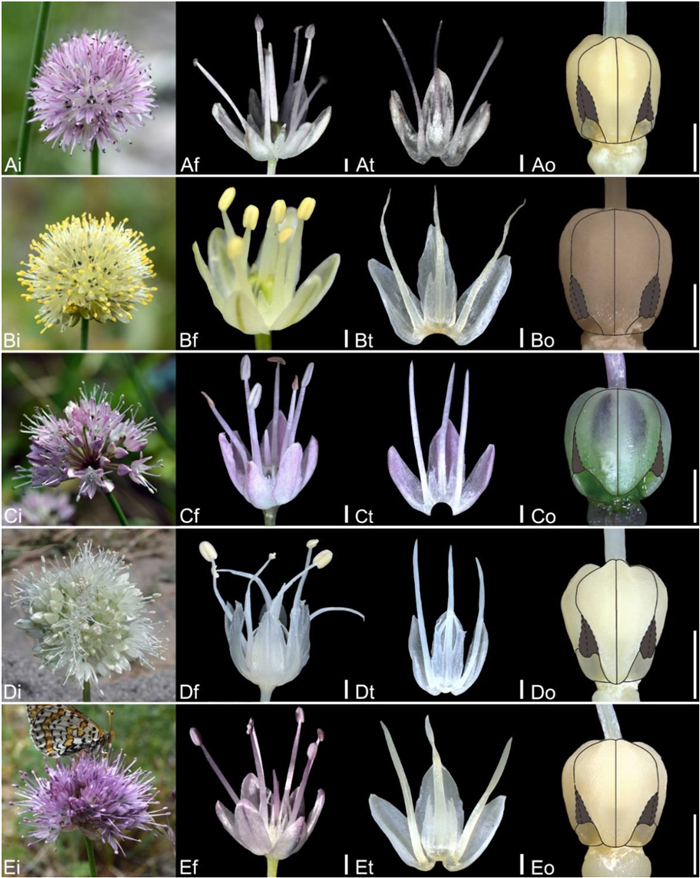
| Fig. 11 Floral morphology in Allium (A–C) Subg. Polyprason (A) A. petraeum (B) A. subtilissimum (C) A. tianschanicum (D–E) Subg. Reticulatobulbosa (D) A. jodanthum (E) A. amphibolum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
| Fig. 12 Floral morphology in Allium (A–E) Subg. Reticulatobulbosa (A) A. clathratum (B) A. flavidum (C) A. koreanum (D) A. leucocephalum (E) A. malyschevii (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
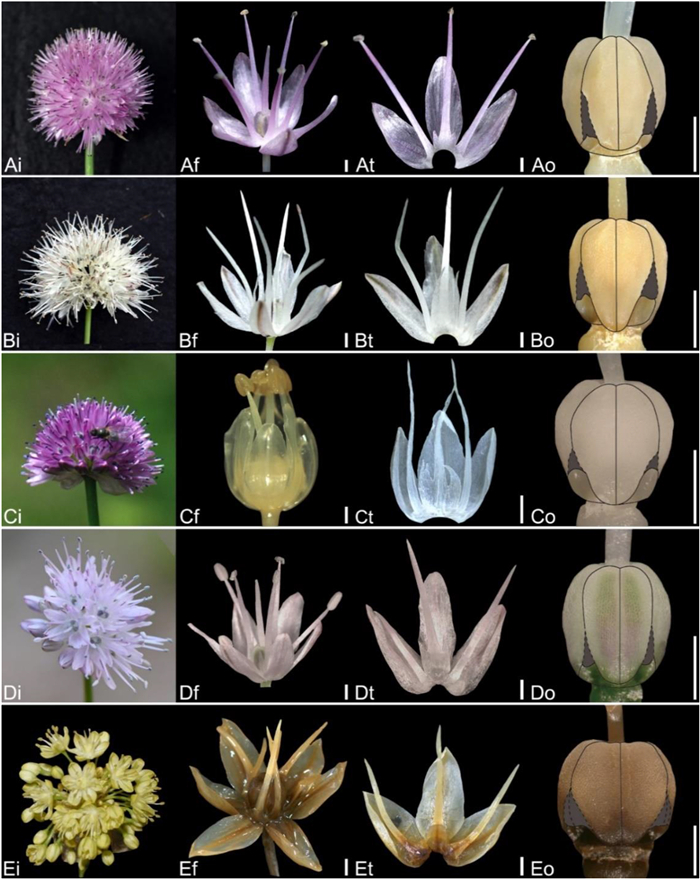
| Fig. 13 Floral morphology in Allium (A–E) Subg. Reticulatobulbosa (A) A. pumilum (B) A. schischkinii (C) A. splendens (D) A. ubsicola (E) A. scabriscapum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |

|
| Fig. 14 Floral morphology in Allium (A–E) Subg. Rhizirideum (A) A. eduardii (B) A. austrosibiricum (C) A. dumebuchum (D) A. minus (E) A. prostratum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
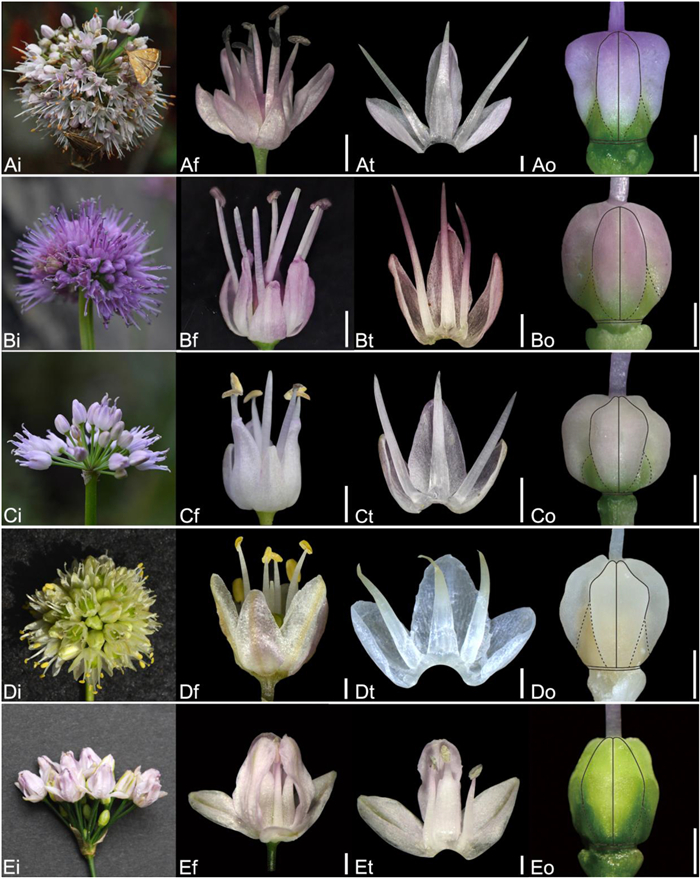
| Fig. 15 Floral morphology in Allium (A–E) Subg. Rhizirideum (A) A. senescens (B) A. spirale (C) A. spurium (D) A. tuvinicum (E) A. bidentatum (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
|
| Fig. 16 Floral morphology in Allium (A–E) Subg. Rhizirideum (A) A. mongolicum (B) A. polyrhizum (C) A. anisopodium (D) A. tenuissimum (E) A. vodopjanovae (i – inflorescence, f – flower, t – tepal and filament arrangement, o – ovary). Scale bars = 1 mm. |
The perianth shapes are typically categorized as campanulate, semi-stellate, and stellate (Figs. 2-16; Table S2).
3.1.3. Tepal shape and sizeThe tepals of Allium species comprise inner and outer parts (Figs. S1–S6). In general, tepal shapes (inner and outer) can be categorized as ovate (Allium condensatum, Fig. S1X), lanceolate (A. karelinii, Fig. S2D), elliptic (A. giganthum, Fig. S2H), oblong (A. monanthum, Fig. S2IM), linear (A. fetisowii, Fig. S2E), or obovate (A. anisopodium, Fig. S3O). Tepal apex is categorized as rounded (A. oschaninii, Fig. S1W), obtuse (A. koreanum, Fig. S2W), acute (A. scabriscapum, Fig. S3B), mucronate (A. oreoprasum, Fig. S1P), acuminate (A. kirilovii, Fig. S2N), retuse (A. sabulosum, Fig. S1J), or truncate (A. vodopjanovae, Fig. S3Q). Detailed information on the shapes of each species can be found in the Supplementary data.
We measured inner and outer tepals for each taxon (Table S3). Based on the mean value (mean ± standard deviation), inner tepal length/width ratio ranged from 1.48 ± 0.16 to 6.94 ± 1.66, with Allium taguetii having the smallest (1.23; Fig. S1AA) and A. karataviense having the biggest (10.68; Fig. S2G). Similarly, outer tepal length/width ratio ranged from 1.34 ± 0.07 to 7.4 ± 1.88, with A. taguetii having the smallest (1.14; Fig. S4AA) and A. karataviense having the biggest (10.4; Fig. S5G). However, the length of inner tepals ranged from 2.72 ± 0.26 to 11.64 ± 0.85 mm, with A. logicuspis having the shortest (2.46 mm; Fig. S1D) and A. schoenoprasum var. schoenoprasum having the longest (13.46 mm; Fig. S2C). The inner tepal width ranged from 1.01 ± 0.1 to 4.00 ± 0.3 mm, with A. fetisowii having the narrowest (0.74 mm; Fig. S2E) and A. semenovii having the widest (5.19 mm; Fig. S1S). In addition, the outer tepal length ranged from 2.69 ± 0.12 to 14.16 ± 1.18 mm, with A. sabulosum having the shortest (2.32 mm; Fig. S4J) and A. semenovii having the longest (15.96 mm; Fig. S4S). Finally, the outer tepal width ranged from 0.85 ± 0.07 to 4.22 ± 0.38 mm, A. logicuspis having the narrowest (0.73 mm; Fig. S4D) and A. semenovii having the widest (4.88 mm; Fig. S4S).
3.1.4. StamenThe relative length of the filament with perianth can be categorized into four types: non-exserted (e.g. Allium griffithianum, Fig. 3A), slightly non-exserted (e.g. A. maximowiczii, Fig. 8A), slightly exserted (e.g. A. subtilissimum, Fig. 11B), and exserted (e.g. A. leucocephalum, Fig. 12D). The filament shapes are represented as triangular (e.g. A. filidens, Fig. 2B), narrowly triangular (e.g. A. sabulosum, Fig. 4A), broadened for length (e.g. A. bidentatum, Fig. 15E), subulate (e.g. A. thunbergii var. thunbergii, Fig. 7E), and subulate from a triangular base (e.g. A. stipitatum, Fig. 9B). In addition, the margin of inner filament was entire (e.g. A. macrostemon, Fig. 2E) or toothed (e.g. A. malyschevii, Fig. 12E). Based on the length/width ratio, the shape of anthers was classified into three types: ovate (e.g. A. vodopjanovae), elliptic (e.g. A. prostratum), and oblong (e.g. A. ulleungense). More detailed information on the stamen is presented in Figs. 2-16 and Table S2.
We measured two filaments (inner and outer) for each taxon, which are summarized in Table S3. Based on the mean value (mean ± standard deviation), the length of inner filament ranged from 2.21 ± 0.14 to 12.79 ± 0.73 mm, with Allium griffithianum having the shortest (1.31 mm; Fig. 3A) and A. altaicum having the longest (13.96 mm; Fig. 6A). Similarly, the length of outer filament ranged from 1.83 ± 0.21 to 10.37 ± 1.21 mm, with A. griffithianum having the shortest (1.31 mm; Fig. 3A) and A. altaicum having the longest (13.42 mm; Fig. 6A). The anther length ranged from 0.88 ± 0.06 to 2.48 ± 0.19 mm, with A. monanthum having the shortest (0.74 mm) and A. ramosum having the longest (2.71 mm); whereas the anther width ranged from 0.49 ± 0.04 to 1.03 ± 0.06 mm, with A. monanthum having the narrowest (0.36 mm) and A. pseudojaponicum having the widest (1.18 mm).
3.1.5. Pistil shape and sizeThe shape of ovaries can be divided in eight types: ellipsoid, subglobose, globose, oblate spheroid, ovoid, obovoid, obconical, subcubical (Figs. 2-16). Two types of ovarian processes, with and without hood-like appendages, were observed (Figs. 2-16). The number of ovules per locule was typically two. Some exceptions include zero or one in the male of Allium monanthum, one ovule in Allium microdictyon, A. ulleungense, two to four in A. ramosum, and four to six in A. fetisowii. In contrast, the style of A. semenovii and A. monanthum is trigonous (Fig. 5E and 9D), while the remaining species are terete. Three types of stigmas were observed in the species studied: slightly three-cleft in A. oreoprasum (Fig. 5B), three-cleft in A. semenovii and A. monanthum (Fig. 5E and 9D), and smooth, the most common stigma type.
We measured the length and width of the ovary for each species (Table S3). Based on the mean value (mean ± standard deviation), the length of the ovary ranged from 1.2 ± 0.18 to 3.6 ± 0.15 mm, with Allium anisotepalum having the shortest (0.99 mm; Fig. 4C) and A. ulleungense having the longest (4.1 mm; Fig. 5A). The width of the ovary ranged from 1.24 ± 0.09 to 3.51 ± 0.25 mm, with A. anisotepalum having the narrowest (1.06 mm; Fig. 4C) and A. karataviense having the widest (4.36 mm; Fig. 9C).
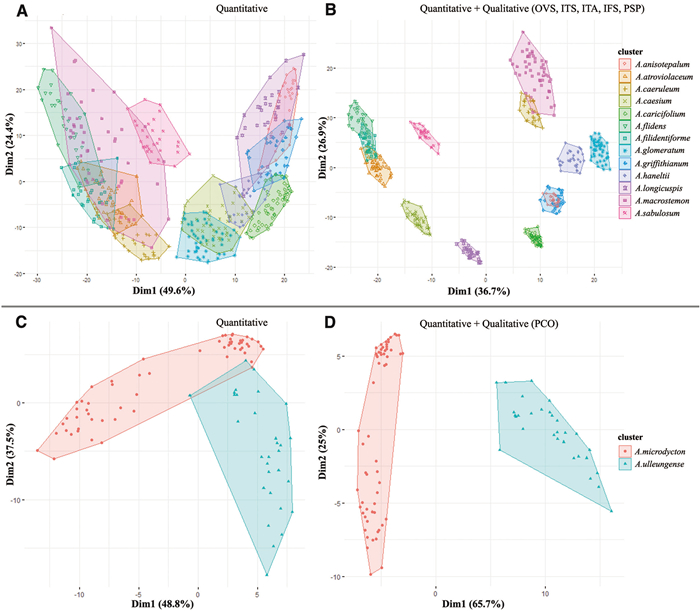
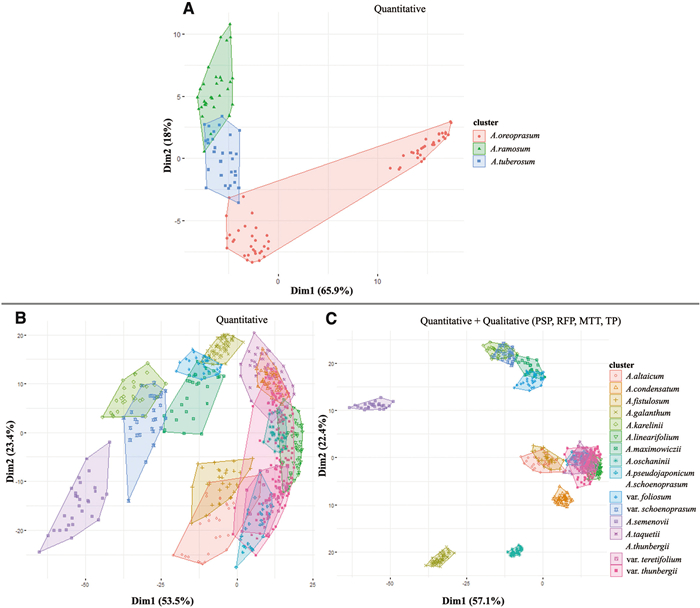
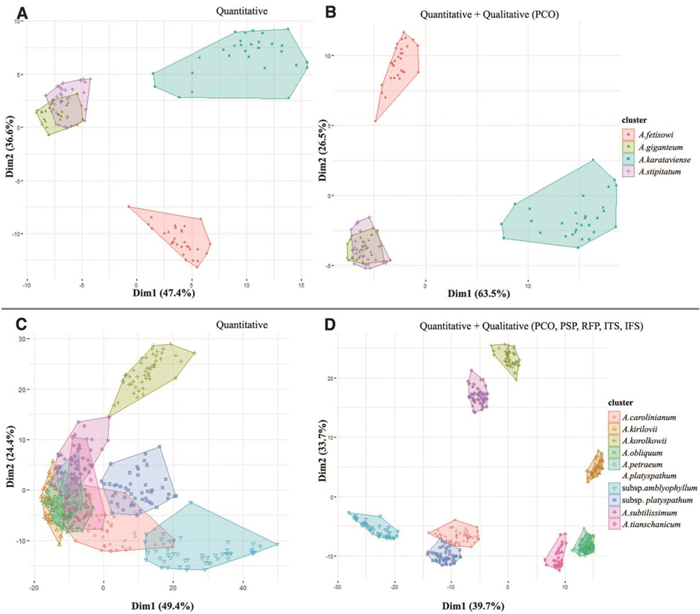
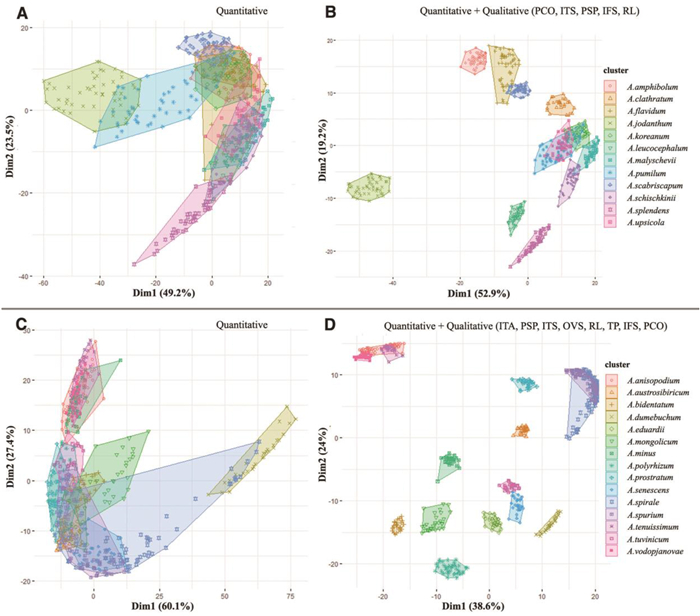
3.2. Statistical analysis at the subgenus levelCluster analysis was conducted using 11 quantitative and 17 qualitative floral morphological characteristics. We present the two different cluster plots: one based on only quantitative data, and one based on combined quantitative and selected qualitative characteristics at the species level within each subgenus of Allium (Figs. 17-20). According to quantitative and qualitative results, the species are relatively well separated from each other in the subgenera Anguinum (Fig. 17C and D), Butomissa (Fig. 18A), and Melanocrommym (Fig. 19A and B). Further, species are not well distinguished based on quantitative characteristics in subgenera Allium (Fig. 17A), Cepa (Fig. 18B), Polyprason (Fig. 19C), Reticulatobulbosa (Fig. 20A), and Rhizirideum (Fig. 20C). However, species within the four subgenera, which were difficult to distinguish only by quantitative characteristics, were more clearly distinguished when analyzed with qualitative characteristics (Fig. 17B, 18C, 19D, and 20B and D).
|
| Fig. 17 Cluster analyses at the specific level within subgenera (A, B) Subg. Allium (A) cluster plot with only quantitative factors (B) cluster plot with combined quantitative and qualitative factors strongly associated with clustering (C, D) Subg. Anguinum (C) cluster plot with only quantitative factors (D) cluster plot with combined quantitative and qualitative factors strongly associated with clustering. |
|
| Fig. 18 Cluster analyses at the specific level within subgenera (A) Subg. Butomissa: cluster plot with only quantitative factors (B, C) Subg. Cepa (B) cluster plot with only quantitative factors (C) cluster plot with combined quantitative and qualitative factors strongly associated with clustering. |
|
| Fig. 19 Cluster analyses at the specific level within subgenera (A, B) Subg. Melanocrommyum (A) cluster plot with only quantitative factors (B) cluster plot with combined quantitative and qualitative factors strongly associated with clustering (C, D) Subg. Polyprason (C) cluster plot with only quantitative factors (D) cluster plot with combined quantitative and qualitative factors strongly associated with clustering. |
|
| Fig. 20 Cluster analyses at the specific level within subgenera (A, B) Subg. Reticulatobulbosa (A) cluster plot with only quantitative factors (B) cluster plot with combined quantitative and qualitative factors strongly associated with clustering (C, D) Subg. Rhizirideum (C) cluster plot with only quantitative factors (D) cluster plot with combined quantitative and qualitative factors strongly associated with clustering. |
According to statistical analysis, flower morphology is not sufficiently significant to distinguish each subgenus accessed in this study (Fig. S8). However, evidently, the flower morphology is the key diagnoses for the species within each subgenus; shape and size of floral organs such as perianth, tepals, filament, and pistils are useful diagnostic characteristics at the specific level (Figs. 17-20). In addition, the flower character has been consistently noted to be the most important key characteristics to distinguish species in the genus Allium (Choi and Oh, 2011; Choi et al., 2012; Xie et al., 2020c; Jang et al., 2021; Bartolucci et al., 2022). In this study, we present several important diagnoses between closely related taxa at the section and subgenus levels.
3.3.1. Subgenus AlliumThe subgenus Allium is a large group including over 375 taxa divided into 19 sections, which is placed in the third evolutionary line (Friesen et al., 2006; Khassanov, 2018; Yang et al., 2023). However, a detailed phylogenetic classification of this taxonomic group remains incomplete (Khassanov, 2018; Munavvarov et al., 2022). In this study, 15 accessions of 13 species representing eight sections (Allium, Avulsea, Caerulea, Eremoprasum, Haneltia, Macrostemon, Minuta, and Pallasia) were investigated. The quantitative cluster plot revealed that 75.3% of the total variation is not adequately separated from each other (Fig. 17A). However, the combined cluster plot, which included five important qualitative characteristics (ovary shape, inner tepal shape, inner tepal apex, inner filament shape and perianth shape), indicated that species were almost separated from one another in 74% of the total variation (Fig. 17B). In this subgenus, some species in the combined cluster plot were clustered overlapping. For example, Allium atroviolaceum, A. filidens, and A. filidentiforme (all species belongs to sect. Allium) are closely placed (Fig. 17B), but are distinguished mostly by perianth color, the relative length of filament with perianth and tepal size (Fig. 2A–C). Previous research has suggested that A. filidens s. str. is a rather polymorphic taxon with respect to the form of flowers and tepals (Khassanov, 1997). Moreover, A. anisotepalum (sect. Minuta; Fig. 4C) and A. griffithianum (sect. Avulsea; Fig. 3A) were mixed in the combined cluster plot (Fig. 17B), but the former is distinguished by the lanceolate and purplish pink perianth. Based on our results, both species may belong to the same section because they were not included in the recent phylogenetic studies (Friesen et al., 2006; Li et al., 2010). Recently, a monotypic section Macrostemon of subg. Allium was newly categorized based on a complete chloroplast genome (Yang et al., 2023). According to our results, the flower morphology of A. macrostemon (Fig. 2E) can be clearly distinguished from the other species of subg. Allium (Figs. 2-4), in line with the findings of Yang et al. (2023).
3.3.2. Subgenus Anguinum (G. Don ex Koch) N. FriesenThis subgenus consists of ten species and several varieties and is part of the second of three main evolutionary lines of Allium (Friesen et al., 2006; Li et al., 2010; Choi et al., 2012). Three accessions of two species from sect. Anguinum were studied. Two species (A. microdictyon and A. ulleungense) were well distinguished from each other based on qualitative (86.3% of the total variation) and combined qualitative and selected quantitative characteristics (90.7% of the total variation) (Fig. 17C and D). The perianth color is the most important characteristic among the two species (Fig. 4E and 5A). In addition, the molecular phylogenetic analysis clearly indicated that A. ulleungense is distinct from other species of the subg. Anguinum (Choi et al., 2019; Yang et al., 2023).
3.3.3. Subgenus Butomissa (Salisb.) N. FriesenThe subg. Butomissa is placed in the third evolutionary line within the Allium genus, including only two sections (Friesen et al., 2006). In this study, four accessions of three species from two sections (Austromontana and Butomissa) were investigated. We only provide finding from the quantitative cluster plot because informative qualitative characteristics were not significant (Fig. S7D). Based on the quantitative plot, all taxa are well separated (83.9% of the total variation) among the studied species (Fig. 18A). Additionally, two accessions of A. oreoprasum are separated into sub-cluster owing to size of floral morphology (Fig. 18A).
3.3.4. Subgenus Cepa (Mill.) RadićThis subgenus includes approximately 30 taxa and five sections (Friesen et al., 2006). We investigated 22 accessions of 15 taxa, including two varieties belonging to five sections (Annuloprason, Cepa, Condensatum, Sacculiferum, Schoenoprasum). The quantitative cluster plot revealed approximately 76.9% of the total variation and the species that were generally gathered, excluding sect. Annuloprason (Allium semenovii) and sect. Schoenoprasum (four taxa) (Fig. 18B). However, the combined cluster plot displays 83% of the total variation and the species that were better separated (Fig. 18C). In this subgenus, four quantitative characteristics, including perianth shape, reflexed point of tepal apex, filament margin toothed, and filament toothed position, are most important (Fig. 18C).
In sect. Schoenoprasum, A. karelinii is more similar to A. schoenoprasum var. schoenoprasum than to its variety, A. schoenoprasum var. foliosum, with 9–14 mm long tepals, pedicels usually shorter than perianth, tepals often with a reflexed point at the tip (Vvedensky, 1935; Ohwi, 1965); however, there is a slight difference between them, for example, A. karelinii has a purplish violet perianth (vs reddish purple) and obovate-lanceolate tepals (vs oblong-lanceolate) (Fig. 8C and D).
Molecular studies have failed to clearly distinguish species within sect. Sacculiferum (Choi et al., 2012). We found that floral morphology does not provide a statistically significant approach to delimiting species within this section either (Fig. 7). According to Choi (2009), the floral characteristics of this section show continuous variation among taxa. In addition, the polyploid is especially common in this group (Baasanmunkh et al., 2018). The taxa of sect. Sacculiferum can be distinguished by features such as the leaf blade (the texture, the color, the shape in cross-section) and the position of scape from bulbs during development (Kim et al., 2021). However, the main characteristics for classification are limited to vegetative organs that are affected by the environment. In addition, it has been difficult to distinguish between two species in this section: A. pseudojaponicum (Fig. 7B) and A. thunbergii (Fig. 7D and E), and their reproductive characteristics are not distinctly discontinuous (Choi, 2009). Similarly, Baasanmunkh et al. (2018) also suggested that Allium sacculiferum be considered as a synonym of A. thunbergii, because it does not clearly fulfill the requirements of any species concept. Therefore, further comprehensive studies of sect. Sacculiferum are needed to better understand their taxonomic rank.
3.3.5. Subgenus Melanocrommyum (Webb & Berth.) RouyThis subgenus comprises over 150 species and represents one of the largest subgenera in Allium (Gurushidze et al., 2008; Fritsch, 2012; Friesen et al., 2021). In this study, four species belonging to four sections (Compactoprason, Longibidentata, Megaloprason, and Miniprason) were investigated. The quantitative (84% of the total variation) and combined cluster plots (90% of the total variation) separated A. karataviense (sect. Miniprason) from A. fetisowii (sect. Longibidentata), while Allium giganteum (sect. Compactoprason) and A. stipitatum (sect. Megaloprason) remained overlapping (Fig. 19A and B). The most important characteristic among the studied species of this subgenus is perianth color (Fig. S7).
3.3.6. Subgenus Polyprason RadićThe subg. Polyprason belongs to the third evolutionary line, which includes over 50 species and four sections (Friesen et al., 2006; Bartolucci et al., 2022). Nine taxa (including one subspecies) belonging to two sections (Falcatifolia and Oreiprason) were investigated. The quantitative cluster plot shows approximately 73.8% of the total variation, wherein species were not clearly separated, except for Allium korolkowii (Fig. 19C). For the studied taxa here, 73.4% of the total variation can be explained by quantitative characteristics and five important qualitative characteristics, specifically, perianth shape, perianth color, inner tepal shape, reflexed point of tepal apex, and inner filament shape, except for in A. obliquum and A. petraeum (sect. Oreiprason). The flower morphology of both species is quite similar (Figs. 10E and 11A).
3.3.7. Subgenus Reticulatobulbosa (Kamelin) N. FriesenThis subgenus comprises over 80 species belonging to four sections and is placed in the third evolutionary line (Friesen et al., 2006; Li et al., 2010). In this study, 12 species belonging to three sections (Campanulata, Reticulatobulbosa, and Scabriscapum) were investigated. The quantitative cluster plot showed approximately 72.7% of the total variation where species are generally gathered (Fig. 20A); in contrast, the combined quantitative and qualitative cluster plot showed 79.1% of the total variation wherein most species are well separated (Fig. 20B). In this subgenus, the most important characteristics among the studied taxa are perianth shape, perianth color, inner tepal shape, filament relative length compared with perianth, and inner filament shape (Fig. S7). Notably, in subg. Reticulatobulbosa, Allium koreanum and A. malyschevii (sect. Reticulatobulbosa) have similar floral morphology; however, it differs by the color and shape of perianth (Fig. 12C and E).
3.3.8. Subgenus Rhizirideum (G. Don ex Koch) WendelboThe subgenus Rhizirideum includes over 40 species belonging to six sections and is placed in the third evolutionary line (Friesen et al., 2006, 2020; Jang et al., 2021). In this study, 19 accessions of 15 species belonging to four sections (Caespitosoprason, Eduardia, Rhizirideum, and Tenuissima) were investigated. Based on a quantitative cluster plot, almost all species (87.5% of the total variation) were distributed on the left side except Allium dumebuchum (Fig. 20C). The combined cluster plot shows that 62.6% of the total variation was well separated, except A. spirale, A. spurium (sect. Rhizirideum), A. tenuissimum, A. anisopodium, and A. vodopjanovae (sect. Tenuissima) (Fig. 20D). In this subgenus, the most important characteristics are perianth shape and color, inner tepal apex and shape, filament relative length, inner filament shape eight, filament toothed position, and ovary shape (Fig. S7).
In sect. Tenuissima, the flower morphology of Allium tenuissimum, A. anisopodium, and A. vodopjanovae are remarkably similar and all have obovoid inner tepals, non-exserted filaments, and an ovoid ovary; however, they are slightly distinguished by shape, size, and apex of outer tepals (Fig. 16C–E). In addition, these species are possibly inter-species hybridizations in the wild (Friesen, personal observation).
3.4. Morphological characteristics of selected flowers traits in the phylogenetic treePrevious phylogenetic studies have shown that Allium is monophyletic and includes three main evolutionary lines (Friesen et al., 2006; Li et al., 2010). However, phylogenetic evidence has been unable to confirm that species from subgenera Allium, Cepa, Reticulatobulbosa, and Polyrason (i.e., species in the third evolutionary line) are monophyletic (Friesen et al., 2006; Li et al., 2010; Xie et al., 2020a; Yang et al., 2023). Species in these subgenera differ in a variety of ways (Khan et al., 2022). In this study, we examined how five important morphological traits of flowers differ between these species and whether those traits mapped onto a phylogenetic tree (Fig. 21). Our findings indicate that the morphological characters of flowers also do not highlight monophyletic problems in the above-mentioned subgenera (Fig. 21). There is also no consensus on the phylogenetic significance of testa cell characteristics and whether they have been reflected in the different evolutionary levels recognized in Allium (see Yusupov et al., 2022). However, pistil characteristics have been strongly correlated with three major evolutionary lines (Fig. 21). For instance, except for subgenera Butomissa and Rhizirideum, members of the third evolutionary lineage possess hood-like appendages in the ovary (Fig. 21). Ovule numbers are key taxonomic characters for subg. Angunium species, which is placed in the second evolutionary line and which have only one ovule per locule, as expected in all the taxa studied (Fig. 21). Reticulate-fibrous bulb tunics of Allium eduardii (subg. Rhizirideum) are similar to those of subg. Reticulatobulbosa, but are distinct from those subg. Reticulatobulbosa, which have ovaries without appendages. A. monanthum and A. semenovii, which are placed at the first and third evolutionary lines, have only trigonous ovary styles and 3-cleft stigma among the taxa studied, respectively (Fig. 21). Therefore, the pistil characters of Allium semevonii require further study. Furthermore, this study has shown that the diversity and richness of pistil morphology (e.g., the shape of style and ovary) are useful for elucidating the specific species of Allium.

|
| Fig. 21 Selected flower morphological characteristics on phylogenetic tree. Phylogenetic tree was constructed with Bayesian inference posterior probability/maximum parsimony, which are given on each branch; maximum likelihood is shown below branches. Asterisks indicate maximum support values, bold asterisks indicate all maximum values of each values, and values lower than 50% are excluded. Ⅰ–Ⅲ and branch colors represent major clades. |
This comparative study on floral morphology of 74 Allium taxa confirmed that floral morphology provides useful taxonomic information for species delimitation in Allium. The quantitative characteristics of floral traits, which refer to their size, have been used to identify Allium species. However, we observed high variability in the size of the floral factors, showing a wide range of clusters and overlapping with other groups in the cluster plot with only quantitative factors. Therefore, a combination of qualitative and quantitative factors is required to address the taxonomy of Allium. In this study, we determined which qualitative characteristics influence clustering in each subgenus. Four quantitative characteristics—ovary shape, perianth color, perianth shape, and inner tepal apex—were strongly associated with clustering among the studied subgenera. In particular, perianth color was shown to be a key characteristic within subg. Melanocrommyum, Polyprason, and Reticulatobulbosa. Ovary shape, perianth shape, and inner tepal apex were significant characteristics within subg. Allium, Cepa, Rhizirideum. Our results also indicated that flower morphology of several species within various sections are not clearly distinguished, such as sect. Sacculiferum (subg. Cepa) and sect. Tenuissima (subg. Rhizirideum). Despite their importance, floral studies have been limited because observations on fresh material must be made in a specific season, and dry specimens do not provide satisfactory results (Pandey et al., 2022). Our findings suggest that understanding Allium phylogeny requires that more attention be paid to floral organs during field expeditions. In particular, inflorescences must be photographed in the field using a color chart and be preserved in 70% ethanol for subsequent measurement. The present study provides a basis for future studies on infrageneric relationships and evolutionary trends in Allium and other genera with a complicated taxonomy.
AcknowledgmentsOur study was supported by research grants from the Korea National Arboretum (Grant No. KNA1-1-26, 20-1) and the Mid-level professor Financial Program at Changwon National University in 2023. We would like to thank the three anonymous reviewers for valuable comments and further suggestions which have considerably improved the study.
Author contributions
HJC designed research. HJC, SB, KT and ZY collected samples. JEJ, SB, NN, SOY and HJC analyzed data. SB and JEJ writing – original draft. All the authors revised and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.06.009.
Armağan, M., 2021. Allium muratozelii (Amaryllidaceae), a new species from Turkey. Phytotaxa, 498: 255-264. DOI:10.11646/phytotaxa.498.4.3 |
Aryakia, E., Karimi, H.R., Nagvahi, M.R., et al., 2016. Morphological characterization of intra-and interspecific diversity in some Iranian wild Allium species. Euphytica, 211: 185-200. DOI:10.1007/s10681-016-1729-8 |
Baasanmunkh, S., Jang, J.E., Duchoslav, M., et al., 2018. Cytotype distribution and ecology of Allium thunbergii (= A. sacculiferum) with a special reference to South Korean populations. Korean J. Plant Taxon., 48: 278-288. DOI:10.11110/kjpt.2018.48.4.278 |
Baasanmunkh, S., Lee, J.K., Jang, J.E., et al., 2020. Seed morphology of Allium L. (Amaryllidaceae) from central Asian countries and its taxonomic implications. Plants, 9: 1239. DOI:10.3390/plants9091239 |
Baasanmunkh, S., Choi, H.J., Oyuntsetseg, B., et al., 2021. Seed testa sculpture of species of Allium L. (Amaryllidaceae) and its taxonomic implications. Turczaninowia, 24: 154-161. DOI:10.14258/turczaninowia.24.1.17 |
Bartolucci, F., Locchi, M., Castro, O.D., et al., 2022. Allium ducissae (A. subgen. Polyprason, Amaryllidaceae) a new species from the Central Apennines (Italy). Plants, 11: 426. DOI:10.3390/plants11030426 |
Celep, F., Koyuncu, M., Fritsch, R.M., et al., 2012. Taxonomic importance of seed morphology in Allium (Amaryllidaceae). Syst. Bot., 37: 893-912. DOI:10.1600/036364412X656563 |
Choi, H.J., 2009. Systematics of the Genus Allium (Alliaceae) in Korea and Northeastern China. Ph. D. thesis, The Chungbuk National University, Cheongju, Republic of Korea.
|
Choi, H.J., Oh, B.U., 2011. A partial revision of Allium (Amaryllidaceae) in Korea and north-eastern China. Bot. J. Linn. Soc., 167: 153-211. DOI:10.1111/j.1095-8339.2011.01166.x |
Choi, H.J., Davis, A.R., Cota-Sanchez, J.H., 2011. Comparative floral structure of four new world Allium (Amaryllidaceae) species. Syst. Bot., 36: 870-882. DOI:10.1600/036364411X604895 |
Choi, H.J., Giussani, L.M., Jang, C.G., et al., 2012. Systematics of disjunct northeastern Asian and northern north American Allium (Amaryllidaceae). Botany, 90: 491-508. DOI:10.1139/b2012-031 |
Choi, H.J., Yang, S., Yang, J.C., et al., 2019. Allium ulleungense (Amaryllidaceae), a new species endemic to Ulleungdo Island, Korea. Korean J. Plant Taxon., 49: 294-299. DOI:10.11110/kjpt.2019.49.4.294 |
Friesen, N., Pollner, S., Bachmann, K., et al., 1999. RAPDs and noncoding chloroplast DNA reveal a single origin of the cultivated Allium fistulosum from A. altaicum (Alliaceae). Am. J. Bot., 86: 554-562. DOI:10.2307/2656817 |
Friesen, N., Fritsch, R.M., Blattner, F.R., 2006. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso, 22: 372-395. DOI:10.5642/aliso.20062201.31 |
Friesen, N., Smirnov, S.V., Shmakov, A.I., et al., 2020. Allium species of section Rhizomatosa, early members of the Central Asian steppe vegetation. Flora, 263: 151536. DOI:10.1016/j.flora.2019.151536 |
Friesen, N., Smirnov, S.V., Leweke, M., et al., 2021. Taxonomy and phylogenetics of Allium section Decipientia (Amaryllidaceae): morphological characteristics do not reflect the evolutionary history revealed by molecular markers. Bot. J. Linn. Soc., 197: 190-228. DOI:10.1093/botlinnean/boab023 |
Friesen, N., Grützmacher, L., Skaptsov, M., et al., 2022. Allium pallasii and A. caricifolium — surprisingly diverse old steppe species, showing a clear geographical barrier in the Area of Lake Zaysan. Plants, 11: 1465. DOI:10.3390/plants11111465 |
Fritsch, R.M., 1992. Sepal nectarines in the genus Allium. In: Hanelt, P., Hammer, K., Knupffer, H. (Eds.) The Genus Allium - Taxonomic Problems and Genetic Resources (Proceedings of an International Symposium held at Gatersleben, June 11-13, 1991). IPK, Gatersleben, pp. 77-85.
|
Fritsch, R.M., 2012. Illustrated key to the sections and subsections and brief general circumscription of Allium subg. Melanocrommyum. Phyton, 52: 1-176. |
Fritsch, R.M., Friesen, N., 2002. Evolution, domestication, and taxonomy. In: Rabinovich, H.D., Currah, L. (Eds), Allium Crop Science: Recent Advances. CABI Publishing, Wallingford, pp. 5-30.
|
Govaerts, R., Kington, S., Friesen, N., et al., 2021. World Checklist of Amaryllidaceae family. Facilitated by the Royal Botanic Gardens, Kew, UK. http://apps.kew.org/wcsp/.
|
Gregory, M., Fritsch, R.M., Friesen, N., et al., 1998. Nomenclator Alliorum: Allium Names and Synonyms-A World Guide. Kew, Richmond.
|
Gurushidze, M., Fritsch, R.M., Blattner, F.R., 2008. Phylogenetic analysis of Allium subg. Melanocrommyum infers cryptic species and demands a new sectional classification. Mol. Phylogenet. Evol., 49: 991-1007. |
Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser., 4: 95-98. |
Han, T.S., Zheng, Q.J., Onstein, R.E., et al., 2020. Polyploidy promotes species diversification of Allium through ecological shifts. New Phytol., 225: 571-583. DOI:10.1111/nph.16098 |
Hanelt, P., Schulze-Motel, J., Fritsch, R.M., et al., 1992. Infrageneric grouping of Allium-the Gatersleben approach. In: Hanelt, P., Hammer, K., Knupffer, H., (Eds), The Genus Allium - Taxonomic Problems and Genetic Resources (Proceedings of an International Symposium held at Gatersleben, June 11-13, 1991). IPK, Gatersleben, pp. 107-123.
|
Herden, T., Hanelt, P., Friesen, N., 2016. Phylogeny of Allium L. Subgenus Anguinum (G. Don. ex WDJ koch) N. Friesen (Amaryllidaceae). Mol. Phylogenet. Evol., 95: 79-93. DOI:10.1016/j.ympev.2015.11.004 |
Jang, J.E., Park, J.S., Jung, J.Y., et al., 2021. Notes on Allium section Rhizirideum (Amaryllidaceae) in South Korea and northeastern China: with a new species from Ulleungdo Island. PhytoKeys, 176: 1-19. DOI:10.3897/phytokeys.176.63378 |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Khan, N., Fritsch, R.M., Sultan, A., et al., 2021. Allium (Amaryllidaceae) species in Pakistan: two new records and a new species from Zhob (Balochistan). Pakistan J. Bot., 53: 1759-1765. |
Khan, N., Friesen, N., Sultan, A., et al., 2022. Allium sulaimanicum: a new Allium species and section from Pakistan. Front. Plant Sci., 13: 1020440. DOI:10.3389/fpls.2022.1020440 |
Khassanov, F.O., 1997. Conspectus of the wild growing Allium species of Middle Asia. In: Ozuturk, M., secmen, O., Gork, G. (Eds), Plant life in Southwest and Central Asia. Ege University Press, Izmir, pp. 141-159.
|
Khassanov, F.O., 2018. Taxonomical and Ethnobotanical Aspects of Allium Species from Middle Asia with Particular Reference to Subgenus Allium. In: Shigyo, M., Khar, A., Abdelrahman, M. (Eds), The Allium Genomes. Compendium of Plant Genomes. Springer, Cham, pp. 11-21. https://doi.org/10.1007/978-3-319-95825-5_2.
|
Khassanov, F.O., Fritsch, R.M., 1994. New taxa in Allium L. subg. Melanocrommyum (Webb & Berth.) Rouy from central Asia. Linz. Biol. Beitr., 26: 965-990. |
Khorasani, M., Saeidi Mehrvarz, S., Zarre, S., 2018. Bulb tunic anatomy and its taxonomic implication in Allium L. (Amaryllidaceae: Allioideae). Plant Biosyst., 152: 1311-1328. DOI:10.1080/11263504.2018.1448013 |
Kim, Y.M., Lee, J., Choi, H.J., 2021. Allium stenodon (= A. baekdusanense), a neglected member among the Korean flora. Korean J. Plant Taxon., 51: 141-146. DOI:10.11110/kjpt.2021.51.2.141 |
Koçyiğit, M., Seregin, A.P., Özhatay, N., et al., 2016. Allium urusakiorum (Amaryllidaceae), a new member of the Balkan clade of the section Oreiprason from European Turkey. Phytotaxa, 275: 228-242. DOI:10.11646/phytotaxa.275.3.2 |
Li, Q.Q., Zhou, S.D., He, X.J., et al., 2010. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann. Bot., 106: 709-773. DOI:10.1093/aob/mcq177 |
Li, M.J., Tan, J.B., Xie, D.F., et al., 2016. Revisiting the evolutionary events in Allium subgenus Cyathophora (Amaryllidaceae): insights into the effect of the Hengduan Mountains Region (HMR) uplift and Quaternary climatic fluctuations to the environmental changes in the Qinghai-Tibet Plateau. Mol. Phylogenet. Evol., 94: 802-813. DOI:10.1016/j.ympev.2015.10.002 |
Li, M.J., Yu, H.X., Guo, X.L., et al., 2021. Out of the Qinghai–Tibetan plateau and rapid radiation across eurasia for Allium section Daghestanica (Amaryllidaceae). AoB Plants, 13: plab017. DOI:10.1093/aobpla/plab017 |
Mashayekhi, S., Columbus, J.T., 2014. Evolution of leaf blade anatomy in Allium (Amaryllidaceae) subgenus Amerallium with a focus on the North American species. Am. J. Bot., 101: 63-85. DOI:10.3732/ajb.1300053 |
Mes, T.H.M., Friesen, N., Fritsch, R.M., et al., 1997. Criteria for sampling in Allium based on chloroplast DNA PCR-RFLPs. Syst. Bot., 22: 701-712. DOI:10.2307/2419436 |
Munavvarov, A., Yusupov, Z., Ergashov, I., et al., 2022. Complete chloroplast genomes of ten species from subgenus Allium (Allium, Amaryllidaceae). Plant Divers. Central Asia, 2: 67-81. |
Namin, H.H., Mehrvarz, S.S., Zarre, S., et al., 2009. Pollen morphology of selected species of Allium (Alliaceae) distributed in Iran. Nord. J. Bot., 27: 54-60. DOI:10.1111/j.1756-1051.2009.00319.x |
Nguyen, N.H., Driscoll, H.E., Specht, C.D., 2008. A molecular phylogeny of the wild onions (Allium; Alliaceae) with a focus on the western North American center of diversity. Mol. Phylogenet. Evol., 47: 1157-1172. DOI:10.1016/j.ympev.2007.12.006 |
Ohwi, J., 1965. Allium L. In: Meyer, F.G., Walker, E.H. (Eds), Flora of Japan. Smithsonian Inst., Washington, pp. 294-296.
|
Pandey, A., Malav, P.K., Rai, K.M., et al., 2022. Genus Allium L. of the Indian Region: A Field Guide for Germplasm Collection and Identification. ICAR-National Bureau of Plant Genetic Resources, New Delhi, India.
|
Pandey, A., Rai, K.M., Malav, P.K., et al., 2021. Allium neglectum (Amaryllidaceae): a new species under subg. Rhizirideum from Uttarakhand Himalaya, India. PhytoKeys, 183: 77-93. DOI:10.3897/phytokeys.183.65433 |
Peruzzi, L., Carta, A., Altinordu, F., 2016. Chromosome diversity and evolution in Allium (Allioideae, Amaryllidaceae). Plant Biosyst., 151: 212-220. |
R Core Team, 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
|
Rambaut, A., 2012. FigTree v1. 4. Molecular evolution, phylogenetics and epidemiology. University of Edinburgh, Institute of Evolutionary Biology.
|
Ronquist, F., Teslenko, M., Van Der Mark, P., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Royal Horticultural Society, 2015. RHS Colour Chart. Sixth Edition. The Royal Horticultural Society, London.
|
Seregin, A.P., Anačkov, G., Friesen, N., 2015. Molecular and morphological revision of the Allium saxatile group (Amaryllidaceae): geographical isolation as the driving force of underestimated speciation. Bot. J. Linn. Soc., 178: 67-101. DOI:10.1111/boj.12269 |
Sinitsyna, T.A., Herden, T., Friesen, N., 2016. Dated phylogeny and biogeography of the eurasian Allium section Rhizirideum (Amaryllidaceae). Plant Syst. Evol., 302: 1311-1328. DOI:10.1007/s00606-016-1333-3 |
Stamatakis, A., 2006. RAcML-VL-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Swofford, D.L., Documentation, B., 1989. Phylogenetic Analysis Using Parsimony. Illinois Natural History Survey, Champaign.
|
Thompson, J.D., Gibson, T.J., Plewniak, F., et al., 1994. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25: 4876. |
Vvedensky, A.I., 1935. Allium L. In: Komarov, V.L. (Ed), Flora of the U.S.S. R, vol. 4. Botanical Institute of Academy of Science, Leningrad, pp. 112-280.
|
Xie, D.F., Tan, J.B., Yu, Y., et al., 2020a. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot., 125: 1039-1055. DOI:10.1093/aob/mcaa024 |
Xie, D.F., Yu, Y., Wen, J., et al., 2020b. Phylogeny and highland adaptation of Chinese species in Allium section Daghestanica (Amaryllidaceae) revealed by transcriptome sequencing. Mol. Phylogenet. Evol., 146: 106737. DOI:10.1016/j.ympev.2020.106737 |
Xie, D.F., Xie, F.M., Jia, S.B., et al., 2020c. Allium xinlongense (Amaryllidaceae, Allioideae), a new species from western Sichuan. Phytotaxa, 432: 274-282. DOI:10.11646/phytotaxa.432.3.4 |
Xu, J.M., Kamelin, R.V., 2000. Allium L. In: Wu, Z.Y., Raven, P.H. (Eds), Flora of China, vol. 24. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, pp. 165-202.
|
Yang, J.Y., Kim, S.H., Gol, H.Y., et al., 2023. New insights into the phylogenetic relationships among wild onions (Allium, Amaryllidaceae), with special emphasis on the subgenera Angunium and Rhizirideum, as revealed by plastid genomes. Front. Plant Sci., 14: 1124277. DOI:10.3389/fpls.2023.1124277 |
Yusupov, Z., Ergashov, I., Volis, S., et al., 2022. Seed macro- and micro- morphology in Allium (Amaryllidaceae) and its phylogenetic significance. Ann. Bot., 129: 869-911. DOI:10.1093/aob/mcac067 |
Zuraw, B., Weryszko-Chmielewska, E., Laskowska, H., et al., 2009. The structure of septal nectaries and nectar presentation in the flowers of Allium aflatunense B. Fedtsch. Acta Agrobot., 62: 31-41. |
Zuraw, B., Weryszko-Chmielewska, E., Laskowska, H., et al., 2010. The location of nectaries and nectar secretion in the flowers of Allium giganteum Regel. Acta Agrobot., 63: 33-40. |



