b. Key Laboratory of Economic Plants and Biotechnology, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China;
c. Yunnan Key Laboratory for Wild Plant Resources, Kunming 650201, Yunnan, China;
d. Kunming University, Institute of Agriculture and Life Sciences, Kunming 650214, Yunnan, China
Biodiversity is declining at an unprecedented rate and plant diversity is increasingly threatened by fragmentation of once-continuous habitats and populations (Pimm et al., 2014; Ceballos et al., 2015). As an essential part of biodiversity, genetic diversity confers population-level resilience to ecological perturbation and critical for adaptation to the future climatic environmental change. Thus, it is prudent to quantify the architecture of genetic variation over populations of a threatened species and prevent as much within-species diversity loss as possible. The inferences obtained from population genetics analyses could ultimately help in guiding effectively genetic rescue, which is now becoming widely accepted as a biodiversity conservation approach to conserve small and declining populations (Whiteley et al., 2015; Bell et al., 2019). However, it may be challenging to delineate conservation and management units, and further impede genetic rescue, especially when population differentiation is subtle in species with an extremely small number of populations (Fraser and Bernatchez, 2001; Frankham, 2010).
Generally, there are several reasons concerning with subtle population structure revealed, among which sampling strategy and the chosen genetic markers may be the two most probable impacts on inferring intraspecific genetic differentiation (Younger et al., 2017). From a conservation perspective, sampling an appreciable number of individuals from each location in the species' range is important, because sampling strategy could affect a comprehensive understanding of its genetic status and correctly assessing evolutionary potential. For instances, natural populations in the center may represent genetic admixture (hybrid population), and a source of novel diversity contributed to species' adaptation to ongoing climate change (e.g., Thompson et al., 2010; Xu et al., 2017; Li et al., 2020). Overlooking such populations would inevitably limit the ability to detect the population structure in the distribution range (Papuga et al., 2018). Moreover, populations close to geographic range limits, may contain low levels of genetic variation but display micro-ecological niche differences from those in central populations (Eckert et al., 2008; Papuga et al., 2018). These populations are important for species' survival and long-term evolution, and thus should not be omitted but deserve conservation priority when implementing conservation requirements (Hampe and Petit, 2005).
If the reason of the limited ability to detect population structure based on the sampling strategies could be excluded, the subtle population structure may be attributed to the features of near-neutral markers employed. Currently, single- and multi-locus molecular markers provide most of the genetic data available to inform conservation decisions (see Nielsen et al., 2020). Yet, the constraints of limited numbers of markers and/or low mutation rate markers have been increasingly observed, which perhaps hinder evaluating weak genetic differentiation among populations and consequently confound conservation efforts (Ouborg et al., 2010). Under such circumstances, population genomic data with greater resolution are required to evaluate the impact of genetic processes and anthropogenic factors on the genetic variation pattern, and to delineate conservation units especially for rare and endangered species with limited distribution ranges (Ouborg et al., 2010; Funk et al., 2012; Yang et al., 2018; Iannucci et al., 2021).
In the last decade, developments in next generation sequencing technologies and advances in bioinformatics tools, have made it possible to step up from single- and multi-locus molecular assays to genome-wide estimates of genetic variation and address previously intractable questions in conservation genetics (Ouborg et al., 2010; Andrews et al., 2016; Cilingir et al., 2017). Among a variety of molecular techniques available for population genomic studies, restriction site-associated DNA sequencing (RAD-seq), is a flexible and low-cost approach, which allows a comprehensive description of the patterns of genetic variation among individuals, and investigation of population structure estimated for multiple polymorphisms filtered from genomes (Puritz et al., 2012; Andrews et al., 2016; Nielsen et al., 2020). RAD-seq better detects low levels of genetic structure than traditional genetic markers do, such as for dispersive marine fish (D'Aloia et al., 2020) or rare plant species with small population sizes (e.g., Lee et al., 2020; Liu et al., 2020; Chen et al., 2021).
Paeonia L. (Paeoniaceae), contains globally renowned ornamentals and traditional medicinal plants, but wild species of tree peonies in China are now threatened to some extent (Hong et al., 2017; Zhang et al., 2019; Yang et al., 2020b). Despite growing efforts in Paeonia conservation, the available genomic resources within Paeonia are scarce, probably due to the large genome size, high-complexity and heterozygosity (Lv et al., 2020). These characteristics of genome has hitherto restricted population genetic analyses of Paeonia species to chloroplast DNA, microsatellite markers (see Fan et al., 2020) and the nuclear gene (Zhao et al., 2021). RAD-seq has only been recently applied in construction of a high-density genetic map for Paeonia suffruticosa (Li et al., 2019).
Paeonia ludlowii, is a threatened species, distributed in the Yarlung Zangbo Valley, which is an important evolutionary hotspot belonging to the Qinghai-Tibet Plateau (QTP) (Yu et al., 2019a). This species is an essential peony genetic resource because of its tall stature and pure yellow flowers within Paeonia (Hong, 1997; Yang et al., 2020b) (Fig. 1), and also of medicinal values for the unique chemical components such as paeonolum in roots (Zhang et al., 2019). Additionally, recent research has found P. ludlowii is an important source of oil with beneficial properties for human health, and flowers and seed oils of P. ludlowii could be potential food resources in human diets (Li and Wang, 2019). In contrast to the long evolutionary history of Paeoniaceae (Wikstrom et al., 2001), it has been proposed that some extant tree peony species, such as P. qiui, P. jishanensis, P. rockii and P. ludlowii, distributed in China probably originated during the late Pleistocene, indicating diversification processes within these species in a very short time frame (Zhang et al., 2018; Xu et al., 2019; Zhao et al., 2021). Now P. ludlowii is restricted to Nyingchi and Shannan regions, and extant populations occupy larger sized habitat in Nyingchi than that of the Shannan region, but where the populations have both become fragmented by local urbanization and farming (Hong et al., 2017). P. ludlowii has been included in List of National Key Protected Wild Plants in China (http://www.forestry.gov.cn/, 2021) and categorized as Plant Species with Extremely Small Populations (PSESP) (Yang et al., 2020a). Hence, providing a framework for managing genetic resources of P. ludlowii in conservation programs has become urgent.

|
| Fig. 1 The habitat and morphogical characteristics of Paeonia ludlowii: (a) habitat; (b) flower and pollinated insect; (c) fruit; (d) seeds (Photographs were taken by Jian Liu, Zi Wang and Huihui Xi). |
Previous studies of variation in Paeonia ludlowii using uniparentally inherited chloroplast DNA and parentally nuclear genetic data (microsatellite markers and the nuclear gene) found genetic similarity within populations and no statistically genetic differentiation across the Nyingchi region (Zhang et al., 2018; Zhao et al., 2021). Unfortunately, the sampling without populations in the Shannan region could not adequately explore the differentiation, because Shannan region contains the peripheral population, which may represent a source of novel diversity having adaptive potential for future climate changes (Hampe and Petit, 2005; Thompson et al., 2010). One more recently empirical work employing microsatellite markers, in a thorough sampling of the population of P. ludlowii, showed relatively lower levels of within-population genetic diversity in Shannan region, and also exceptionally low differentiation among populations (Xue et al., 2021). However, the underlying causes of shaping the observed genetic patterns remain still unclear. In this work, 40 samples were collected from the remaining wild populations of P. ludlowii, across its distribution range. We employed genome-wide single nucleotide polymorphisms (SNPs) derived from RAD-seq, to further investigate the magnitude and spatial distribution of the genetic diversity in fragmented populations, to assess the possible occurrence of genetic differentiation patterns previously undetected, and to reconstruct the demographic history in order to get a more complete picture of how evolutionary processes shaping genetic variation in P. ludlowii. All these results would ultimately help to inform genetic management of the remnant populations.
2. Materials and methods 2.1. Sample collection and sequencingLeaf tissue was collected from Paeonia ludlowii during summers of 2018–2020. All samples were dried on silica gel immediately and then stored at −20 ℃. According to the recent field survey, the number of remaining populations is small. And previous analyses based on cpDNA, SSR markers and the nuclear gene all showed no genetic variation within all sampled populations distributed in Nyingchi region (Zhang et al., 2018; Zhao et al., 2021). As a result, sites with more than about 25 km apart were chosen to maximize representation across all known locations but cover its distribution range. Four populations distributed across the two sides of Yarlung Zangbo River were sampled, three (JRK, BJ and NY) of which were located along the midstream of in Nyingchi region and one (ZB) in Shannan region (Fig. 2a). Fruits containing seeds of P. ludlowii were also collected and stored in flowerpot filled with fine sand, and then the status of rooting were observed per month. After rooting, the plantlets were transferred to a plastic pot filled with soil and humus in a greenhouse. The average temperature and the relative air humidity of the greenhouse were controlled at 20 ℃ and 70%, respectively.
|
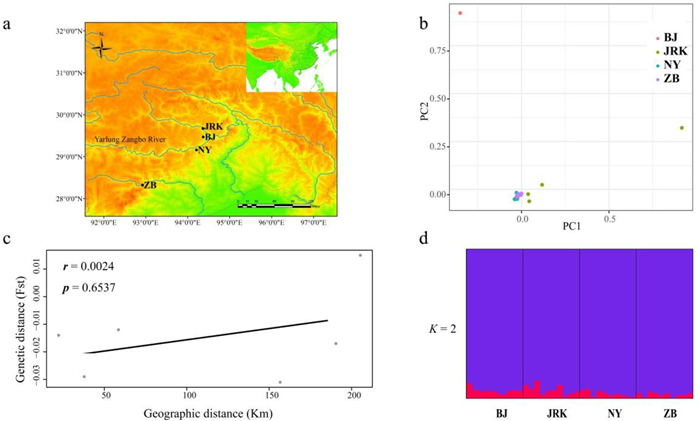
| Fig. 2 (a) Geographic distribution of sampled Paeonia ludlowii populations. (b) Principal component analyses (PCA) assessing all individuals of Paeonia ludlowii. (c) Correlation of pairwise geographic distance vs. pairwise genetic distance Fst of Paeonia ludlowii populations calculated in Arlequin. (d) STRUCTURE analysis containing all Paeonia ludlowii individuals, showing ancestry coefficients for K = 2. |
To obtain estimates of genome-wide patterns of genetic differentiation, we used Illumina sequencing of restriction-site associated DNA tags (RAD-seq). Genomic DNA was extracted from 200 mg of silica-dried leaves using the modified CTAB protocol (Doyle, 1991). DNA purity, concentration, and integrity were assessed using a Nanodrop spectrophotometer, Qubit and agarose gel electrophoresis. For each individual, 1 μg of genomic DNA was used for DNA digestion with EcoRI enzyme and RAD library construction following Baird et al. (2008), including P1 adapter ligation, fragments selection, purification, P2 adapter ligation and RAD-tag amplification. The qualified libraries were sequenced by Illumina Hiseq 2500 platform and 150 bp pair-end reads were generated. Libraries were pooled to a target of 4 Gb raw data per individual.
2.2. Quality filtering and SNP callingA de novo pipeline in STACKS v.2.48 (Rochette et al., 2019) was performed to call SNPs. Raw reads were initially checked by FastQC v.0.11.9 (Andrews, 2019), demultiplexed and filtered using process radtags module. Perl wrapped denovo.map.pl was used for executing the modules in stacks. Following to the suggested protocol, a range of parameter values with m, M and n values ranging from 2 to 4 were investigated. In order to increase coverage depth and maximize loci, three parameters most influence the number and error rate of SNPs were set as m = 4, M = 2 and n = 2. Specifically, RAD loci within individuals were identified using ustacks with a minimum depth of 4 reads to form a stack and the maximum nucleotide mismatch allowed between 2 stacks. Using cstacks and sstacks, RAD loci from individuals were then combined into a catalogue, allowing for a maximum of 2 nucleotide mismatches between stacks. In the populations module, SNPs with minor allele frequency lower than 0.01 and loci with an observed heterozygosity with 0.5 were removed; the first SNP per RAD-tag was chosen; a locus must have been present in all populations to be included in the final data set (-p 4); and a locus must have been genotyped in at least 80% of individuals per population to be included. PDGSpider v.2.1.1.5 was used to transform files into different format files required by downstream analyses (Lischer and Excoffier, 2012).
2.3. Population genomics analysesWe tested for signatures of selection using the FST-outlier method implemented in Bayescan v.2.01 using the default settings (Foll and Gaggiotti, 2008). This approach uses differences in allele frequency to identify candidate loci under selection by decomposing FST coefficients into population and loci components. And models with and without selections were evaluated by a reversible jump MCMC and posterior probabilities of the parameters under different models are calculated. A threshold value to detect selection was set using a conservative maximum false discovery rate (the expected proportion of false positives) of 0.05. It has been demonstrated that the Bayescan is one of the most commonly used and powerful approaches in detecting selection signatures (Ahrens et al., 2018). To assess the genetic diversity of P. ludlowii, observed heterozygosity (Ho), expected heterozygosity (He), nucleotide diversity (π) and the inbreeding coefficient (FIS) were calculated with populations module in STACKS v.2.48 (Rochette et al., 2019).
Using the program Arlequin v.3.5 (Excoffier et al., 2005), AMOVA (analysis of molecular variance) quantified the proportion of variation at the species level based on SNPs data with 10, 000 permutations and allowed 0.05 missing level per site. We also calculated pairwise genetic differentiation FST among the studied populations, and assessed the significance of differentiation while accounting for pairwise comparisons (p < 0.05). The relationship between genetic and geographic distance (isolation by distance, IBD) was tested by population pairs. Geographic distances were calculated with the program GenAlex v.6.1 (Peakall and Smouse, 2006) based on the coordinate information. We used FST calculated above as genetic distance and performed Mantel test on distance matrices with 10, 000 permutations within the mantal.randtest function in package ade4 executed with R v.4.0.1 (Dray et al., 2007).
STRUCTURE v.2.3.4 (Pritchard et al., 2000) was used to infer population structure and identify possible genetic mixture among the populations of Paeonia ludlowii. The admixture ancestry model was implemented, exploring values of K = 1–6 with 10 interactions per K value. Each run was composed 200, 000 generations following a burn-in of 100, 000 generations. In order to explore the existing population structure as far as possible, we did this both with and without sampling locations (Nyingchi and Shannan regions) supplied as prior information. Adding sampling locations may be useful for detecting structure when it is subtle. The optimal K value was determined using the methods of Evanno in Structure Harvester (Evanno et al., 2005). Interactions of the optimal K value were combined and averaged by CLUMPP (Jakobsson and Rosenberg, 2007). Structure results were visualized using DISTRUCT (Rosenberg, 2004). Complementary to the Bayesian clustering, we also performed principal component analysis (PCA) as implemented in PLINK v.1.9 (Chang et al., 2015) and R v.4.0.1, to visualize genetic structure along the first two principle components.
2.4. Demographic history inferencesThe stairway plot method (Liu and Fu, 2015), was used to reconstruct continuous changes in population effective size over time based on the site frequency spectrum (SFS). Since the populations were not very distinctly structured, according to the above-mentioned genetic analyses (see results), we joint all P. ludlowii population together as a whole, and SNPs recorded at a rate of more than 25% among all samples were extracted, and the minimum allele frequency was set to 0.01. To reduce the biased estimates of SFS due to the missing data, we chose to project the full data set down to a smaller number of samples thereby maximizing the number of segregating sites in the SFS (Gutenkunst et al., 2009; Terhorst and Song et al., 2015). The folded SFS was computed from the downsampled diploid samples for P. ludlowii using the python script easySFS (https://github.com/isaacovercast/easySFS). Because there are no reliable fossil information for P. ludlowii and the accurate evolutionary rates for RAD loci is unknown. The mutation rate was estimated from the observed sequence divergence (d) of ATP gene utilized in our previous study between P. ludlowii and P. delavayi, whose splitting (T) has occurred between 5 and 6 million years ago, based on recent calibrations analyses of fossils related with Paeoniaceae (Zhou et al., 2021). We employed Tamura and Nei's distance (Tamura and Nei, 1993) as a measure of sequence divergence. The d-value between the two species was calculated as 0.023 in MEGA 7.0.18 (Kumar et al., 2016). Then we applied the formula μ = d/2T (Nei and Kumar, 2000), μ was then approximated between 1.92–2.30 × 10−9, and used to represent the mutation rate of RAD loci. According to the long-term observation of Ni and Wang (2009), it often takes 7–12 years for P. ludlowii to firstly flowers in the wild due to its seed dormancy and vegetative reproduction by rhizomes (Hao et al., 2014), and the average generation time was set to ten years, which was generally assumed for other Paeonia species (Xu et al., 2019). A two-epoch model was built, the suggested parameters according to the manual was set, and 67% of segregating sites used for training and 200 bootstraps. We also used NeEstimator v.2.1 to estimate contemporary effective population sizes for each population and P. ludlowii under the linkage disequilibrium method with the random mating model and the minimum allele frequency cutoff of 1/(2n) (n = sample size) (Do et al., 2014).
3. Results 3.1. Identification of SNPsWe obtained 164.3 Gb data containing 547, 647, 817 raw reads for 40 individuals of Paeonia ludlowii. After quality filtering, the averaged 12.8 million reads per sample (range 10.32–15.8 million) were processed for all individuals. According to the pre-tests, the minimum number of identical, raw reads required to create a stack (m ranging from 2 to 4), was set to 4. The depths of coverage for processed samples ranged from 11.33x (JRK01) ~22.98x (ZB02), with a mean of 17.41x. When m was set to 4, results from de novo assemblies showed no significant differences in the number of loci and SNPs depending on the set of parameters used. The mismatch within individuals (M) and the mismatch between individuals (n) were both set to be 2. After our restrictive filtering (p = 4, r = 0.8), we obtained 5908 RAD loci containing 560 variant sites. The mean proportion of per individual missing data was less than 5% and the missing data per SNP was less than 20%. From these, we did not detect candidate outliers with Bayescan, and therefore all variant sites were putatively neutral and used for the population genomics analyses.
3.2. Genetic diversityAt the species level, the genetic diversity statistics of Ho, He, and π were all small (Ho: 0.0320, He: 0.0315 and π: 0.0319) (Table 1). At the population level, the range of Ho, He and π was small (Ho: 0.0196–0.0415; He: 0.0177–0.0386; π: 0.0187–0.0408), and the overall genetic diversity of the three statistics showed the same trend and was ranked as follows: JRK > BJ > NY > ZB (Table 1). The inbreeding coefficient at the species level (FIS = 0.0026) was close to zero, and all populations also displayed no evidence of inbreeding (Table 1). Private alleles were not uniformly distributed across populations, spanning a wide range (75–183). JRK and BJ showed the highest fraction of private polymorphisms (33.04% and 32.32% of the total SNP variation, respectively). Proportions of private SNPs were lower in NY population (16.79%) and lowest in ZB population (13.39%) (Table 1).
| code | location | n | latitude (N°) | longitude (E°) | He | Ho | π | private allele | FIS | Ne (95% CI) |
| JRK | Jiarika, Nyingchi | 10 | 29.67 | 94.36 | 0.0386 | 0.0415 | 0.0408 | 181 | −0.0027 | 45.6 (29.8–90.3) |
| BJ | Bujiu, Nyingchi | 10 | 29.47 | 94.37 | 0.0353 | 0.0374 | 0.0373 | 183 | −0.0002 | Inf (Inf–Inf) |
| NY | Nanyi, Nyingchi | 10 | 29.16 | 94.21 | 0.0208 | 0.0226 | 0.0220 | 94 | −0.0018 | 16.6 (11.5–26.7) |
| ZB | Zhunba, Shannan | 10 | 28.32 | 92.92 | 0.0177 | 0.0196 | 0.0187 | 75 | −0.0023 | 229.2 (37.2-Inf) |
| total | 40 | 0.0315 | 0.0320 | 0.0319 | 0.0026 | 48.8 (45.8–52.2) | ||||
| n, number of individuals sequenced; He, expected heterozygosity; Ho, observed heterozygosity; π, nucleotide diversity; FIS, inbreeding coefficient; Ne (95% CI), contemporary effective population size estimates with 95% confidence intervals. Inf estimates refer to Ne 'infinite'. | ||||||||||
We found extremely low levels of pairwise population differentiation for Paeonia ludlowii, and values ranged from 0.0004 to 0.0087 (Table 2), with only estimates of pairwise population differentiation between JRK and BJ showing significant difference (p < 0.05). Similarly, the hierarchical AMOVA revealed considerably more genetic variation within populations (99.63%) than among populations (0.37%) (p > 0.05) (Table 3). When considering Nyingchi and Shannan two regions, the divergence were even lower among groups, with higher divergences also restricted to the comparisons within populations (data not shown). The result of PCA analysis using all individuals showed no clear separation of individuals from the sampling regions (Fig. 2b). There was no very strong correlation between pairwise geographical and genetic distances detected overall in the study area (Mantel r = 0.0024, p = 0.6537) (Fig. 2c). The STRUCTURE analysis showed that a model of two clusters (K = 2) was most likely structure for the data set (Fig. S1), and no matter whether the sampling locations were added or not. In the second-best model of K = 3 (data not shown), the genetic compositions of three clusters were all similar, which corresponded to the results of K = 2 and also indicated little genetic differentiation among the geographic regions (Fig. 2d).
| Population | JRK | BJ | NY | ZB |
| JRK | 0 | 0.0156 | 0.3682 | 0.1426 |
| BJ | 0.0087 | 0 | 0.4502 | 0.1895 |
| NY | 0.0004 | 0.0006 | 0 | 0.3945 |
| ZB | 0.0057 | 0.0029 | 0.0020 | 0 |
| Source of variation | d.f. | Percentage of variation | Fixation indices |
| Among populations | 3 | 0.37 | 0.0037 |
| Within populations | 76 | 99.63 | 0.9963 |
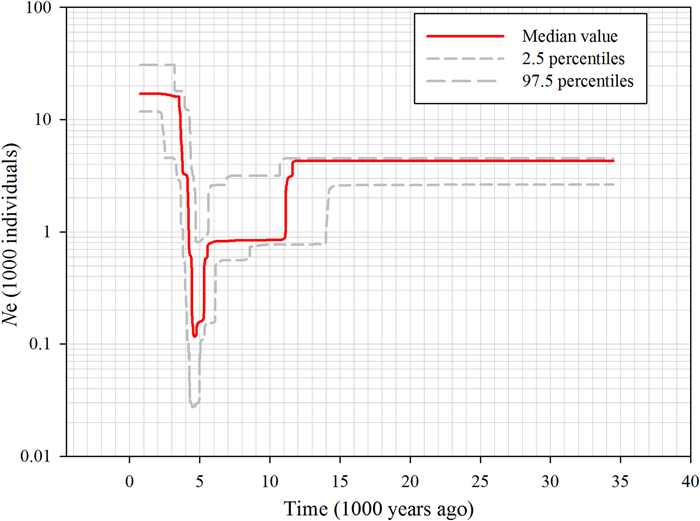
We estimated past effective population size (Ne) for the entire Paeonia ludlowii population in our study using stairway plot. Based on the two different mutation rates, we obtained similar change trends of effective population sizes from about 35, 000 years ago to present, thus one of the results was shown (Fig. 3). Between 15, 000 and 5000 years before the present, the decrease in Ne appeared progressive, starting at least 12, 000 years ago, following with a briefly constant period, and then an acceleration of the decline visible between 6000 and 5000 years ago. During this recent period, Ne suffered a sharp reduction from about 1000 to 100 individuals. Following the recent bottleneck, we observed that between 4500 and 3500 years before the present, the effective population size exhibited a trend of recovery. Additionally, we estimated contemporary effective population size of 48.8 (95% CI: 45.8–52.2) for the full P. ludlowii individuals, and only observed Ne in two populations (JRK and NY) in Nyingchi region. The estimated Ne of JRK was 45.6 (95% CI: 29.8–90.3), larger than that of NY (16.6, 95% CI: 11.5–26.7) (Table 1).
|
| Fig. 3 Inference of demographic history of the entire Paeonia ludlowii population using Stairway plot. The red line represents the median effective population size (Ne) and the grey dash lines represent the 2.5 and 97.5 percentiles of Ne, respectively. |
In this study, our work provided the first estimation of the population genomic status of the threatened tree peony (Paeonia ludlowii) restrictedly distributed in the evolutionary hotspot region within QTP. By employing RAD-sequencing and thorough sampling, we were able to detect the subtle genetic structure within P. ludlowii. Different from previously reported, our results indicated low but varied levels of genetic diversity within populations. Moreover, subtle population subdivisions were revealed, likely reflecting the effects of ancestral polymorphisms rather than currently genetic connectivity. Demographic inference indicated that P. ludlowii probably experienced a temporal history of sharp population decline during the period of Last Glacial Maximum, and a subsequent bottleneck event resulting from prehistoric human activities on the Qinghai-Tibet Plateau. All these events, together with current habitat fragment and excavation might contribute to the endangered status of P. ludlowii. The genetic data here reported could also provide essential source of information for conserving this locally threatened species.
4.1. Low but varied levels of intra-population genetic diversityEstimates of genetic diversity (He, Ho and π) showed that the genetic diversity of Paeonia ludlowii was low for all populations (Table 1). For examples, genetic diversity values (π) were lower than estimates reported for endangered species with limited distribution ranges, such as perennial herb (Viola uliginosa: 0.044) (Lee et al., 2020), shrub (Rhododendron cyanocarpum: 0.0702) (Liu et al., 2020) or trees (Horsfieldia tetratepala: 0.096 and Phoebe zhennan: 0.158) (Xiao et al., 2020; Cai et al., 2021), in which RAD-seq was also used for population genomics analyses. Low genomic diversity has commonly been the product of a combination of historical events, recent bottlenecks and gene flow. Species with short evolutionary time is inclined to accumulate limited genetic variation (Marques et al., 2019). This may also be the case for P. ludlowii, which originated recently due to the effect of Quaternary climatic changes and not already contained high genetic diversity (Zhang et al., 2018; Zhao et al., 2021). Additionally, local urbanization-related disturbance such as road construction, selective farming and excavation for medicinal usage has-modified locally ecological and genetic processes, perhaps resulting in population declines, further aggravating the loss of genetic diversity and impeding effective gene flow via seeds and pollen (Young et al., 1996; Aguilar et al., 2008; Banks et al., 2013). However, this postulation should been taken with caution, as the genetic consequences of habitat fragmentation on species are complicated, especially for species with differences in time since fragmentation, rarity status and life-history characteristics (Aguilar et al., 2008; Vranckx et al., 2012). Actually, for a specific species, it is not easy to distinguish pre-existing genetic diversity from those resulting from recent fragmentation, until the accurate estimates of genetic variation in natural pre-fragmented populations can be made (e.g., Jensen et al., 2018; Llorens et al., 2018). For P. ludlowii, the characteristics of sexual propagation and the near zero estimates of inbreeding coefficient, seemed unfavorable for it to counteract to the effects of fragmentation. Conversely, a lifespan of greater than 10 years in the field may help to cope with the immediate effects on genetic variation (Ni and Wang, 2009), if fragmentation is a relatively event (Yao et al., 2007). It is reasonable to believe that, regardless of whether P. ludlowii could be resilient to fragmentation (see discussion below), P. ludlowii maintains a relatively low genetic diversity. This pattern is consistent with the expectation for species that are rare, geographically restricted and has limited seed dispersal (Hamrick and Godt, 1996).
However, unlike the patterns of genetic variation showing highly genetic uniformity within populations based on cpDNA, SSR markers and the nuclear gene (Zhang et al., 2018; Zhao et al., 2021), we found that each population possessed an own genotype profile and different levels of genetic diversity. In particular, the population JRK on the edge of the distribution of Paeonia ludlowii in Nyingchi region, showed higher genetic diversity values (He: 0.0386; Ho: 0.0415; π: 0.0408) than the population ZB located in Shannan region (He: 0.0177; Ho: 0.0196; π: 0.0187) and adjacent populations distributed in the same region (Table 1). Meanwhile, in addition to the higher genetic diversity in population JRK, the number of private alleles endemic to JRK also showed the highest proportion. That is, genetic diversity decreased towards the limit of the distribution of P. ludlowii (Fig. 2a). A possible explanation for this pattern may be the differences in population sizes among the populations. According to the field survey, population ZB exhibited scarcity of small individuals in Shannan region, suggesting weaker capacities of seedling establishment in comparison with those in Nyingchi region. And this finding further indicated that the conditions here were not very suitable for population inhabiting of P. ludlowii, where micro-environment differed from those in Nyingchi region. Comparatively, the larger atmosphere humid, medium light intensity and good soil water holding capacity in Nyingchi region may be more favorable for seed germination and seedling regeneration for P. ludlowii, and consequently P. ludlowii was restrictively distributed in Shannan region and could not expand south (Ni and Wang, 2009; Hong et al., 2017). Moreover, when considering the parapatric distribution of its ancestor-like species P. delavayi, the possibility of gene flow between P. ludlowii and P. delavayi increasing genetic diversity may not be completely excluded. According to our recent survey on the geographic boundary between the two species, there are only about 11 km of isolation barriers, thus this distance may not enough to impede completely interspecific gene exchange via generalist insect-pollination such as bees and flies (Tang et al., 2021). Although gene flow between this progenitor–derivative pair were not detected in previous studies, perhaps due to the resolution ability based on the limited number of molecular markers, no convincing evidence is available about the extent of reproductive isolation between them either (Shuai and Zang, 2016; Yang et al., 2020b). Therefore, this theme needed further investigation using more genome-wide variation involved in reproductive isolation to help understanding how variation evolve over the course of speciation process (Stankowski and Ravinet, 2021).
4.2. Subtle genetic structureAlthough habitat fragmentation is commonly expected to increase inter-population genetic divergence of plant populations, some empirical studies have demonstrated that anthropogenic fragmentation may not alter pre-existing patterns of differentiation (e.g., Changiostyrax dolichocarpa, Yao et al., 2007; Prunus africana, Farwig et al., 2008; Grevillea caleyi, Llorens et al., 2018). In accordance with previous results, Paeonia ludlowii exhibited weak genetic structure at the species level (Fst = 0.0037) (Table 3), as shown in multiple genetic clustering analyses (Fig. 2b and d). There were two possible alternative scenarios to interpret the observed genetic differentiation. First, the lack of genetic differentiation between populations could be the results of contemporary gene flow via long distance dispersal among populations. Second, rather than contemporary genetic connectivity, the observed genetic similarities could also be attributed to the result of shared ancestry among now isolated populations, which have not been isolated for an enough time to diverge genetically. When the fruit of P. ludlowii become ripe, big seeds usually drop down. The occurrence probabilities of effective dispersal by animals, wind and water are low, and fruit ingestion by animals or transport by mice have been seldom reported; as a result, seedlings are usually distributed around the maternal plant and the distribution pattern of P. ludlowii population is collective (Zhang, 2008) (Fig. 1). In addition, the main pollination insects for P. ludlowii were Apidae and Syrphidae (Shuai and Zang, 2016), which are not expected to travel long-distance pollen dispersal but rather forge primarily within patches in comparison with butterflies and large moths (see ref. there in Barthelmess et al., 2006). Given the poor dispersal ability of P. ludlowii via insect-pollination and/or gravity-dispersed seeds between small populations with more than averaged 110 km distances, we consider the second scenario, that the current separated populations of P. ludlowii were connected by historical gene flow, to be the most likely explanation for the genetic similarity found here. No significant correlation between geographic distance and genetic distance (Fig. 2c), also suggested unequilibrium population structure, and indicated that divergence was largely driven by effects of genetic drift over time, instead of currently gene flow (Hutchison and Templeton, 1999). Accordingly, we could assume that the establishment of P. ludlowii took place when the habitat was still contiguous, and extensive exchange could take place, but the directly disturbance effects on genetic differentiation among populations through its influence on genetic drift and migration has not been so impressive during a short time scale (Banks et al., 2013). This explanation may be particularly likely for long lifespans (15–20 years) species such as P. ludlowii (Zhang et al., 2018). Moreover, our finding of low, yet statistically significant genetic differentiation between the two closest populations (JRK and BJ) (Table 2), did not reflect genetic connections related with geographic proximity, but indicated currently considerable restriction of gene flow. These results may provide further support for the persistence of shared ancestry rather than gene flow.
4.3. Demographic historyPaeonia ludlowii is a relatively young species by ecological specialization and has a restricted distribution range in Yarlung Zangbo valley on the QTP (Zhang et al., 2018; Zhao et al., 2021). In addition to the short evolutionary time, historical demographic processes such as population expansion or bottlenecks and decline are also known to exert a fundamental influence on the particular genetic diversity (Hewitt, 2000). The first predominant decline event, occurring about 12, 000 years ago, was observed in P. ludlowii. The timing of this decline was coincided with the Baiyu (the last) Glaciation during 10, 000–70, 000 years ago on the QTP (Zheng et al., 2002). During the last glaciation period, the temperature and rainfall fluctuations have influenced the vegetation distribution range on the QTP, and led to the observed decrease of genetic diversity. Our result suggested this occurrence of population decline of P. ludlowii, was also consistent with previous ecological modeling results, indicating a few suitable habitats on the QTP at the LGM compared to the present (Zhang et al., 2018). Additionally, we also found the evidence of a more recent bottleneck in Ne from 1000 to 100 occurring approximately 5000 years ago (Fig. 3), which might relate to the greater anthropogenic activities beginning for this region. The fossil evidence showed that modern human occupied the interior region of the QTP about 40 thousand years ago and even in a much earlier time frame (Zhang et al., 2020). Similarly, pollen records of human activities in the Southeast region on the QTP has uncovered evidence that the effects of environmental disturbances emerged due to the enlarged range of human activities since Late Holocene, resulting in the loss of forest and habitat fragment (Chen et al., 2020). All these events probably contributed to the history of long-term decline in Ne in P. ludlowii. Although P. ludlowii had shown some recovery in the subsequent time, it seemed tough for such a species with long-time lifespan to accumulate more mutations to increase genetic diversity over the short hundreds of years. We also acknowledge that the exact timing and magnitude of changes in Ne of P. ludlowii should be interpreted with some cautions. One the one hand, SFS-based approaches typically require very large numbers of segregating sites for accurate inference (Barley et al., 2022), although we applied a less stringent filter approach, the number of SNP markers employed in our study may still be inadequate for the huge genome size of P. ludlowii. Applying a limited amount of segregating sites may probably underestimate the time points. An additional concern with the demographic inference approach we used is that it depends on the mutation rate and generation time to convert the scaled parameter estimates to absolute units of Ne and years. The mutation rate and generation times are seldom known with precision, and especially it is not possible to accurately estimate the evolutionary rates for RAD loci. As a result, for example, if a higher mutation rate was assumed, the value of Ne would be decreased and move the timing of events forward; likewise, a longer generation time would push back the timing of demographic events. However, irrespective of the precise mutation rate and generation time assumed, when we used another dataset with a much stringent filter in which just the SNPs recorded at a rate of more than 50% among all samples were kept, no apparent discrepancy but a similar trend of Ne was obtained (data not shown). Ultimately, therefore, we believe the demographic trends appear robust, and could provide some power for demographic inferences.
4.4. Conservation implicationsThis study improves the genetic characterization of the endangered tree peony (Paeonia ludlowii) covering its natural distribution in Tibet as a basis to assist conservation and management. According to the previous and our field survey in the last years, the number of adult plants within populations could seldom exceed 500; the natural recruitment of population is poor and the population size is declining due to low seed setting percentage, high mortality of young seedling as well as disturbances of human activities (Yang et al., 2007). Further, together with the small number of remaining populations, the relatively low diversity found in all P. ludlowii populations revealed by our different molecular markers suggest that this attribute may have negligible effects on long-term population viability. Meanwhile, our genetically based estimate of small contemporary Ne contrasts with the roughly estimated population size of mature individuals for P. ludlowii (Table 1). This could indicate that, on the one hand, population sample might not capture the full spectrum of genetic diversity within P. ludlowii. Such disparities is not unexpected to be found and genetically based Ne estimates are often substantially smaller than population census size estimates due to sampling, changes to population sizes and impact of inbreeding (Frankham, 1995). On the other hand, the low ratio between the two estimates offers additional support for strong declines of P. ludlowii, making them further potentially vulnerable to deleterious effects of genetic drift due to habitat fragment and excavation in the long term. Therefore, taking all the characteristics into consideration, P. ludlowii should still be categorized as a PSESP taxon that is in urgent need of rescue (Ma et al., 2013; Yang et al., 2020a). Such a small number of isolated populations should deserve both ex-situ and in-situ protection (Crane, 2020). According to the results of our experiment, P. ludlowii has a high seed germination rate, and the transplanted plant could bloom in botanical garden. The representativeness of the genetic diversity captured in the wild for population reinforcement and ex situ conservation in germplasms (seeds) should be considered for all population, considering their existed private alleles. Conserving these germplasms in botanical garden could facilitate scientific research, public outreach and education programs, especially for the need of developing economic and medicinal values for genetic breeding for scarce cultivars with yellow flowers, analyses of seed oil composition and chemical components extraction from P. ludlowii, and ultimately reduce the need of wild resources of P. ludlowii. Meanwhile, given the poor capacity of natural recruitment of P. ludlowii, re-introduction and restoration programs are also necessary for increasing population sizes in the wild.
Except for ex-situ conservation programs, it is essential to protect Paeonia ludlowii in their native habitats, where they could continue to provide diverse ecosystem services. Despite the plant uniqueness and diversity in Yarlung Zangbo Valley, its current nature reserve of diversity remains incomplete (Liu et al., 2003; Yu et al., 2019b). Massive recent anthropogenic habitat loss and fragmentation may lead to further decrease of the natural distribution of P. ludlowii. Currently, only population NY was relatively well protected in Scenic Area, and all the rest were not located in the range of natural reserves. Therefore, more in-situ conservation sites should be established and new law and policies should also be formulated to avoid illegal excavation.
Furthermore, genetic rescue could be attempted as a biodiversity conservation approach, in which restoring gene flow into these small, isolated population can alleviate genetic load and increase persistence probability (Whiteley et al., 2015). For populations within Nyingchi region, restoring gene flow among these populations distributed in similar environmental conditions may aid increases in population growth rate. We recommend that landscape management aimed at maintaining connectivity in the natural habitat would be useful for protecting populations with relatively large population sizes (e.g, JRK, BD and NY in Nyingchi region). Additionally, large populations with diverse genetic composition could likely be the sources for genetic rescue of smaller and less genetically diverse population (Bell et al., 2019). The isolated population ZB in Shannan region, is vulnerable to extinction through increasing farming and would need some reinforcement actions with individuals from the same or nearby populations. On the other hand, obviously the population ZB in Shannan region had the lowest nucleotide diversity. Introducing seedlings germinated from seeds produced in population NY, which was least genetically different from ZB, may help to increase the genetic diversity and decrease the probability of population extinction. But such decisions should be made with caution, because the two regions hold differences in several environmental factors such as atmosphere humid, light intensity and soil water holding capacity (Ni and Wang, 2009), and admixtures of populations with regional adaptation may bring the risk of outbreeding depression (Frankham et al., 2011), although this does not appear to be a common contribution to extinction (Bell et al., 2019). Future research using more powerful genomic methods searching for adaptive variation and selective breeding should be encouraged towards a sustainable conservation of this plant genetic resources (Fraser and Bernatchez, 2001; Frankham, 2010).
AcknowledgmentsWe are grateful to Dr. Jian Liu, Dr. Xiu-Yan Feng and Dr. Hui-Hui Xi for help in sampling. We thank Dr. Yue-Zhi Pan and Dr. Ning-Ning Zhang for assistant on data analysis. This work was supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0502), the National Natural Science Foundation of China (NSFC, 31400324), Independent research project of Yunnan Provincial Key Laboratory of Wild Resources Plant Research (E03K581261) and National Key Research Development Program of China (2022YFC2601200).
Author contributions
X.G. designated the study and managed the project. Y.J.Z. prepared DNA samples, sequencing, performed population genomics data analyses and wrote the manuscript. G.S.Y modified and improved the manuscript.
Declaration of competing interest
We declare the work described has not been published previously and not under consideration for publication elsewhere. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.07.002.
Aguilar, R., Quesada, M., Ashworth, L., et al., 2008. Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol. Ecol., 17: 5177-5188. DOI:10.1111/j.1365-294X.2008.03971.x |
Ahrens, C.W., Rymer, P.D., Stow, A., et al., 2018. The search for loci under selection: trends, biases and progress. Mol. Ecol., 27: 1342-1356. DOI:10.1111/mec.14549 |
Andrews, S., 2019. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
|
Andrews, K.R., Good, J.M., Miller, M.R., et al., 2016. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet., 17: 81-92. DOI:10.1038/nrg.2015.28 |
Baird, N.A., Etter, P.D., Atwood, T.S., et al., 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One, 3: e3376. DOI:10.1371/journal.pone.0003376 |
Banks, S.C., Cary, G.J., Smith, A.L., et al., 2013. How does ecological disturbance influence genetic diversity?. Trends Ecol. Evol., 28: 670-679. DOI:10.1016/j.tree.2013.08.005 |
Barley, A.J., Cordes, J.E., Walker, J.M., et al., 2022. Genetic diversity and the origins of parthenogenesis in the teiid lizard Aspidoscelis laredoensis. Mol. Ecol., 31: 266-278. DOI:10.1111/mec.16213 |
Barthelmess, E.L., Richards, C.M., McCauley, D.E., 2006. Relative effects of nocturnal vs diurnal pollinators and distance on gene flow in small Silene alba populations. New Phytol., 169: 689-698. DOI:10.1111/j.1469-8137.2005.01580.x |
Bell, D.A., Robinson, Z.L., Funk, W.C., et al., 2019. The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol., 34: 1070-1079. DOI:10.1016/j.tree.2019.06.006 |
Cai, C., Xiao, J., Ci, X., et al., 2021. Genetic diversity of Horsfieldia tetratepala (Myristicaceae), an endangered plant species with extremely small populations to China: implications for its conservation. Plant Syst. Evol., 307: 1-12. DOI:10.1007/s00606-021-01774-z |
Ceballos, G., Ehrlich, P.R., Barnosky, A.D., et al., 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci. Adv., 1: e1400253. DOI:10.1126/sciadv.1400253 |
Chang, C.C., Chow, C.C., Tellier, L., et al., 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4: 7. DOI:10.1186/s13742-015-0047-8 |
Chen, S.F., Guo, W., Chen, Z.X., et al., 2021. Strong genetic structure observed in Primulina danxiaensis, a small herb endemic to Mount Danxia with Extremely Small Populations. Front. Genet., 12: 722149. DOI:10.3389/fgene.2021.722149 |
Chen, X.L., Hou, G.L., Jin, S.M., et al., 2020. The pollen records of human activities in Qinghai-Tibet Plateau during the Middle and late Holocene. Earth Environ., 48: 643-651. DOI:10.4171/rmi/1212 |
Cilingir, F.G., Rheindt, F.E., Garg, K.M., et al., 2017. Conservation genomics of the endangered Burmese roofed turtle. Conserv. Biol., 31: 1469-1476. DOI:10.1111/cobi.12921 |
Crane, P., 2020. Conserving our global botanical heritage: the PSESP plant conservation program. Plant Diver., 42: 319-322. DOI:10.1016/j.pld.2020.06.007 |
D'Aloia, C.C., Andres, J.A., Bogdanowicz, S.M., et al., 2020. Unraveling hierarchical genetic structure in a marine metapopulation: a comparison of three high-throughput genotyping approaches. Mol. Ecol., 29: 2189-2203. DOI:10.1111/mec.15405 |
Do, C., Waples, R.S., Peel, D., et al., 2014. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour., 14: 209-214. DOI:10.1111/1755-0998.12157 |
Doyle, J., 1991. DNA protocols for plantsdCTAB total DNA isolation. In: Hewitt, G.M., Johnston, A. (Eds.), Molecular Techniques in Taxonomy. Springer, Berlin, pp. 283–293.
|
Dray, S., Dufour, A.B., Chessel, D., 2007. The ade4 package-Ⅱ: two-table and K-table methods. R. News, 7: 47-52. |
Eckert, C.G., Samis, K.E., Lougheed, S.C., 2008. Genetic variation across species' geographical ranges: the central–marginal hypothesis and beyond. Mol. Ecol., 17: 1170-1188. DOI:10.1111/j.1365-294X.2007.03659.x |
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Excoffier, L., Laval, G., Schneider, S., 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinforma. Online., 1: 47-50. |
Fan, Y.M., Wang, Q., Dong, Z.J., et al., 2020. Advances in molecular biology of Paeonia L. Planta, 251: 1-47. DOI:10.1007/s00425-019-03299-9 |
Farwig, N., Braun, C., Böhning-Gaese, K., 2008. Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae). Conserv. Genet., 9: 317-326. DOI:10.1007/s10592-007-9343-x |
Foll, M., Gaggiotti, O., 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics, 180: 977-993. DOI:10.1534/genetics.108.092221 |
Frankham, R., 1995. Effective population-size adult-population size ratios in wildlife - a review. Genet. Res., 66: 95-107. DOI:10.1017/s0016672300034455 |
Frankham, R., 2010. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv., 143: 1919-1927. DOI:10.1016/j.biocon.2010.05.011 |
Frankham, R., Ballou, J.D., Eldridge, M.D.B., et al., 2011. Predicting the probability of outbreeding depression. Conserv. Biol., 25: 465-475. DOI:10.1111/j.1523-1739.2011.01662.x |
Fraser, D.J., Bernatchez, L., 2001. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol. Ecol., 10: 2741-2752. DOI:10.1046/j.1365-294X.2001.t01-1-01411.x |
Funk, W.C., McKay, J.K., Hohenlohe, P.A., et al., 2012. Harnessing genomics for delineating conservation units. Trends Ecol. Evol., 27: 489-496. DOI:10.1016/j.tree.2012.05.012 |
Gutenkunst, R.N., Hernandez, R.D., Williamson, S.H., et al., 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genetics, 5: e1000695. DOI:10.1371/journal.pgen.1000695 |
Hampe, A., Petit, R.J., 2005. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett., 8: 461-467. DOI:10.1111/j.1461-0248.2005.00739.x |
Hamrick, J.L., Godt, M.J.W., 1996. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci., 351: 1291-1298. DOI:10.1098/rstb.1996.0112 |
Hao, H.P., He, Z., Li, H., et al., 2014. Effect of root length on epicotyl dormancy release in seeds of Paeonia ludlowii, Tibetan peony. Ann. Bot., 113: 443-452. DOI:10.1093/aob/mct273 |
Hewitt, G., 2000. The genetic legacy of the Quaternary ice ages. Nature, 405: 907-913. DOI:10.1038/35016000 |
Hong, D.Y., 1997. Paeonia (Paeoniaceae) in Xizang (Tibet). Novon: 156-161. DOI:10.2307/3392188 |
Hong, D.Y., Zhou, S.L., He, X.J., et al., 2017. Current status of wild tree peony species with special reference to conservation. Biodivers. Sci., 25: 781-793. DOI:10.17520/biods.2017129 |
Hutchison, D.W., Templeton, A.R., 1999. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution, 53: 1898-1914. DOI:10.1111/j.1558-5646.1999.tb04571.x |
Iannucci, A., Benazzo, A., Natali, C., et al., 2021. Population structure, genomic diversity and demographic history of Komodo dragons inferred from whole-genome sequencing. Mol. Ecol., 30: 6309-6324. DOI:10.1111/mec.16121 |
Jakobsson, M., Rosenberg, N.A., 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23: 1801-1806. DOI:10.1093/bioinformatics/btm233 |
Jensen, E.L., Edwards, D.L., Garrick, R.C., et al., 2018. Population genomics through time provides insights into the consequences of decline and rapid demographic recovery through head-starting in a Galapagos giant tortoise. Evol. Appl., 11: 1811-1821. DOI:10.1111/eva.12682 |
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Li, J.L., Milne, R.I., Ru, D.F., et al., 2020. Allopatric divergence and hybridization within Cupressus chengiana (Cupressaceae), a threatened conifer in the northern Hengduan Mountains of western China. Mol. Ecol., 29: 1250-1266. DOI:10.1111/mec.15407 |
Li, J., Wang, Z.H., 2019. Nutrients, fatty acid composition and antioxidant activity of the flowers and seed oils in wild populations of Paeonia ludlowii. Emir. J. Food Agric., 31: 206-213. DOI:10.9755/ejfa.2019.v31.i3.1922 |
Li, S.M., Lv, S.Z., Yu, K., et al., 2019. Construction of a high-density genetic map of tree peony (Paeonia suffruticosa Andr. Moutan) using restriction site associated DNA sequencing (RADseq) approach. Tree Genet. Genomes, 15: 63. DOI:10.1007/s11295-019-1367-0 |
Lee, K.M., Ranta, P., Saarikivi, J., et al., 2020. Using genomic information for management planning of an endangered perennial, Viola uliginosa. Ecol. Evol., 10: 2638-2649. DOI:10.1002/ece3.6093 |
Lischer, H.E.L., Excoffier, L., 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 28: 298-299. DOI:10.1093/bioinformatics/btr642 |
Liu, X.M., Fu, Y.X., 2015. Exploring population size changes using SNP frequency spectra. Nat. Genet., 47: 555-559. DOI:10.1038/ng.3254 |
Liu, J.G., Ouyang, Z.Y., Pimm, S.L., et al., 2003. Protecting China's biodiversity. Science, 300: 1240-1241. DOI:10.1126/science.1078868 |
Liu, D.T., Zhang, L., Wang, J.H., et al., 2020. Conservation genomics of a threatened Rhododendron: contrasting patterns of population structure revealed from neutral and selected SNPs. Front. Genet., 11: 757. DOI:10.3389/fgene.2020.00757 |
Llorens, T.M., Ayre, D.J., Whelan, R.J., 2018. Anthropogenic fragmentation may not alter pre-existing patterns of genetic diversity and differentiation in perennial shrubs. Mol. Ecol., 27: 1541-1555. DOI:10.1111/mec.14552 |
Lv, S.Z., Cheng, S., Wang, Z.Y., et al., 2020. Draft genome of the famous ornamental plant Paeonia suffruticosa. Ecol. Evol., 10: 4518-4530. DOI:10.1002/ece3.5965 |
Ma, Y.P., Chen, G., Grumbine, R.E., et al., 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers. Conserv., 22: 803-809. DOI:10.1007/s10531-013-0434-3 |
Marques, D.A., Meier, J.I., Seehausen, O., 2019. A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol., 34: 531-544. DOI:10.1016/j.tree.2019.02.008 |
Nei, M., Kumar, S., 2000. Molecular Evolution and Phylogenetics. Oxford University, New York.
|
Ni, S.W., Wang, L.Y., 2009. Introduction and Ex-Situ Conservation of Paeonia delavayi, Paeonia lutea, Paeonia ludlowii. Beijing Forestry University.
|
Nielsen, E.S., Beger, M., Henriques, R., et al., 2020. A comparison of genetic and genomic approaches to represent evolutionary potential in conservation planning. Biol. Conserv., 251: 108770. DOI:10.1016/j.biocon.2020.108770 |
Ouborg, N.J., Pertoldi, C., Loeschcke, V., et al., 2010. Conservation genetics in transition to conservation genomics. Trends Genet., 26: 177-187. DOI:10.1016/j.tig.2010.01.001 |
Papuga, G., Gauthier, P., Pons, V., et al., 2018. Ecological niche differentiation in peripheral populations: a comparative analysis of eleven Mediterranean plant species. Ecography, 41: 1650-1664. DOI:10.1111/ecog.03331 |
Peakall, R., Smouse, P.E., 2006. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes, 6: 288-295. DOI:10.1111/j.1471-8286.2005.01155.x |
Pimm, S.L., Jenkins, C.N., Abell, R., et al., 2014. The biodiversity of species and their rates of extinction, distribution, and protection. Science, 344: 1246752. DOI:10.1126/science.1246752 |
Pritchard, J.K., Stephens, M., Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1111/j.1471-8286.2007.01758.x |
Puritz, J.B., Addison, J.A., Toonen, R.J., 2012. Next-generation phylogeography: a targeted approach for multilocus sequencing of non-model organisms. PLoS One, 7: e34241. DOI:10.1371/journal.pone.0034241 |
Rochette, N.C., Rivera-Colon, A.G., Catchen, J.M., 2019. Stacks 2: analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol., 28: 4737-4754. DOI:10.1111/mec.15253 |
Rosenberg, N.A., 2004. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes, 4: 137-138. DOI:10.1046/j.1471-8286.2003.00566.x |
Shuai, Y.T., Zang, J.C., 2016. Paeonid ludlowii and Paeonia delavayi flower characteristics and change of flower-visiting insects and phenotypic selection. Southwest China J. Agric. Sci., 29: 2714-2719. |
Stankowski, S., Ravinet, M., 2021. Defining the speciation continuum. Evolution, 75: 1256-1273. DOI:10.1111/evo.14215 |
Tamura, K., Nei, M., 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol., 10: 512-526. DOI:10.1093/oxfordjournals.molbev.a040023 |
Tang, Y., Yuan, T., Chen, T.Q., 2021. Flowering characteristics and breeding system of Paeonia ludlowii. Acta Bot. Boreal.-Occident. Sin., 41: 782-794. |
Terhorst, J., Song, Y.S., 2015. The sample frequency spectrum. Proc. Natl. Acad. Sci. U.S.A., 112: 7677-7682. DOI:10.1073/pnas.1503717112 |
Thompson, J.D., Gaudeul, M., Debussche, M.A.X., 2010. Conservation value of sites of hybridization in peripheral populations of rare plant species. Conserv. Biol., 24: 236-245. DOI:10.1111/j.1523-1739.2009.01304.x |
Vranckx, G., Jacquemyn, H., Muys, B., et al., 2012. Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv. Biol., 26: 228-237. DOI:10.1111/j.1523-1739.2011.01778.x |
Whiteley, A.R., Fitzpatrick, S.W., Funk, W.C., et al., 2015. Genetic rescue to the rescue. Trends Ecol. Evol., 30: 42-49. DOI:10.1016/j.tree.2014.10.009 |
Wikstrom, N., Savolainen, V., Chase, M.W., 2001. Evolution of the angiosperms: calibrating the family tree. Proc. R. Soc. B-Biol. Sci., 268: 2211-2220. DOI:10.1098/rspb.2001.1782 |
Xu, X.X., Cheng, F.Y., Peng, L.P., et al., 2019. Late Pleistocene speciation of three closely related tree peonies endemic to the Qinling-Daba Mountains, a major glacial refugium in Central China. Ecol. Evol., 9: 7528-7548. DOI:10.1002/ece3.5284 |
Xiao, J.H., Ding, X., Li, L., et al., 2020. Miocene diversification of a golden-thread nanmu tree species (Phoebe zhennan, Lauraceae) around the Sichuan Basin shaped by the East Asian monsoon. Ecol. Evol., 10: 10543-10557. DOI:10.1002/ece3.6710 |
Xu, T.T., Wang, Q., Olson, M.S., et al., 2017. Allopatric divergence, demographic history, and conservation implications of an endangered conifer Cupressus chengiana in the eastern Qinghai-Tibet Plateau. Tree Genet. Genomes, 13: 100. DOI:10.1007/s11295-017-1183-3 |
Xue, Y.Q., Liu, R., Xue, J.Q., et al., 2021. Genetic diversity and relatedness analysis of nine wild species of tree peony based on simple sequence repeats markers. Hortic. Plant J., 7: 579-588. DOI:10.1016/j.hpj.2021.05.004 |
Yang, J., Cai, L., Liu, D., et al., 2020a. China's conservation program on plant species with extremely small populations (PSESP): progress and perspectives. Biol. Conserv., 244: 108535. DOI:10.1016/j.biocon.2020.108535 |
Yang, Y., Ma, T., Wang, Z., et al., 2018. Genomic effects of population collapse in a critically endangered ironwood tree Ostrya rehderiana. Nat. Commun., 9: 5449. DOI:10.1038/s41467-018-07913-4 |
Yang, Y., Sun, M., Li, S., et al., 2020b. Germplasm resources and genetic breeding of Paeonia: a systematic review. Hortic. Res., 7: 107. DOI:10.1038/s41438-020-0332-2 |
Yang, X.L., Wang, Q.J., Lan, X.Z., et al., 2007. Numeric dynamics of the endangered plant population of Paeonia ludlowii. Acta Ecol. Sin., 27: 1242-1247. |
Yao, X.H., Ye, Q.G., Kang, M., et al., 2007. Microsatellite analysis reveals interpopulation differentiation and gene flow in the endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China. New Phytol., 176: 472-480. DOI:10.1111/j.1469-8137.2007.02175.x |
Young, A., Boyle, T., Brown, T., 1996. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol., 11: 413-418. DOI:10.1016/0169-5347(96)10045-8 |
Younger, J.L., Clucas, G.V., Kao, D.M., et al., 2017. The challenges of detecting subtle population structure and its importance for the conservation of emperor penguins. Mol. Ecol., 26: 3883-3897. DOI:10.1111/mec.14172 |
Yu, H.B., Deane, D.C., Sui, X.H., et al., 2019a. Testing multiple hypotheses for the high endemic plant diversity of the Tibetan Plateau. Glob. Ecol. Biogeogr., 28: 131-144. DOI:10.1111/geb.12827 |
Yu, H.B., Favre, A., Sui, X.H., et al., 2019b. Mapping the genetic patterns of plants in the region of the Qinghai-Tibet Plateau: implications for conservation strategies. Divers. Distrib., 25: 310-324. DOI:10.1111/ddi.12847 |
Zhang, L., 2008. Population Characteristics and Seed's Biology of Paeonia ludlowii. Master, Beijing Forestry University.
|
Zhang, J.M., Lopez-Pujol, J., Gong, X., et al., 2018. Population genetic dynamics of Himalayan-Hengduan tree peonies, Paeonia subsect. Delavayanae. Mol. Phylogenet. Evol., 125: 62-77. DOI:10.1016/j.ympev.2018.03.003 |
Zhang, D.J., Shen, X.K., Cheng, T., et al., 2020. New advances in the study of prehistoric human activity on the Tibetan Plateau. Chin. Sci. Bull., 65: 475-482. DOI:10.1360/tb-2019-0382 |
Zhang, X., Zhai, Y., Yuan, J., et al., 2019. New insights into Paeoniaceae used as medicinal plants in China. Sci. Rep., 9: 18469. DOI:10.1038/s41598-019-54863-y |
Zhao, Y.J., Yin, G.S., Pan, Y.Z., et al., 2021. Climatic refugia and geographical isolation contribute to the speciation and genetic divergence in Himalayan-Hengduan tree peonies (Paeonia delavayi and Paeonia ludlowii). Front. Genet., 11: 595334. DOI:10.3389/fgene.2020.595334 |
Zheng, B.X., Xu, Q.Q., Shen, Y.Q., 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quat. Int.: 97–98, 93–101. DOI:10.1016/S1040-6182(02)00054-X |
Zhou, S.L., Xu, C., Liu, J., et al., 2021. Out of the Pan-Himalaya: evolutionary history of the Paeoniaceae revealed by phylogenomics. J. Syst. Evol., 59: 1170-1182. DOI:10.1111/jse.12688 |



