b. Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China
The plastid is widely present in algae and plants with important functions in the process of photosynthesis, carbon fixation, and stress response (Shi et al., 2022). Despite the consistency between plastid genomes in plants, size variation of the plastid genome, gene loss, structure changes, and pseudogenes have been frequently observed (Ivanova et al., 2017). Plastid genome has currently shown a wide application in research of phylogeny, populations and biogeography in combination with nucleus genome (Wang et al., 2021). As is known, the plastid originates from cyanobacteria through two independent secondary endosymbiosis and has its own genetic replication mechanism (Howe et al., 2003). Thus, plastids have been suggested not to be transmitted according to the rules of Mendelian genetics, but generally demonstrate uniparental inheritance mode (Birky, 1995). Concerning the uniparental inheritance of plastids, research have been mainly conducted on species of angiosperms, but few on gymnosperms or ferns (Kita et al., 2005; Li et al., 2013).
Uniparental inheritance potentially evolved from relaxed organelle inheritance patterns because it mitigates the spread of cytoplasmic components. Three possible patterns were suggested for plastid inheritance, including maternally, paternally and biparentally (Reboud and Zeyl, 1994; Kormutak et al., 2018). Angiosperms seem to display mainly a maternal transmission of its plastids (Greiner et al., 2015). In most gymnosperms, the plastid transmission is considered to occur exclusively by paternal inheritance. Cryptomeria japonica was the first gymnosperm known to inherit its plastid genome from its male parent (Ohba et al., 1971). Most conifers exhibit exclusively or predominantly paternal inheritance of both plastids and mitochondrias (Mogensen, 1996; Jansen and Ruhlman 2012; Lubna et al., 2021). However, there are a few exceptions. Several gymnosperms, including Ginkgo, are supposed to probably exhibit maternal inheritance of both plastids and mitochondrias. A few of studies have been carried out for plastid inheritance of gymnosperm species, most of which are based on cytological analysis (Mogensen, 1996; Guo et al., 2005; Zhong et al., 2011). So far, the information seems to be fragmentary and no strong evidence is available specifically for Ginkgo.
Ginkgo biloba L. is a sole representative of Ginkgoales - one of the eight extant gymnosperm orders (Yang et al., 2022). This species shows a geographical distribution range in broadleaved forest of both subtropical and temperate zones in Eastern Asia. It is widely cultivated all over the world due to its highly horticultural, medicinal and ecological values. Several glacial refugial of G. biloba were identified in subtropical evergreen broad forest in China (Gong et al., 2008; Zhao et al., 2019). This species is a dioecious tree plant, with separate male and female individuals. The characteristics of separate male and female individuals produced by dioecy could rule out the possibility of self-pollination and show advantages in studying plastid genetic research (Zhai and Sun, 2015). During genetic crosses, different parents were selected for artificial pollination to produce hybrid offsprings, then genetic similarities and differences between the parents and offsprings were compared to explore their organelle inheritance more distinctively. A few of research have been conducted on G. biloba since its first plastid genome was assembled and annotated (Lin et al., 2012; Zhou et al., 2020; Yang et al., 2021). However, previous research only focused on structural comparison and gene composition of the plastid genome, which aimed to develop molecular markers, analyze plant phylogeny, or compare genomics. So far, no molecular evidence is available to reveal the inheritance mode of the plastid genome of G. biloba.
Traditional methods to investigate organelle inheritance involve hybridization experiment, using electron microscopy or DAPI fluorescence microscopy, as well as molecular markers, such as restriction fragment length polymorphism (RFLPs), and simple sequence repeats (SSRs) (Zhong et al., 2011). Most recently, the development of next-generation sequencing technologies has stimulated a rapid and successful achievement in the database of plastid genomes (Wang et al., 2018). Thus, genomic data demonstrates a strong potentiality to explore the molecular mechanism of organelle inheritance mode (Villanueva-Corrales et al., 2021). In the current study, we conducted artificial pollination for three crosses of Ginkgo biloba. Using next generation sequencing, plastid genomes of all the parents and offsprings were investigated and compared, showing strong genomic evidence on maternal inheritance mode.
Two female and three male trees of Ginkgo biloba were selected as candidates for genetic crosses. Artificial pollinations were conducted in the county of Pingtian, Nanxiong, Guangdong Province, in southern China, which is suggested as one of the glacial refugia with a large population of Ginkgo trees (Zhao et al., 2019). Two female Ginkgo trees over 100 years, showing an average diameter at breast height (DBH) more than 80 cm and an average height of ~20 m, are recorded to possess high seed production. Thus, they were chosen as maternal origin. Three old male Ginkgo trees, estimated to be over 100 years, showing an average DBH more than 80 cm and an average height of ~20 m, were selected as paternal origin. In April, 2016, the branches of the female trees were wrapped with parchment paper and the male flowers from the nearby male trees were all removed before flowering in order to avoid any pollen pollution. Artificial pollinations were conducted for three crosses. For each cross, seeds were collected and germinated. The accession information for each cross and the corresponding offsprings in the current study were listed in Table S1.
Fresh leaves were collected from the seedlings in the spring. Leaf tissue was ground in tubes with glass beads with the tissue homogenizer Tissuelyser-96 (Shanghai Jingxin Industrial Development Co., Ltd). Total genomic DNA was extracted with modified cetyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle, 1987). An Illumina HiSeq2000 sequencer was used to sequence paired-end (PE) sequencing libraries with an average 300 bp insert length. Over 10 million clean reads were passed through quality control with a 150 bp each read length.
We assembled the plastid genome using GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle) and editing de novo assembly graph using Bandage (Coil et al., 2015). The plastid genome of Ginkgo biloba (MN443423.1) (Yang et al., 2021) was downloaded from NCBI and used as the reference sequence. PGA (https://github.com/quxiaojian/PGA) (Qu et al., 2019), Geneious 9.1.4 (Biomatters Ltd., Auckland, New Zealand), and ARAGORN program were jointly used for annotating the plastid genome in comparison with references. The circular genome map of G. biloba was illustrated with the Organellar Genome DRAW tool (OGDRAW, available online: http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al., 2013).
The plastid genome sequences from the finalized data set of all parents and F1 individuals were aligned with MAFFT v.7.0.0 (Katoh and Standley, 2013) with manual adjustment when necessary. Using DnaSP 6 (Rozas et al. al., 2017), we determined the substitutions numbers and types of the sequences, and also performed comparative analyses of the nucleotide diversity (Pi) among the complete plastid genomes of the parents and offsprings based on a sliding window analysis. All the protein coding genes (PCGs) were extracted and aligned using MAFFT v.7.0.0. Using DnaSP 6 and MEGA v.11.0 (Tamura et al., 2021), we estimated the genetic distance between the parents and offsprings based on Kimura 2-parameter (K2–P's) model.
The original reads were mapped to plastid genome references using BWA-0.7.17-R1188 (Li, 2013) to detect the SNPs between the parents and the offsprings. The generated bam files were sorted and variant calls were performed by Samtools-1.7 and Bcftools v.1.9, respectively (Danecek et al., 2021; Li, 2009, 2011). Sequence similarities between the parents and offsprings were used as reference indicators. To identify SNPs among parents and F1 individuals, we inspected the alignment results in Geneious and generated haplotype files using DnaSP 6. All the SNPs were statistic to identify the polymorphism between the parents and offsprings.
All parents and F1 individuals were used to reconstruct phylogenetic relationships in order to trace plastid genome inheritance. Cycas revoluta (JN867588) was used as outgroup. Maximum parsimony (MP) method was applied for phylogenetic analyses by PAUP* v.4.0 (Swofford, 2002). Bootstrap values were calculated in PAUP* with 1000 bootstrap replicates. Neighbor-Joining (NJ) method was also used to conduct phylogenetic analysis among all parents and F1 individuals by MEGA v.11.0 (Tamura et al., 2021). Additionally, using GetOrganelle pipeline, we assembled nuclear ribosomal RNA sequences (18S-ITS1-5.8S-ITS2-26S) and extracted the ITS regions, which were further used to reconstruct phylogenetic trees as a control.
A total number of 2 × 150 bp pair-end reads of 6, 504, 575–35, 285, 882 were produced with 1.95–10.59 Gb of clean data. All reads data were deposited in the NCBI Sequence Read Archive (SRA) (Table S3). The size of the complete plastid genome is from 156, 970 bp to 156, 999 bp, which is supposed to be smaller than cycads ranging from 161, 815 to 166, 341 bp (Wu and Chaw, 2015). The plastid genome displays a typical quadripartite structure, including a pair of IRs of 17, 733 bp each in length, separated by LSC region ranging from 99, 248 bp to 99, 267 bp and SSC region from 22, 257 bp to 22, 266 bp (Fig. 1 and Table S2). The GC content of the plastid genome is 39.6%, within that range of gymnosperms from 34.3% to 40.11% (Wu and Chaw, 2015). The plastid genome encodes 134 predicted functional genes when duplicated genes in the IR regions were only counted once. A total of 85 PCGs, 41 tRNA genes and eight rRNA genes are identified among all the individuals (Table S2). The remaining non-coding regions include introns, intergenic spacers, and pseudogenes. Each of the four genes (two PCGs and two tRNA genes) contains only one intron.
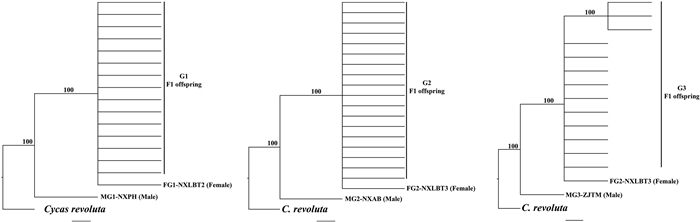
|
| Fig. 1 Phylogenomic analysis for all parents and F1 offsprings in three genetic crosses (G1, G2 and G3) of Ginkgo bilboa based on Maximum Parsimony (MP) method. Cycas revoluta was used as an outgroup. Numbers above the branches represent bootstrap values. |
The IRs can be normally identified by a central unit of eight rRNA genes including rrn4.5, rrn5, rrn16 and rrn23. In Ginkgo biloba, the IRs were detected to be composed of 13 genes, including three PCGs, six tRNA genes, and four rRNA genes. In comparison with other gymnosperms, the length of IRs in G. biloba is shorter than Cycas revoluta (25, 066 bp; JN867588), Nothotsuga longibracteata (25, 918 bp) and Ephedra sinica (20, 743 bp), but longer than Cycas taitungensis (15, 830 bp) and some conifers that lost IRs. Due to the lack of ycf2 gene, the IRs in G. biloba are relatively shorter than that of the most angiosperms. Consequently, the LSC/IR junction region, which is supposed to retain the complete ycf2 gene and the adjoining psbA or rpl23-rps3 gene cluster in order, has been changed and produces a pseudolized Ψrpl23 gene.
The average nucleotide variability (Pi) value was estimated to be 0.75 × 10−5 among parents and offsprings based on the comparative analysis with DnaSP 6 (Fig. S2). The nucleotide variability (Pi) is overall low with only eight genes displaying relatively high values (Pi > 0.0001), which are psbK-I, trnG-R, infA-rps8, ycf2, trnL-ndhB, ndhA intron, ycf1, and ndhB-trnL. The first four loci are present in the LSC, while two genes ndhA intron and ycf1 in SSC. Therefore, the highest variation was found in the LSC region with Pi ranging from 0.0001 to 0.0014, followed by the SSC region (Fig. S2). The IRs had much lower nucleotide diversity, each of which possesses only one gene showing Pi > 0.0001. Those eight highly variable loci are thus suggested as phylogenetic informative markers for population research of Ginkgo biloba.
Based on the K2–P's parameter model, we calculated the genetic distance among the parents and offsprings using 86 PCGs. The K2–P's genetic distance is major generated by that between male parent and offsprings, with the value of (0.06–1.27) × 10−4 among the three genetic crosses (Table 1). The K2–P's genetic distance between female parent and offsprings is extremely weak with overall values to be 0. We aligned the plastid genome sequences for all Ginkgo parents and off springs to detect the SNPs and check the numbers of offsprings that are consistent with female or male parent. Notably, all offsprings show SNPs identical to their female parent (Table 1). Consequently, all offsprings possess the same haplotypes with female parent based on three genetic crosses (Table S4).
| Crosses | Parents (F) | Parents (M) | K2–P's genetic distance (× 10−4) | No. of SNPs | |||||||
| F1 vs. F | F1 vs. M | M | F | Introns | Exon | Intergenic | Total | ||||
| G1 | FG1-NXLBT2 | MG1-NXPH | 0 | 0.57 | 0 | 9 | 1 | 4 | 4 | 9 | |
| G2 | FG2-NXLBT3 | MG2-NXAB | 0 | 1.27 | 0 | 20 | 0 | 1 | 19 | 20 | |
| G3 | FG2-NXLBT3 | MG3-ZJTM | 0 | 0.06 | 0 | 1 | 1 | 0 | 0 | 1 | |
| Average | 0 | 0.63 | 0 | 30 | 2 | 5 | 23 | 30 | |||
| M: male; F: female; F1: offsprings. | |||||||||||
We performed phylogenetic analyses for each cross, in order to detect the evolutionary relationship of the parent and offsprings based on MP and NJ trees (Figs. 1, S3 and S4). In each cross, all F1 individuals grouped with their female parent, forming a monophyly with a high bootstrap value, while the male parent was divergent from them based on MP and NJ analysis (Fig. 1). As for the control based on ITS data in G1 and G2 crosses, some individuals of the F1 individuals grouped with female parent, while others with male parents. No result was shown for G3 cross as there is no variation detected in ITS among the individuals.
The plastid genome of Ginkgo biloba is revealed to be a circular molecule about 156, 978 bp with a pair of IRs (35, 466 bp) separated by large single-copy (LSC: 99, 254 bp) and small single-copy (SSC: 22, 258 bp) regions with GC content 39.6% (Fig. S1), which is consistent with the result given by Lin et al. (2012) and Lubna et al. (2021), although the total lengths differ slightly. The plastid genome of G. biloba is characterized by the shortened IR region due to the complete loss of ycf2, which is about 7, 269 bp in length (Lin et al., 2012; Lubna et al., 2021). This gene is detected to be informative at population level with relatively high nucleotide diversity (Pi > 0.0001). It is also suggested to be an information phylogenetic marker for ferns and gymnosperms, as it shows two copies in the IR regions in Cycas and Genetum, but only one copy in LSC or SSC region in G. biloba and Pinus, respectively (Lin et al., 2012). Therefore, the evolution of ycf2 gene demonstrates to be potential in phylogenetic analysis and speciation of plant species.
In the current study, altogether 134 genes were identified including 85 PCGs, 41 tRNAs and four rRNAs. The total number of the genes is consistent with the previous research (Lin et al., 2012). However, when we compared with the data set in GenBank deposited by Lin et al. (2012), about ten genes were missing in the annotations of the plastid genome. After annotations for all genes, the gene rpl23 was also revealed to be pseudolized. This gene is detected to be pseudo due to the truncated 5' region as in comparison with Cycas (Lin et al., 2012; Lubna et al., 2021). The gene rpl23 is commonly found in angiosperms, but lost in some gymnosperms, therefore, the pseudo rpl23 demonstrates strong evolutionary implications in plant species as well.
As is known, the inheritance of plastid genome differs from that of the nuclear genome. Maternal inheritance, which is the most common form of seed plant organelle transmission, demonstrates the transfer of chloroplasts from the female parent to the progeny. Maternal inheritance of chloroplast occurs when the organelles are only present in the cytoplasm of the egg cells during the fertilization of eggs in seed plants (Vaughn et al., 1980). Though paternal inheritance of plastid genome is widely distributed in gymnosperms, several species have been reported to show maternal inheritance, including Ginkgo, Gnetum, and Cycas (Neale et al., 1989; Mogensen, 1996). However, the evidence for uniparental inheritance of gymnosperms is mostly based on cytological method previously. The current study is further confirmed plastid genome inheritance in Ginkgo based on genomic data using next generation sequencing.
Concerning the investigations of organelle inheritance of plant species, traditional methods including hybridization experiment, electron microscopy, DAPI fluorescence microscopy or genetic markers demonstrate several disadvantages. First, the cytological methods only show the process of the transfer of chloroplasts without any further investigation in the offsprings. No molecular evidence is available to prove the phylogenetic relationship among the parents and offsprings. Second, the traditional molecular markers, such as PCR-RFLP or SSRs, are less informative and effective than SNPs based on genomic data. With the development of next-generation sequencing technologies, a rapid and successful achievement has been promoted in the database of genomic data, among which is plastid genome (Wang et al., 2018). Genomic data shows a strong potentiality to be applied in the exploration of molecular mechanism of organelle inheritance (Villanueva-Corrales et al., 2021). Additionally, artificial crosses help to control the origins of male and female parents, which make possible to compare the genomic constituent among parents and offsprings. The combination of manually genetic crosses and chloroplast genomic data is an efficient way to investigate the inheritance mode of the chloroplasts in land plants. In our study, strong molecular evidence has been provided for maternal inheritance mode of its plastid genomes. Therefore, we suggested artificial crosses together with subsequent verification of SNPs among parents and progenies to be a recommendable way to directly infer organelle genome inheritance in land plants.
AcknowledgementsWe are very grateful to Miss Juan Zhou and Dr. Wanzhen Liu for their assistance in field work. This work is supported by the National Natural Science Foundation of China (32270218 and 31970231).
Data availability
The high-quality sequencing data of current study were deposited in the NCBI under Bioproject accession number PRJNA866875. Other supporting data were provided within the article.
Author contributions
W.G. was the principal investigator and wrote the manuscript. R.Z. contributed mainly to field work, including sample collections and artificial pollinations. M.F. and H.K. contributed equally to lab work, data analysis, and figures. M.F. edited the manuscript and had discussion with H.K. and M.L.
Declaration of competing interest
The authors declared no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.09.001.
Birky, C.J., 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. U.S.A., 92: 11331-11338. DOI:10.1073/pnas.92.25.11331 |
Coil, D., Jospin, G., Darling, A.E., 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics, 31: 587-589. DOI:10.1093/bioinformatics/btu661 |
Danecek, P., Bonfield, J.K., Liddle, J., et al., 2021. Twelve years of SAMtools and BCFtools. GigaScience, 10: 1-4. DOI:10.14232/ejqtde.2021.1.24 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phyto. Chem. Bull., 19: 11-15. |
Gong, W., Zeng, Z., Chen, Y., et al., 2008. Glacial refugia of Ginkgo biloba and human impact on its genetic diversity: evidence from chloroplast DNA. J. Integr. Plant Biol., 50: 368-374. DOI:10.1111/j.1744-7909.2007.00375.x |
Greiner, S., Sobanski, J., Bock, R., 2015. Why are most organelle genomes transmitted maternally?. Bioessays, 37: 80-94. DOI:10.1002/bies.201400110 |
Guo, F., Hu, S., Yuan, Z., et al., 2005. Paternal cytoplasmic transmission in Chinese pine (Pinus tabulaeformis). Protoplasma, 225: 5-14. DOI:10.1007/s00709-005-0088-4 |
Howe, C.J., Barbrook, A.C., Koumandou, V.L., et al., 2003. Evolution of the chloroplast genome. Phil. Trans. Roy. Soc. Lond. B, 358: 99-107. DOI:10.1098/rstb.2002.1176 |
Ivanova, Z., Sablok, G., Daskalova, E., et al., 2017. Chloroplast genome analysis of resurrection Tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci., 8: 204. |
Jansen, R.K., Ruhlman, T.A., 2012. Plastid genomes of seed plants. In: Genomics of Chloroplasts and Mitochondria, Advances in Photosynthesis and Respiration. Springer, Dordrecht, The Netherlands, pp. 103-126.
|
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kita, K., Kurashige, Y., Yukawa, K., et al., 2005. Plastid inheritance and plastome-genome incompatibility of intergeneric hybrids between Menziesia and Rhododendron. J. Jpn. Soc. Hortic. Sci., 74: 318-323. DOI:10.2503/jjshs.74.318 |
Kormutak, A., Galgoci, M., Sukenikova, D., et al., 2018. Maternal inheritance of chloroplast DNA in Pinus mugo Turra: a case study of Pinus mugo × Pinus sylvestris crossing. Plant Syst. Evol., 304: 71-76. DOI:10.1007/s00606-017-1449-0 |
Li, H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27: 2987-2993. DOI:10.1093/bioinformatics/btr509 |
Li, H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics. https://ui.adsabs.harvard.edu/abs/2013arXiv1303.
|
Li, H., Handsaker, B., Wysoker, A., et al., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25: 2078-2079. DOI:10.1093/bioinformatics/btp352 |
Lin, C., Wu, C., Huang, Y., et al., 2012. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evol., 4: 374-381. DOI:10.1093/gbe/evs021 |
Lohse, M., Drechsel, O., Kahlau, S., et al., 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res., 41: 575-581. DOI:10.1093/nar/gks1075 |
Lubna, A.S., Khan, A.L., Jan, R., et al., 2021. The dynamic history of gymnosperm plastomes: insights from structural characterization, comparative analysis, phylogenomics, and time divergence. Plant Genome, 14: e20130. DOI:10.1002/tpg2.20130 |
Mogensen, H.L., 1996. The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot., 83: 383-404. DOI:10.1002/j.1537-2197.1996.tb12718.x |
Neale, D.B., Marshall, K.A., Sederoff, R.R., 1989. Chloroplast and mitochondrial DNA are paternally inherited in Sequoia sempervirens D. Don Endl. Proc. Natl. Acad. Sci. U.S.A., 86: 9347-9349. DOI:10.1073/pnas.86.23.9347 |
Ohba, K., Iwakawa, M., Okada, Y., et al., 1971. Paternal transmission of a plastid anomaly in some reciprocal crosses of Sugi, Cryptomeria japonica D. Don. Silvae Genet., 20: 101-107. |
Qu, X., Moore, M.J., Li, D., et al., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. DOI:10.1186/s13007-019-0435-7 |
Reboud, X., Zeyl, C., 1994. Organelle inheritance in plants. Heredity, 72: 132-140. DOI:10.1038/hdy.1994.19 |
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J.C., et al., 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol., 34: 3299-3302. DOI:10.1093/molbev/msx248 |
Shi, Y., Ke, X., Yang, X., et al., 2022. Plants response to light stress. J. Genet. Genomics, 49: 735-747. DOI:10.1016/j.jgg.2022.04.017 |
Swofford, D.L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). version 4.
|
Tamura, K., Stecher, G., Kumar, S., 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol., 38: 3022-3027. DOI:10.1093/molbev/msab120 |
Vaughn, K.C., Debonte, L.R., Wilson, K.G., et al., 1980. Organelle alteration as a mechanism for maternal inheritance. Science, 208: 196-198. DOI:10.1126/science.208.4440.196 |
Villanueva-Corrales, S., García-Botero, C., Garcés-Cardona, F., et al., 2021. The complete chloroplast genome of Plukenetia volubilis provides insights into the organelle inheritance. Front. Plant Sci., 12: 667060. DOI:10.3389/fpls.2021.667060 |
Wang, H., Park, S.Y., Lee, A.R., et al., 2018. Next-generation sequencing yields the complete chloroplast genome of C. goeringii acc. smg222 and phylogenetic analysis. Mitochondrial DNA B Resour., 3: 215-216. DOI:10.1080/23802359.2018.1437812 |
Wang, Y., Wang, S., Liu, Y., et al., 2021. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics, 22: 103. DOI:10.1186/s12864-021-07394-8 |
Wu, C., Chaw, S., 2015. Evolutionary stasis in Cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol. Evol., 7: 2000-2009. DOI:10.1093/gbe/evv125 |
Yang, X., Zhou, T., Su, X., et al., 2021. Structural characterization and comparative analysis of the chloroplast genome of Ginkgo biloba and other gymnosperms. J. For. Res., 32: 765-778. DOI:10.1007/s11676-019-01088-4 |
Yang, Y., Ferguson, D.K., Liu, B., et al., 2022. Recent advances on phylogenomics of gymnosperms and a new classification. Plant Divers., 44: 340-350. DOI:10.1016/j.pld.2022.05.003 |
Zhai, F., Sun, Z., 2015. Progress in study on sexual differences of woody dioecious plants. Sci. Silvae Sin., 51: 110-116. |
Zhao, Y., Fan, G., Yin, P., et al., 2019. Resequencing 545 ginkgo genomes across the world reveals the evolutionary history of the living fossil. Nat. Commun., 10: 4201. DOI:10.1038/s41467-019-12133-5 |
Zhong, Z., Li, N., Qian, D., et al., 2011. Maternal inheritance of plastids and mitochondria in Cycas L. (Cycadaceae). Mol. Genet. Genom., 286: 411-416. DOI:10.1007/s00438-011-0653-9 |
Zhou, X., Liao, Y., Kim, S., et al., 2020. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep., 10: 13723. DOI:10.1038/s41598-020-69305-3 |



