b. Wolfson College, Oxford University, Oxford, UK;
c. Institute of Biomedical and Environmental Science & Technology, School of Life Sciences, University of Bedfordshire, Park Square, Luton, UK;
d. School of Life Sciences, Shanxi University, Taiyuan, China;
e. College of Horticulture and Landscape, Yunnan Agricultural University, Kunming, China
High-quality fruit has become a priority for many growers and consumers as horticulture has improved. Fruit quality is usually evaluated based on its color, appearance, flavor, texture, and nutritional value (Barrett et al., 2010). Among them, fruit flavor dominates fruit evaluation. The flavor is composed of two parts, aroma and taste. Fruit aromas are primarily produced by volatile compounds (El Hadi et al., 2013), and the taste of fruits are affected by sugars and acids, which are the main determinants of fruit taste (Barrett et al., 2010). Some secondary metabolites also affect the quality of fruits, such as flavonoids, terpenoids and other compounds (El Hadi et al., 2013; Liu et al., 2022). These not only provide flavor for fruits, but also participate in the growth and development of plants, and help plants resist the damage caused by adverse environments such as ultraviolet radiation, frost and drought (Patrick et al., 2013; Pichersky and Raguso, 2018; Treutter, 2006). At the same time, these compounds also play an important role in human health. For example, flavonoids are antioxidants, inhibiting obesity, and protecting heart health (Khan et al., 2021; Oliveira et al., 2022). Terpenoids are also reported to be beneficial because of their anti-inflammatory and anti-cancer effects (Kiyama, 2017).
Hawthorn is an important medicinal and edible fruit tree in the Rosaceae family, it is widely distributed in Asia, Europe, and America (Phipps et al., 1990). The Hawthorn genus (Crataegus) contains over 200 species, among which ~20 species are distributed across China (Gu and Spongberg, 2003). As a food, hawthorn is widely used in the food industry because of its special sour taste and rich nutritional value. Hawthorn also has significant value as traditional Chinese medicine, and it is often used to treat indigestion, abdominal pain, hyperlipidemia, and other diseases (Commission, 2020). Hawthorn fruit is rich in flavonoids, organic acids, terpenoids, and vitamin C, and has preventive effects on various diseases and is beneficial to human health (Gundogdu et al., 2014; Orhan, 2018; Zhang et al., 2022). For example, studies have shown that hawthorn has the effect of preventing cardiac disease, preventing atherosclerosis, anti-microbial activity, anti-inflammatory, anti-diabetic, stomach and liver protection (Alirezalu et al., 2020; Orhan, 2018; Zhang et al., 2022). Therefore, hawthorn has become one of the indispensable fruits in health care.
The hawthorns used for cultivation in China are mainly Crataegus pinnatifida, C. hupehensis, C. scabrifolia, and C. bretschneideri (Du et al., 2019; Hu et al., 2021). Research on hawthorn currently focuses mainly on cultivated hawthorn (C. pinnatifida var. major) with less research on other wild species. However, studies have shown that different hawthorn species have different contents of organic acids, sugars, flavonoids, and other compounds (Gundogdu et al., 2014; Lund et al., 2020). Therefore, research on other hawthorn species is also of great potential significance. Our study focused on Crataegus chungtienensis, an endemic species of southwest China, which is mainly distributed at an altitude of about 3500 m (Gu and Spongberg, 2003). Compared with the cultivated hawthorn, the fruit of the C. chungtienensis has obvious differences in appearance and taste. Ripe C. chungtienensis fruits are red, 0.6 cm in diameter, without brown spots on the surface, high edible ratio, and taste sweet and almost not sour (Fig. S1) (Gu and Spongberg, 2003). The special flavor of C. chungtienensis gives it a special value in research and utilization.
In recent years, multi-omics combined analysis methods including transcriptomics and metabolomics have been widely used in the study of fruit flavor metabolic pathways and the identification of regulatory genes (Gong et al., 2021; Peng et al., 2022; Wang et al., 2022). In the present study, leaves, flowers, young and mature fruits of Crataegus chungtienensis were selected as the research material. The changes in genes related to flavonoids, terpenoids, saccharides, and organic acid synthesis pathways during the fruit development of C. chungtienensis were analyzed by transcriptome data. Furthermore, metabolome data was used to analyze flavonoids, terpenoids, and flavor-related metabolites in young and mature fruits of C. chungtienensis. Finally, through integrated analysis of metabolome and transcriptome, we studied the changes of gene expression and metabolites from young to mature fruit and provide insights into the regulatory networks related to the formation of fruit quality in C. chungtienensis. Overall, we have provided metabolite and gene data that relate to the fruit quality formation in C. chungtienensis, as well as a theoretical basis for improving the use of C. chungtienensis.
2. Materials and methods 2.1. Plant materialsThe materials were collected from the same Crataegus chungtienensis tree planted in the Shangri-La Alpine Botanical Garden. The collected samples were divided into two parts. One for preparative transcriptome sequencing, including leaves (LE), flowers (FL), young fruits (YF), and mature fruits (MF), with three biological replicates per sample. Another for metabolite extraction, included young fruits (YF) and mature fruits (MF), with five biological replicates per sample. The details of leaves, flowers, young fruits and mature fruits collection are as follows: tender leaf and blooming flowers were collected on May 23, 2021, young fruits were collected on July 12, 2021 (50 days after flowering), and mature fruits were collected on September 29, 2021 (120 days after flowering). The fresh material collected was immediately frozen in liquid nitrogen and stored in an ultra-low temperature freezer at −80 ℃.
2.2. RNA-seq processing and data analysisTotal RNA was extracted and purified from the above samples using RNAprep Pure Plant Plus Kit (TIANGEN, China). The extracted RNA was tested for purity, concentration, and integrity. After the samples were qualified, the mRNA was isolated and purified by Oligo (dt) for the construction of the cDNA library. Illumina Novaseq 6000 sequencing was performed after the library was qualified. The original data obtained were filtered to remove primer and connector sequences, sequences with fragment length < 50 bp, sequences with N base ratio > 10%, low-quality reads, and low-quality bases. The similarity between clean data and the reference genome (C. pinnatifida var. major) was compared using hisat2 (Kim et al., 2019; Zhang et al., 2022). RSeQC software (Wang et al., 2012) was used to evaluate the quality of transcriptome data, and to analyze the sequencing data after passing the quality evaluation. TPM was used to estimate gene expression level. DESeq (Love et al., 2014) was used for differential expression analysis between samples. The fold change of expression was calculated between two samples. Using P-value ≤ 0.05, Q-value ≤ 0.05, and |log2(Fold Change)| > 1 as screening criteria, genes satisfying all the above criteria were considered as differentially expressed genes. Eggnog (Huerta-Cepas et al., 2019) was used to construct GO and KEGG databases, and then the R package clusterProfiler was used to conduct GO and KEGG enrichment analysis on DEGs.
2.3. Metabolome analysisAfter the freeze-dried samples were crushed, the extraction solution (methanol/water = 3:1) was added and vortexed for 30 s. Then the samples were placed in an ice-water bath for ultrasound extraction, and the samples extracted overnight on a 4 ℃ blender. The samples were centrifuged at 4 ℃ for 15 min, and the supernatant filtered through the microporous membrane. The supernatant was diluted with a methanol/water mixture and vortexed for 30 s. Finally, the prepared samples were stored at −80 ℃.
The UHPLC separation was carried out using an EXIONLC System (Sciex). The mobile phase A was 0.1% formic acid in water, and the mobile phase B was acetonitrile. The column model was Waters Acquity UPLC HSS T3 1.8 μm 2.1 × 100 mm. The column temperature was set at 40 ℃. The auto-sampler temperature was set at 4 ℃ and the injection volume was 2 μl. The liquid chromatography mobile phase condition settings are shown in Table S7. A SCIEX 6500 QTRAP + triple quadrupole mass spectrometer equipped with IonDrive Turbo V ESI ion source was used for mass spectrometry in multiple reaction monitoring (MRM) mode. The ion source parameters were as follows: IonSpray Voltage: +5500/-4500 V, Curtain Gas: 35 psi, Temperature: 400 ℃, Ion Source Gas 1:60 psi, Ion Source Gas 2:60 psi, DP: ±100 V. SCIEX Analyst Work Station Software (V.1.6.3) was employed for MRM data acquisition and processing. Metabolite information was matched in the self-established database according to the data detected by MRM (Q1, Q3, and RT). During the analysis, isotopic signals, repeated signals containing K+ ions, and NH14+ ions, and repeated signals of fragments ions which themselves are substances with larger molecular weights were removed. The characteristic ions of each substance were screened through the triple quadrupole mass spectrometer, and the signal intensity of the characteristic ions was obtained in the detector. Then the chromatographic peak integration and correction were carried out, and the relative content of metabolites was calculated. After obtaining the collated data, SIMCA (V.16.0.2) software was used for analysis, including PCA and OPLS-DA. Based on the OPLS-DA result, the metabolites of P-value < 0.05 and VIP > 1 were regarded as the DAMs.
2.4. qRT-PCR validationWe randomly selected 11 genes related to fruit flavor, flavonoids and terpenoids for qRT-PCR analysis according to the fold change of genes. β-Actin from Malus baccata (Genome Database for Rosaceae, ID: MABA012705_v1.0) was used as a reference gene, and all primers used in this study are listed in Table S8. According to the instructions of RNAprep Pure Plant Plus Kit (TIANGEN, China), total RNA was extracted from young and mature fruits of Crataegus chungtienensis, and three biological repeats were performed on different samples. cDNA was synthesized using FastKing gDNA Dispelling RT SuperMix (TIANGEN, China). The total volume of the amplification system was 20 μl, including 10 μl 2 × SuperReal PreMix Plus, 0.6 μl upstream Primer with 10 μm, 0.6 μl down-stream primer with 10 μm, 1 μl diluted cDNA, 0.4 μl 50 × POX Reference Dye and 7.4 μl double-distilled water. The PCR reaction program was set at 95 ℃ for 15 min, followed by 40 cycles at 95 ℃ for 10 s and 60 ℃ for 20 s, with three repeats for each gene.
3. Results 3.1. RNA-seq analysis and DEGs identificationWe conducted transcriptome sequencing of leaves, flowers, young fruits and mature fruits (LE, FL, YF, and MF) of Crataegus chungtienensis with three biological replicates per sample (Table S1). In total, we obtained 71.06 GB of clean data (BioProject no. PRJNA946384 in NCBI SRA database) and the percentages of Q30 bases were all greater than 92%. The mapping rates obtained by comparison with a reference genome (C. pinnatifida var. major) (Zhang et al., 2022) were greater than 85%. Calculation of transcripts per million (TPM) were used as a measure of gene expression. All the genes were grouped into nine clusters using k-means cluster analysis to understand the patterns of gene expression in different periods of C. chungtienensis growth. The gene expression of classes 1, 2, 3, and 4 showed an upward trend with fruit ripening, while the gene expression of classes 5, 6, 7, 8, and 9 decreased with fruit ripening (Fig. 1A). Differentially expressed genes (DEGs) were screened using TPM value, and genes satisfying P-value ≤ 0.05, Q-value ≤ 0.05, and |log2(Fold Change)| > 1 were considered as differentially expressed genes. The results showed that 13, 795, 13, 722, and 10, 896 DEGs were detected in LE vs. FL, FL vs. YF, and YF vs. MF, respectively (Fig. 1B).
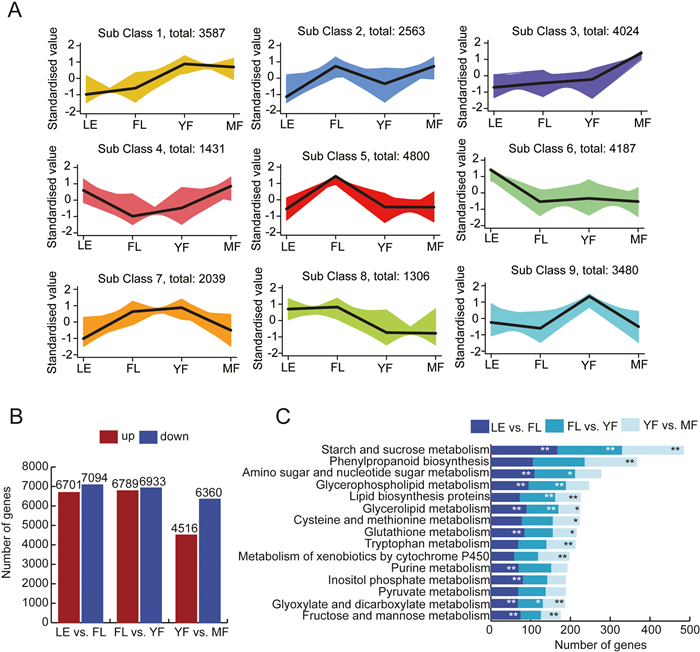
|
| Fig. 1 Transcriptome analysis of different periods during Crataegus chungtienensis fruit development. (A) The K-means analysis of all genes. The black line in the figure represents the average pattern of all genes in each category, and different colors represent different expression trends. (B) Number of differentially expressed genes in LE vs. FL, FL vs. YF and YF vs. MF. (C) The top 15 KEGG metabolic and biosynthetic pathways with the most genes. In the figure, ** represents P-value < 0.01, and * represents P-value < 0.05. |
According to Gene Ontology (GO) functional classification, 'plastid organization' was the most important GO term in FL vs. LE (Fig. S2A), 'cellular carbohydrate metabolism process' was the most important GO term in YF vs. FL (Fig. S2B), and 'secondary metabolic process' was the most important GO term in MF vs. YF (Fig. S2C). DEGs of LE vs. FL, FL vs. YF, and YF vs. MF mapped to 105, 105, and 103 Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic and biological pathways, respectively (Table S2). Among the three groups, the metabolic pathways with the most genes were 'starch and sucrose metabolism', followed by 'phenylpropanoid biosynthesis' and 'amino sugar and nucleotide sugar metabolism' (Fig. 1C).
3.2. Metabolomic profilingA total of 849 metabolites were identified from YF and MF by UHPLC-MS (Table S3). The most detected metabolites were flavonoids (142), followed by terpenoids (92) (Table S4A). Principal component analysis (PCA) was used to reveal the overall metabolite differences between the different groups. The PCA results showed that all samples were divided into two clusters, which indicated that there were distinct metabolite differences in YF and MF (Fig. 2A). To further mine the differentiated metabolites, orthogonal partial least squares discriminant analysis (OPLS-DA) was performed on the identified metabolites, with Variable Importance in the Projection (VIP) > 1 and P-value < 0.05 as the screening criteria for differentially accumulated metabolites (DAMs). A total of 192 DAMs were screened, of which 40 were up-regulated and 152 were down-regulated. Among them, the metabolites which significantly increased in MF included maltotetraose, and stachyose (Fig. 2B). The results of KEGG enrichment analysis showed that the main enrichment pathways of DAMs were 'Aminoacyl-tRNA biosynthesis' 'Phenylalanine metabolism' and 'Glycine, serine and threonine metabolism'. In addition, we observed that 135 DAMs and 1810 DEGs mapped to 50 identical KEGG pathways, which included 'Flavone and flavonol biosynthesis' 'Flavonoid biosynthesis' 'Lysine biosynthesis' and 'Phenylpropanoid biosynthesis' (Fig. 2C).
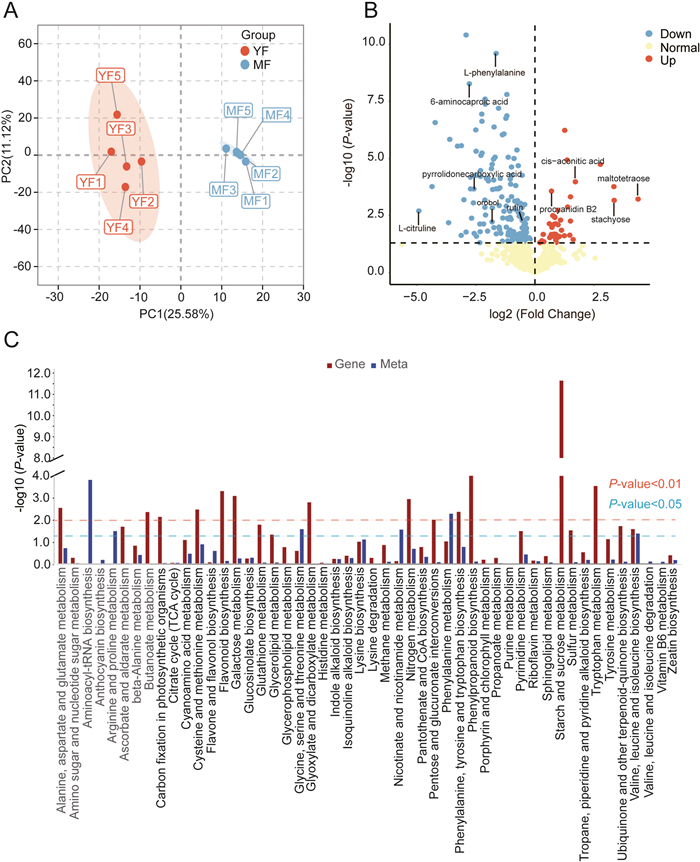
|
| Fig. 2 Metabolomic analysis of young and mature fruits of Crataegus chungtienensis. (A) PCA score plots for all samples, YF: young fruit, MF: mature fruits. (B) Volcano plot of DAMs for YF vs. MF, and some important DAMs are noted in figure. (C) DEGs and DAMs in YF vs. MF are co-enriched into the KEGG pathway. The abscissa represents the metabolic pathway and the ordinate represents the degree of enrichment. |
Sugars and organic acids are the main metabolites affecting fruit flavor. In this study, we screened a total of 98 metabolites related to fruit flavor, including 20 sugars and 78 organic acids (Table S4A). Among these metabolites, 28 metabolites were differentially accumulated (10 up-regulated and 18 down-regulated), including six saccharides and 22 organic acids. In addition, maltotetraose and stachyose were the metabolites that increased the most after ripening, with a 20.54- and 10.20-fold increase respectively, indicating that they may be important sugar components affecting the flavor changes of Crataegus chungtienensis. It was followed by cis-Aconitic acid with a 3.27-fold increase. 6-Aminocaproic acid and pyrrolidonecarboxylic acid were significantly reduced after fruit ripening. These differential metabolites may be important factors causing the flavor changes of C. chungtienensis fruit during ripening (Table S4B).
We screened 128 genes in galactose metabolism, glycolysis/gluconeogenesis and the TCA cycle of Crataegus chungtienensis (LE, FL, YF, and MF), including 99 DEGs. The heat map results of these DEGs show that a, b, c, and d were highly expressed in FL, MF, YF, and LE respectively (Fig. S3A). To further analyze the connections between the transcriptome and metabolome in the pathways of fruit flavor formation, we used the cor function in R to calculate the Pearson correlation coefficient (PCC) of DEGs (YF and MF) and DAMs (three biological replicates were randomly selected) related to galactose metabolism, glycolysis/gluconeogenesis and TCA cycle biosynthesis, and selected DEGs and DAMs with P-value < 0.05 and PCC > 0.8 to draw a network diagram (Fig. 3B). These pathways contained three DAMs and 43 DEGs (Table S4B and Table S5). Two DAMs and 14 DEGs are involved in the galactose metabolite pathway. 14 DEGs encode four enzymes: aldose 1-epimerase (GALM), inositol 3-alpha-galactosyltransferase (GOLS), raffinose synthase (RAFS) and stachyose synthetase (STS). The accumulation of the metabolite UDP-galactose decreased after fruit ripening, while the accumulation of Stachyose increased significantly after fruit ripening. In addition, GALM (LG15.1595), GOLS (LG01.497, LG08.42, LG08.54), RAFS (LG05.1274, LG09.2106, LG14.623, LG15.1416), and STS (LG13.2151) genes were up-regulated in MF. The trend of their expression was consistent with the accumulation pattern of stachyose. The Glycolysis/Gluconeogenesis pathway involves 20 DEGs, which encode six enzymes including phosphoglycerate kinase (PGM), 6-phosphofructokinase 1 (PFK), phosphoglycerate kinase (PGK), fructose-bisphosphate aldolase, class I (ALDO), enolase (ENO), pyruvate kinase (PK). Of these DEGs, there are 6 up-regulated genes and 14 down-regulated genes. One DAM and nine DEGs are involved in the TCA cycle pathway. The accumulation of the metabolite cis-aconitic acid was increased in MF. Nine DEGs encode five enzymes, including malate dehydrogenase (MDH1), ATP citrate (pro-S)-lyase (ACLY), aconitate hydratase (ACO), isocitrate dehydrogenase (IDH), 2-oxoglutarate dehydrogenase E1 component (OGDH), genes ACLY (LG06.1388, LG06.968), ACO (LG13.863), and IDI (LG13.265) were positively correlated with cis-Aconitic acid, and genes MDHI (LG02.1037), ACO (LG08.1390), and OGDH (LG12.878) were negatively correlated with cis-Aconitic acid (Fig. 3 and Table S6).

|
| Fig. 3 DEGs and DAMs associated with galactose metabolism, glycolysis/gluconeogenesis, and TCA cycle pathways. (A) Heat map of gene expression involved in galactose metabolism, glycolysis/gluconeogenesis, and TCA cycle synthesis pathways. DAMs in these pathways are color-marked. Blue indicates metabolites that were decreased in MF, and orange indicates metabolites that were increased in MF. (B) Correlation network diagram of DEGs and DAMs. The blue line indicates a negative correlation, and the red line indicates a positive correlation. |
To compare the content changes of flavonoids during fruit ripening, we analyzed the young and mature fruits of Crataegus chungtienensis. A total of 142 flavonoids were detected, including 71 flavones, six anthocyanins, 10 chalcones, 14 flavanones, 11 flavonols, 12 xanthones, and 20 isoflavones (Table S4A). A total of 21 differentially accumulated flavonoids (DAFs) were screened with P-value < 0.05 and VIP > 1 as the threshold for DAFs. Among these DAFs, three up-regulated flavonoids, 2-benzal-4-hydroxyacetophenone, okanin, and procyanidin B2, were increased by 1.84, 1.65, and 1.62 times, respectively, after fruit ripening. The other 18 flavonoids decreased after fruit ripening (Table 1). KEGG pathway analysis showed that 11 DAFs were mapped to the following pathways: isoflavonoid biosynthesis (apigenin, 2′-hydroxygenistein, genistein), anthocyanin biosynthesis (pelargonidin3, 5-di-beta-D-glucoside, Pelargonidin) flavone, flavonol biosynthesis (isovitexin, apigenin, quercitrin, vitexin, rutin), and flavonoid biosynthesis (pelargonidin, apigenin (-)-epicatechin, eriodicty).
| Compounds | Formula | Class Ⅰ | Class Ⅱ | VIP | Fold change | Log2FC | Type |
| Pelargonidin | C15H10O5 | Flavonoids | Anthocyanin | 1.43 | 0.53 | −0.90 | down |
| Pelargonidin 3, 5-di-beta-D-glucoside | C27H31O15 | Flavonoids | Anthocyanin | 1.73 | 0.46 | −1.12 | down |
| 2-Benzal-4-hydroxyacetophenone | C15H12O2 | Flavonoids | Chalcones | 1.26 | 1.84 | 0.88 | up |
| Okanin | C15H12O6 | Flavonoids | Chalcones | 1.72 | 1.65 | 0.72 | up |
| (-)-Epicatechin | C15H14O6 | Flavonoids | Flavanones | 1.75 | 0.69 | −0.54 | down |
| L-Epicatechin | C15H14O6 | Flavonoids | Flavanones | 1.49 | 0.72 | −0.47 | down |
| Procyanidin B2 | C30H26O12 | Flavonoids | Flavanones | 1.83 | 1.62 | 0.70 | up |
| Theaflavine | C29H24O12 | Flavonoids | Flavanones | 1.80 | 0.28 | −1.84 | down |
| 7-Hydroxyflavone | C15H10O3 | Flavonoids | Flavones | 1.72 | 0.32 | −1.63 | down |
| Apigenin | C15H10O5 | Flavonoids | Flavones | 1.70 | 0.61 | −0.71 | down |
| Astilbin | C21H22O11 | Flavonoids | Flavones | 1.51 | 0.38 | −1.38 | down |
| Eriodictyol | C15H12O6 | Flavonoids | Flavones | 1.35 | 0.45 | −1.15 | down |
| Isovitexin | C21H20O10 | Flavonoids | Flavones | 1.76 | 0.49 | −1.04 | down |
| Norwogonin | C15H10O5 | Flavonoids | Flavones | 1.29 | 0.55 | −0.85 | down |
| Quercitrin | C21H20O11 | Flavonoids | Flavones | 1.35 | 0.63 | −0.67 | down |
| Rutin | C27H30O16 | Flavonoids | Flavones | 1.60 | 0.69 | −0.53 | down |
| Vicenin 2 | C27H30O15 | Flavonoids | Flavones | 1.29 | 0.65 | −0.62 | down |
| Vitexin | C21H20O10 | Flavonoids | Flavones | 1.82 | 0.49 | −1.02 | down |
| 2′-Hydroxygenistein | C15H10O6 | Flavonoids | Isoflavones | 1.36 | 0.75 | −0.42 | down |
| Genistein | C15H10O5 | Flavonoids | Isoflavones | 1.53 | 0.53 | −0.91 | down |
| Orobol | C15H10O6 | Flavonoids | Isoflavones | 1.83 | 0.29 | −1.81 | down |
| Artesunate | C19H28O8 | Terpenoids | Sesquiterpenoids | 1.88 | 0.08 | −3.65 | down |
| Eupatolide | C15H20O3 | Terpenoids | Sesquiterpenoids | 1.69 | 0.24 | −2.09 | down |
| Ganoderal A | C30H44O2 | Terpenoids | Triterpenoids | 1.78 | 0.43 | −1.21 | down |
| Chamazulene | C14H16 | Terpenoids | Sesquiterpenoids | 1.23 | 0.67 | −0.59 | down |
| Epitulipinolide | C17H22O4 | Terpenoids | Sesquiterpenoids | 1.29 | 0.74 | −0.44 | down |
| Ganoderol A | C30H46O2 | Terpenoids | Triterpenoids | 1.61 | 2.83 | 1.50 | up |
| 18alpha-Glycyrrhetinic acid | C30H46O4 | Terpenoids | Triterpenoids | 1.44 | 3.07 | 1.62 | up |
To explore the mechanism of flavonoid accumulation during fruit ripening of Crataegus chungtienensis, we analyzed the expression patterns of related genes in flavonoid metabolism. A total of 52 candidate unigenes related to the flavonoid biosynthesis pathway were identified in the four phases of fruit development (LE, FL, YF, and MF) (Table S5). Candidate unigenes related to the flavonoid biosynthetic pathway encode 14 putative enzymes, including phenylalanine ammonia-lyase (PAL), 4-coumarate trans-cinnamate 4-monooxygenase (CYP73A), CoA ligase (4CL), chalcone isomerase (CHI), chalcone synthase (CHS), etc (Table S5). In addition, there were 49 DEGs related to the flavonoid biosynthetic pathway (Table S5). Cluster analysis of the expression trends of DEGs showed that these genes had different expression patterns during the four stages of C. chungtienensis. A, b, c, and d were highly expressed in FL, LE, YF, and MF respectively (Fig. S3B).
To understand the relationship between key genes and metabolites in the flavonoid synthesis pathway from young to mature fruit of Crataegus chungtienensis, we used the cor function in R to calculate the PCC of DEGs and DAFs (three biological replicates were randomly selected) related to flavonoid biosynthesis and selected DEGs and DAFs with P-values < 0.05 and PCC > 0.8 to draw network diagrams (Fig. 4B). In addition, a total of 34 DEGs and 10 DAFs in YF vs. MF were mapped to the KEGG pathway related to flavonoid synthesis, and according to the KEGG pathway, we drew a flavonoid synthesis pathway map. The flavonoid biosynthetic pathway is derived from the phenylpropanoid pathway, and phenylalanine is catalyzed by PAL, C4H, and 4CL enzymes to generate cinnamic acid and p-coumaroyl-CoA. The results showed that two PAL genes were up-regulated and two PAL genes were down-regulated in MF. Two CYP73A genes were up-regulated in MF and four CYP73A genes were down-regulated in MF. Five 4CL genes were up-regulated in MF and two 4CL genes were down-regulated in MF. Among them, the genes with low expression of PAL (LG12.1035 and LG16.738), CYP73A (LG05.1360 and LG11.336), and 4CL (LG04.2810, LG07.200, LG07.201, LG16.1800, and LG08.444) in MF had a strong positive correlation with (-)-epicatechin, apigenin, isovitexin, pelargonidin, pelargonidin 3, 5-di-beta-D-glucoside, rutin and vitexin. CHS is a key gene in the pathway from phenylpropanoid biosynthesis to flavonoid biosynthesis. Our results showed an upward trend in the expression of CHS genes. In MF, the CHI gene was significantly down-regulated, and two CHI genes were highly positively correlated with metabolites ((-)-epicatechin, apigenin, isovitexin, pelargonidin, pelargonidin 3, 5-di-beta-D-glucoside, rutin, vitexin). F3H, DFR, ANS, and F3GT genes were highly expressed in MF. The expression levels of CYP93C and ANR genes were decreased in MF, and the two ANR genes (LG02.225 and LG03.2495) were strongly positively correlated with downstream (-)-epicatechin (Fig. 4 and Table S6).
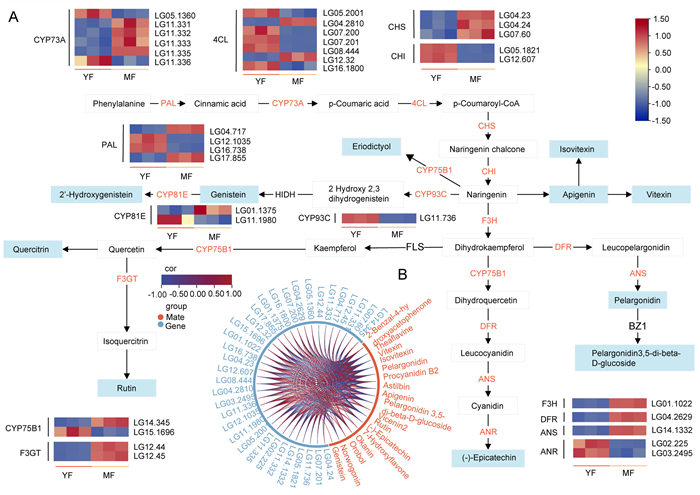
|
| Fig. 4 DEGs and DAMs related to the flavonoid biosynthesis pathway (A) Heat map of gene expression involved in the flavonoids biosynthesis pathway. DAMs in the pathway are color-marked. Blue indicates metabolites that were decreased in MF, and orange indicates metabolites that were increased in MF (B) Correlation network diagram of DEGs and DAMs. The blue line indicates a negative correlation, and the red line indicates a positive correlation. |
A total of 92 terpenoids were identified in the fruits (YF and MF) of Crataegus chungtienensis, including 10 monoterpenoids, 14 diterpenoids, 29 sesquiterpenoids, 34 triterpenoids and five iridoids, of which there were seven differentially accumulated terpenoid metabolites (Table S4A). The accumulation of artesunate, eupatolide, ganoderal A, chamazulene and epitulipinolide decreased after fruit ripening, while the accumulation of ganoderol A and 18alpha-glycyrrhetinic acid increased after fruit ripening (Table 1).
A total of 46 unigenes associated with the terpenoid backbone biosynthesis pathway were identified in four stages (LE, FL, YF, and MF) of Crataegus chungtienensis, these genes regulate 16 enzymes, including acetyl-CoA C-acetyltransferase (ACAT), 1-deoxy-d-xylulose-5-phosphate reductoisomerase (Dxr), 1-deoxy-d-xylulose-5-phosphate synthase (Dxs), farnesyl diphosphate synthase (FDPS), etc (Table S5). Furthermore, there were 28, 24, and 19 DEGs in LE vs. FL, FL vs. YF, and YF vs. MF, respectively (Table S5). Cluster analysis of these DEGs showed that there were three different expression patterns. A, b, and c were highly expressed in FL, LE, and MF respectively (Fig. S3C).
To understand the relationship between differentially accumulated terpenoid metabolites and key genes in terpenoid backbone biosynthetic pathways, we calculated the Pearson correlation coefficient (PCC) between DEGs and differentially accumulated terpenoid metabolites in YF vs. MF using the cor function in R and selected the DEGs and DAMs (three biological replicates are randomly selected) with an absolute value of PCC greater than 0.8 and P-value < 0.05 terpenoid metabolite mapping network (Fig. S4B). The results show a complex regulatory relationship between DEGs (LG01.1562, LG01.2541, LG01.42, LG02.1670, LG03.1111, LG03.665, LG04.1828, LG04.920, LG05.1237, LG05.647, LG06.774, LG08.1234, LG09.1511, LG11.229, LG14.1341, LG15.916, LG16.275, LG16.276, LG01.332) and metabolites (artesunate, cantharidin, epitulipinolide, eupatolide, ganoderol A, and ganoderal A) (Fig. S4 and Table S6).
3.6. qRT-PCR validation of the transcriptome dataIn order to verify the accuracy of transcriptome data, GOLS (LG01.497), RAFS (LG09.2106), ACO (LG13.863), PAL (LG12.1035), CYP73A (LG05.1360), 4CL (LG05.2001), ANS (LG14.1332), ANR (LG03.2495) ACAT (LG08.1234), HMGCS (LG14.1341) and Dxs (LG06.774) were selected for qRT-PCR validation (Fig. 5A). qRT-PCR results showed that the expression trend of these 11 genes in YF vs. MF was consistent with the results of RNA-seq (R = 0.91, P-value < 0.01) (Fig. 5B). The results show that the transcriptome data accurately reflected the expression patterns of most genes in C. chungtienensis.

|
| Fig. 5 The expression pattern of RNA-seq was verified by qRT-PCR. A The relative expression levels of 11 DEGs were statistically analyzed by the -2−ΔΔCT method. B Correlation between qRT-PCR results and RNA-seq data. |
Combinatorial analysis of transcriptome and metabolome is widely used in the study of fruit flavor, which provides important clues for improving fruit flavor and quality (Chen et al., 2021; Peng et al., 2022; Wang et al., 2022). The taste of fruits is composed of flavor, and sweetness and sourness are important factors that affect the taste of fruit. The taste of Crataegus chungtienensis fruit is slightly sweet, and maltotetraose and stachyose were significantly increased after ripening in our study, indicating that they may be important sugar components affecting the flavor change of C. chungtienensis. Maltotetraose is the product of the hydrolysis reaction between maltotetraose amylase and starch (Robyt and Ackerman, 1971). It has a slightly sweet taste, high viscosity, and is a good food additive. In addition, stachyose, a sugar in the raffinose oligosaccharide family (RFO), not only adds sweetness to fruits but also plays an important role in plant growth and stress response (Gil et al., 2012; Hannah et al., 2006). Previous studies found that in frost-tolerant evergreen Labiatae, RFO is involved in carbon translocation and storage (Bachmann and Keller, 1995; Pollock et al., 2010). C. chungtienensis is mainly distributed in alpine areas with an altitude of 2500–3500 m, and where the temperature is low throughout the year. September is the fruiting period of C. chungtienensis (Gu and Spongberg, 2003); as the fruit matures, the temperature gradually decreases. Therefore, the accumulation of stachyose in ripe fruits may be an adaptive response to low temperature. The GOLS is the key enzyme in the synthesis of RFO, and the expression level of GOLS is directly related to the concentration of stachyose in plants (Sengupta et al., 2015). In this study, we screened out seven GOLS genes, in which genes (LG01.497, LG08.42, and LG08.54) were positively correlated with stachyose accumulation, while genes (chr0.2057, LG06.1402, LG07.1277, and LG07.872) were negatively correlated with stachyose accumulation during fruit maturation. In addition, RAFS and STS are also key enzymes in the synthetic pathway of RFO, which catalyze galactinol to produce raffinose and stachyose (Lehle and Tanner, 1973; Peterbauer et al., 1999; Peterbauer and Richter, 1998). In lentil seed development, it was found that the accumulation of RFO corresponded to the accumulation of RAFS and STS transcripts, and the enzymatic activities of RAFS and STS were also consistent with the accumulation of RFO (Kannan et al., 2021). Consistent with previous findings, we screened out four RAFS genes and one STS gene, the expression of these genes was highly positively correlated with the accumulation of stachyose, and they may also play important roles in the biosynthesis and regulation of stachyose. However, their regulation pattern needs further study.
Organic acids are important components in hawthorns (Zhang et al., 2022). In this study, we detected a large number of organic acids in Crataegus chungtienensis fruits, including citric, succinic, ascorbic, quinic, and malic acids, of which citric acid is an important organic acid in several species of hawthorn (Gundogdu et al., 2014; Zhang et al., 2022). Citric acid is isomerized to isocitrate by aconitate hydratase, and cis-aconitic acid is an intermediate product of this reaction. The aconitase reaction establishes a balance between citric acid, cis-aconitic acid and isocitrate (Igamberdiev and Eprintsev, 2016). In citrus aconitate hydratase studies, the CcAco1 and CcAco2 genes are usually induced at the onset of acidity maxima and acid reductions (Terol et al., 2010). In the present study, we screened out two differentially expressed ACO genes, which may have important effects on the acidity changes of C. chungtienensis fruit during ripening.
Flavonoids are one of the most abundant bioactive components in hawthorn (Zhang et al., 2022). A total of 142 flavonoids were detected in the fruit of Crataegus chungtienensis. However, there were differences in flavonoid content between young and mature fruits. Compared with young fruits, the accumulation of several flavonoids decreased in mature fruits. The content of flavonoids in cultivated hawthorns (C. pinnatifida var. major), lemon, raspberry and blackberry fruits also showed the same accumulation pattern (Chen et al., 2021; Wu et al., 2022; Zhang et al., 2022; Zhu et al., 2018). Studies have found that flavonoids have a bitter taste which can protect plants from animal damage by changing the palatability of plants, reducing the nutritional value of fruits, and even may become toxins (Drewnowski, 2001; Harborne and Williams, 2000; Mierziak et al., 2014). Therefore, the accumulation of flavonoids in young fruits of C. chungtienensis may be one of the methods to protect the developing fruit from frugivores. In contrast, the decrease of flavonoids and the increase of flavor-related metabolites (e.g., maltotetraose and stachyose) in C. chungtienensis fruits after ripening may contribute to seed dispersal (Beauchamp, 2016).
The synthetic pathway of flavonoids in plants has been well studied, and the enzyme genes involved in this pathway and their functions have been cloned and verified in some plants (Nabavi et al., 2020; Routaboul et al., 2006). We screened 34 genes related to flavonoids synthesis, among which PAL, CYP73A, and 4CL showed different expression patterns in the fruit development stage of C. chungtienensis. Similar phenomena are found during the development of other fruits, such as the CHS, CHI, and FLS genes of cultivated hawthorn, the PAL, and CHS genes of wild hawthorn (Crataegus maximowiczii) and the CYP73A, FLS, DFR genes of Chinese raspberry (Rubus chingii) (Chen et al., 2021; Zhang et al., 2022; Zhang et al., 2023). The content of flavonoids varies with fruit development, and these DEGs showing different expression patterns in young and mature fruits may be closely related to the dynamic changes of metabolites. The first three steps of the flavonoid synthesis pathway, PAL, CYP73A, and 4CL catalytic pathway are called the general phenylpropane pathway (Dong and Lin, 2021). The general phenylpropane pathway is the common pathway of downstream metabolite flavonoids and lignin, and we screened 17 DEGs in this pathway with high correlation with DAFs. In addition, structural genes in the flavonoids synthesis pathway control the synthesis of flavonoids and play a direct role in this pathway (Liu et al., 2021). In this study, we identified 17 unigenes encoding CHS, CHI, F3H, DFR, ANS, ANR, etc. involved in the synthesis of flavonoids. There is a complex regulatory relationship between these genes and metabolites. CHS represents the beginning of a specific flavonoid pathway (Nabavi et al., 2020), CHS and CHI catalyze p-Coumaroyl-CoA to form naringenin, which as the main metabolite, enters the synthetic pathway of other flavonoids. A previous study found that many genes such as CHS, CHI, F3H, DFR, and ANS in soft-fleshed cultivars of cultivated hawthorns were up-regulated in the late fruit stage (Zhang et al., 2022). The mature fruit of C. chungtienensis is soft-fleshed, and the expression patterns of CHS, F3H, DFR, and ANS genes were similar to soft-fleshed cultivars of cultivated hawthorn. These genes were highly expressed in the ripe fruits of C. chungtienensis.
Hawthorn is a fruit rich in terpenoids (Zhang et al., 2022); in this study, 92 terpenoids were detected from fruit of Crataegus chungtienensis. However, among the differentially accumulated terpenoids, most of the terpenoids decreased after fruit ripening. Several studies found that the terpenoid content of pepper (Zanthoxylum armatum), and finger citron fruits also showed the same accumulation pattern (Hui et al., 2020; Xu et al., 2019). In addition, we screened out 19 DEGs encoding 11 enzymes involved in the terpenoid backbone biosynthesis pathway. The function of some genes has been demonstrated, for example, overexpression of LiDXS from lily increased the content of terpenoids (Zhang et al., 2018). The five DXS genes we screened in C. chungtienensis showed a strong correlation with the differentially accumulated terpenoids, and they may have important effects on changes in the content of terpenoids in C. chungtienensis.
5. ConclusionIn this study, we performed metabolome and transcriptome analyses at different periods during the development of the fruit of Crataegus chungtienensis, and determined genes and metabolites related to the biosynthesis of sugars, organic acids, flavonoids and terpenoids in the fruit of C. chungtienensis. The results showed that maltotetraose, stachyose and cis-Aconitic acid increased significantly after fruit ripening, and they may have important effects on the flavor changes of the fruit of C. chungtienensis. However, most of the differentially accumulated flavonoid and terpenoid metabolites decreased after fruit ripening. Therefore, using unripe C. chungtienensis fruit can obtain more active ingredients such as flavonoids and terpenoids, while the ripe fruit tastes better and is more acceptable. In addition, 43, 34, and 19 DEGs were screened out, highly correlated with differentially accumulated sugar acids, flavonoids, and terpenoids, respectively. We selected 11 key enzyme genes from these DEGs for qRT-PCR validation, and these 11 genes expression patterns were consistent with the transcriptome results. Through the above analysis, we have some comprehension of the biosynthesis pathways of important metabolites in C. chungtienensis and have a new understanding of the formation of fruit quality of C. chungtienensis. These research results will play an important role in promoting the development of the hawthorn industry.
AcknowledgementsExperimental materials and photos of Crataegus chungtienensis were kindly provided by Cong-Wei Yang and Hua-Zhang Wang of Yunnan University of Chinese Medicine. This work was supported by the National Natural Science Foundation of China (32260094, 32060237 to T.Z., 82260739 to G.L., and 32060085 to Q.Q.), and the Major Science and Technology Project of Yunnan Province (202102AE090031) to G.L.
Author contributions
T.Z. and G.L. conceived and designed the study. X.W., D.L., Y.Z., L.J., Q.Q., T.Z., and G.L. prepared the materials and performed the experiments. X.W., D.L., Y.Z., T.Z., Q.Q., and G.L. analyzed the data. X.W., M.J.C.C., and T.Z. verified the results and wrote the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.02.001.
Alirezalu, A., Ahmadi, N., Salehi, P., et al., 2020. Physicochemical characterization, antioxidant activity, and phenolic compounds of hawthorn (Crataegus spp.) fruits species for potential use in food applications. Foods, 9: 436. DOI:10.3390/foods9040436 |
Bachmann, M., Keller, F., 1995. Metabolism of the raffinose family oligosaccharides in leaves of Ajuga reptans L. (Inter- and Intracellular Compartmentation). Plant Physiol., 109: 991-998. DOI:10.1104/pp.109.3.991 |
Barrett, D., Beaulieu, J., Shewfelt, R., 2010. Color, flavor, texture, and nutritional quality of fresh-cut fruits and vegetables: desirable levels, instrumental and sensory measurement, and the effects of processing. Crit. Rev. Food Sci. Nutr., 50: 369-389. DOI:10.1080/10408391003626322 |
Beauchamp, G., 2016. Why do we like sweet taste: a bitter tale?. Physiol. Behav., 164: 432-437. DOI:10.1016/j.physbeh.2016.05.007 |
Chen, Z., Jiang, J., Shu, L., et al., 2021. Combined transcriptomic and metabolic analyses reveal potential mechanism for fruit development and quality control of Chinese raspberry (Rubus chingii Hu). Plant Cell Rep., 40: 1923-1946. DOI:10.1007/s00299-021-02758-6 |
Commission, C.P., 2020. Pharmacopoeia of the People's Republic of China. The Medicine Science and Technology Press of China, Beijing.
|
Dong, N., Lin, H., 2021. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol., 63: 180-209. DOI:10.1111/jipb.13054 |
Drewnowski, A., 2001. The science and complexity of bitter taste. Nutr. Rev., 59: 163-169. DOI:10.1111/j.1753-4887.2001.tb07007.x |
Du, X., Zhang, X., Bu, H., et al., 2019. Molecular analysis of evolution and origins of cultivated hawthorn (Crataegus spp.) and related species in China. Front. Plant Sci., 10: 443. DOI:10.3389/fpls.2019.00443 |
El Hadi, M., Zhang, F., Wu, F., et al., 2013. Advances in fruit aroma volatile research. Molecules, 18: 8200-8229. DOI:10.3390/molecules18078200 |
Gil, L., Ben-Ari, J., Turgeon, R., et al., 2012. Effect of CMV infection and high temperatures on the enzymes involved in raffinose family oligosaccharide biosynthesis in melon plants. J. Plant Physiol., 169: 965-970. DOI:10.1016/j.jplph.2012.02.019 |
Gong, C., Diao, W., Zhu, H., et al., 2021. Metabolome and transcriptome integration reveals insights into flavor formation of 'Crimson' watermelon flesh during fruit development. Front. Plant Sci., 12: 629361. DOI:10.3389/fpls.2021.629361 |
Gu, C., Spongberg, S.A., 2003. Crataegus linnaeus. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds.), Flora of China Vol. 9 (Pittosporaceae through Connaraceae). Science Press, Beijing, pp. 111e117.
|
Gundogdu, M., Ozrenk, K., Ercisli, S., et al., 2014. Organic acids, sugars, vitamin C content and some pomological characteristics of eleven hawthorn species (Crataegus spp.) from Turkey. Biol. Res., 47: 21. DOI:10.1186/0717-6287-47-21 |
Hannah, M., Zuther, E., Buchel, K., et al., 2006. Transport and metabolism of raffinose family oligosaccharides in transgenic potato. J. Exp. Bot., 57: 3801-3811. DOI:10.1093/jxb/erl152 |
Harborne, J.B., Williams, C.A., 2000. Advances in flavonoid research since 1992. Phytochemistry, 55: 481-504. DOI:10.1016/S0031-9422(00)00235-1 |
Hu, G., Wang, Y., Wang, Y., et al., 2021. New insight into the phylogeny and taxonomy of cultivated and related species of Crataegus in China, based on complete chloroplast genome sequencing. Horticulturae, 7: 301. DOI:10.3390/horticulturae7090301 |
Huerta-Cepas, J., Szklarczyk, D., Heller, D., et al., 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res., 47: D309-D314. DOI:10.1093/nar/gky1085 |
Hui, W., Zhao, F., Wang, J., et al., 2020. De novo transcriptome assembly for the five major organs of Zanthoxylum armatum and the identification of genes involved in terpenoid compound and fatty acid metabolism. BMC Genomics, 21: 81. DOI:10.1186/s12864-020-6521-4 |
Igamberdiev, A., Eprintsev, A., 2016. Organic acids: the pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front. Plant Sci., 7: 1042. DOI:10.3389/fpls.2016.01042 |
Kannan, U., Sharma, R., Gangola, M., et al., 2021. Sequential expression of raffinose synthase and stachyose synthase corresponds to successive accumulation of raffinose, stachyose and verbascose in developing seeds of Lens culinaris Medik. J. Plant Physiol., 265: 153494. DOI:10.1016/j.jplph.2021.153494 |
Khan, J., Deb, P., Priya, S., et al., 2021. Dietary flavonoids: cardioprotective potential with antioxidant effects and their pharmacokinetic, toxicological and therapeutic concerns. Molecules, 26: 4021. DOI:10.3390/molecules26134021 |
Kim, D., Paggi, J., Park, C., et al., 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol., 37: 907-915. DOI:10.1038/s41587-019-0201-4 |
Kiyama, R., 2017. Estrogenic terpenes and terpenoids: pathways, functions and applications. Eur. J. Pharmacol., 405–415. DOI:10.1016/j.ejphar.2017.09.049 |
Lehle, L., Tanner, W., 1973. The function of myo-inositol in the biosynthesis of raffinose. Purification and characterization of galactinol: sucrose 6-galactosyltransferase from Vicia faba seeds. Eur. J. Biochem., 38: 103-110. DOI:10.1111/j.1432-1033.1973.tb03039.x |
Liu, S., Grierson, D., Xi, W., 2022. Biosynthesis, distribution, nutritional and organoleptic properties of bitter compounds in fruit and vegetables. Crit. Rev. Food Sci. Nutr., 1–20. DOI:10.1080/10408398.2022.2119930 |
Liu, W., Feng, Y., Yu, S., et al., 2021. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci., 22: 12824. DOI:10.3390/ijms222312824 |
Love, M., Huber, W., Anders, S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15: 550. DOI:10.1186/s13059-014-0550-8 |
Lund, J., Brown, P., Shipley, P., 2020. Quantification of north American and European Crataegus flavonoids by nuclear magnetic resonance spectrometry. Fitoterapia, 143: 104537. DOI:10.1016/j.fitote.2020.104537 |
Mierziak, J., Kostyn, K., Kulma, A., 2014. Flavonoids as important molecules of plant interactions with the environment. Molecules, 19: 16240-16265. DOI:10.3390/molecules191016240 |
Nabavi, S., Šamec, D., Tomczyk, M., et al., 2020. Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnol. Adv., 38: 107316. DOI:10.1016/j.biotechadv.2018.11.005 |
Oliveira, A., de Oliveira E Silva, A., Pereira, R., et al., 2022. Anti-obesity properties and mechanism of action of flavonoids: a review. Crit. Rev. Food Sci. Nutr., 62: 7827-7848. DOI:10.1080/10408398.2021.1919051 |
Orhan, I., 2018. Phytochemical and pharmacological activity profile of Crataegus oxyacantha L. (hawthorn) - a cardiotonic herb. Curr. Med. Chem., 25: 4854-4865. DOI:10.2174/0929867323666160919095519 |
Patrick, J., Botha, F., Birch, R., 2013. Metabolic engineering of sugars and simple sugar derivatives in plants. Plant Biotechnol. J., 11: 142-156. DOI:10.1111/pbi.12002 |
Peng, L., Gao, W., Song, M., et al., 2022. Integrated metabolome and transcriptome analysis of fruit flavor and carotenoids biosynthesis differences between Mature-Green and Tree-Ripe of cv. "Golden Phoenix" mangoes (Mangifera indica L.). Front. Plant Sci., 13: 816492. DOI:10.3389/fpls.2022.816492 |
Peterbauer, T., Mucha, J., Mayer, U., et al., 1999. Stachyose synthesis in seeds of adzuki bean (Vigna angularis): molecular cloning and functional expression of stachyose synthase. Plant J., 20: 509-518. DOI:10.1046/j.1365-313x.1999.00618.x |
Peterbauer, T., Richter, A., 1998. Galactosylononitol and stachyose synthesis in seeds of adzuki bean. Purification and characterization of stachyose synthase. Plant Physiol., 117: 165-172. DOI:10.1104/pp.117.1.165 |
Phipps, J.B., Robertson, K.R., Smith, P.G., et al., 1990. A checklist of the subfamily Maloideae (Rosaceae). Can. J. Bot., 68: 2209-2269. DOI:10.1139/b90-288 |
Pichersky, E., Raguso, R., 2018. Why do plants produce so many terpenoid compounds?. New Phytol., 220: 692-702. DOI:10.1111/nph.14178 |
Pollock, C.J., Lloyd, E.J., Stoddart, J.L., et al., 2010. Growth, photosynthesis and assimilate partitioning in Lolium temulentum exposed to chilling temperatures. Physiol. Plantarum, 59: 257-262. DOI:10.1111/j.1399–3054.1983.tb00768.x |
Robyt, J., Ackerman, R., 1971. Isolation, purification, and characterization of a maltotetraose-producing amylase from Pseudomonas stutzeri. Arch. Biochem. Biophys., 145: 105-114. DOI:10.1016/0003-9861(71)90015-4 |
Routaboul, J., Kerhoas, L., Debeaujon, I., et al., 2006. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta, 224: 96-107. DOI:10.1007/s00425-005-0197-5 |
Sengupta, S., Mukherjee, S., Basak, P., et al., 2015. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci., 6: 656. DOI:10.3389/fpls.2015.00656 |
Terol, J., Soler, G., Talon, M., et al., 2010. The aconitate hydratase family from Citrus. BMC Plant Biol., 10: 222. DOI:10.1186/1471-2229-10-222 |
Treutter, D., 2006. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol., 7: 581-591. DOI:10.1055/s-2005-873009 |
Wang, L., Wang, S., Li, W., 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics, 28: 2184-2185. DOI:10.1093/bioinformatics/bts356 |
Wang, R., Shu, P., Zhang, C., et al., 2022. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol., 233: 373-389. DOI:10.1111/nph.17618 |
Wu, Y., Zhang, C., Huang, Z., et al., 2022. Integrative analysis of the metabolome and transcriptome provides insights into the mechanisms of flavonoid biosynthesis in blackberry. Food Res. Int., 153: 110948. DOI:10.1016/j.foodres.2022.110948 |
Xu, Y., Zhu, C., Xu, C., et al., 2019. Integration of metabolite profiling and transcriptome analysis reveals genes related to volatile terpenoid metabolism in finger citron (C. medica var. sarcodactylis). Molecules, 24: 2564. DOI:10.3390/molecules24142564 |
Zhang, J., Chai, X., Zhao, F., et al., 2022a. Food applications and potential health benefits of hawthorn. Foods, 11: 2861. DOI:10.3390/foods11182861 |
Zhang, T., Qiao, Q., Du, X., et al., 2022b. Cultivated hawthorn (Crataegus pinnatifida var. major) genome sheds light on the evolution of Maleae (apple tribe). J. Integr. Plant Biol., 64: 1487-1501. DOI:10.1111/jipb.13318 |
Zhang, T., Sun, M., Guo, Y., et al., 2018. Overexpression of LiDXS and LiDXR from lily (Lilium 'Siberia') enhances the terpenoid content in tobacco flowers. Front. Plant Sci., 9: 909. DOI:10.3389/fpls.2018.00909 |
Zhang, X., Wang, J., Li, P., et al., 2023. Integrative metabolome and transcriptome analyses reveals the black fruit coloring mechanism of Crataegus maximowiczii C.K. Schneid. Plant Physiol. Biochem., 194: 111-121. DOI:10.1016/j.plaphy.2022.11.008 |
Zhu, C.H., Zhou, X.Y., Li, J.X., et al., 2018. Determination of limonin and flavonoids in the lemon fruit at different development stages. Xiandai Shipin Keji, 34: 246-251. DOI:10.13982/j.mfst.1673-9078.2018.2.038 |



