b. University of Chinese Academy of Sciences, Beijing 100049, China;
c. Vietnam National Museum of Nature, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay 10000, Hanoi, Vietnam;
d. Graduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay 10000, Hanoi, Vietnam
East Asian subtropical/warm-temperate evergreen broadleaved forests (EBLFs) landscapes are prominent, as regions at the similar latitudes (23–35°N) are largely covered by desert or semi-desert (Tang, 2015; Song and Da, 2016). This zonal vegetation develops under a monsoon climate and harbours high biodiversity and endemism, and East Asia was considered a very important center of speciation and evolution, and attracts much attention consequently (Wu, 1980; Axelrod et al., 1996; Wu and Wu, 1996; Song and Da, 2016; Chen et al., 2018).
The East and Southeast Asian flora has been suggested to be ancient (Takhtajan, 1969). Paleovegetational reconstruction in South China clued that EBLFs existed there during the Miocene (Zhao et al., 2004; Jacques et al., 2011; Sun et al., 2011). In addition, molecular phylogenetic analyses on the dominant taxa (such as Fagaceae, Lauraceae, Theaceae, and Magnoliaceae) in this region also suggested that East Asian EBLFs were established mainly around the Oligocene–Miocene (O–M) boundary, possibly affected by the formation and development of the Asian monsoon (Yu et al., 2017; Chen et al., 2018; Deng et al., 2018; Hai et al., 2022; Xiao et al., 2022). However, research on epiphytic orchids (Dendrobium Sw.) indicated that EBLFs may have arisen in mainland Asia since the beginning of the Oligocene (Xiang et al., 2016), which broadly agrees with U–Pb zircon dating of fossil-bearing strata (Tian et al., 2021).
Brandisia Hook. f. & Thomson is one of the representative undergrowth of East Asian subtropical/warm-temperate EBLFs (between 21–30°N and 97–141°E; Figs. 1 and 2; Song and Da, 2016). Brandisia species are shrubs with a more or less liana habit that inhabit mountainous areas at low-medium elevations (500–3000 m), living under forests, along forest edges, mountain slopes, trails and riverbanks. Most Brandisia species are endemic to East Asia, although Brandisia discolor Hook. f. & Thomson can extend into tropical Southeast Asia. The current distribution center is in karst areas in southwestern China (around the Tropic of Cancer, 23.5°N; Fig. 2; Li, 1947; Hong et al., 1998). The continuous karst regions in southwestern China cover a great area (ca. 21–30°N and 100–110°E; Ford and Williams, 2007; Hollingsworth, 2009; Wang et al., 2019), which are characterized by diverse environments and great species diversity and endemism (Davis et al., 1995; Zhu, 2007). Evidence from phylogeny and paleobotany suggested that the karst vegetation formed after the O–M boundary (Li et al., 2022a, 2022b) or even much earlier (at the early Oligocene; Huang et al., 2018; Tian et al., 2021). Moreover, Brandisia is hemiparasitic (Nickrent, 2020), and may need hosts at least in some phases of its lifecycle. Considering the distribution and hemiparasitic habit of Brandisia, it can be considered characteristic of East Asian EBLFs. Therefore, we hypothesized that its evolution and dispersal should be shaped by the historical dynamics of the development of EBLFs in East Asia, especially in the karst regions. Clear taxonomic and phylogenetic contexts are the essential prerequisites for verifying our hypothesis, which have not been fully resolved.

|
| Fig. 1 Photos of Brandisia. A, B. rosea var. flava; B, B. cauliflora; C, B. racemosa; D, B. hancei; E, B. glabrescens; F, B. kwangsiensis; G, B. swinglei; H, B. discolor. I, Hybrid of B. hancei and B. discolor; J1–J2, Flower, corolla and leaves of B. hancei, B. discolor and their hybird. K1–K2, Flower, leaves, and fruits of B. chevalieri; L1–L2, Branch, leaves and fruits of B. scandens. |
|
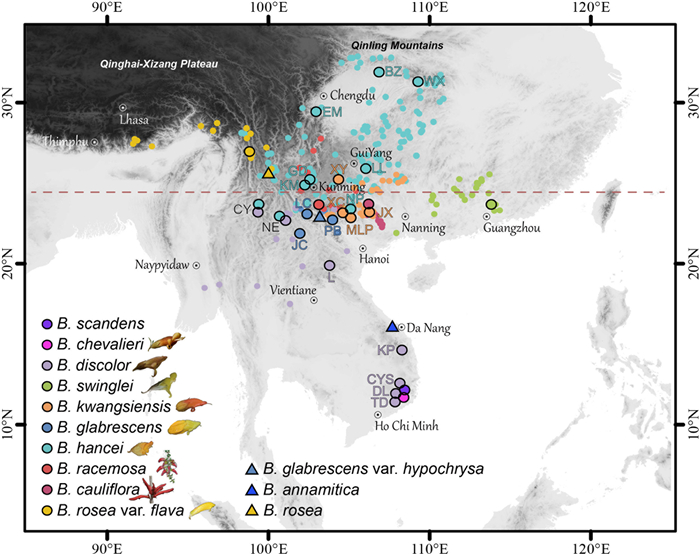
| Fig. 2 Distribution and sampling of Brandisia. Points in different colour show species distribution based on specimen records and our fieldwork. The large points with black outlines show our sampling sites. Letters indicate populations, which are also used in Figs. 3–5. Details regarding collection vouchers can be found in Table S1. Triangles indicate places where species were recorded by specimens but not found personally. The red dashed line shows the Tropic of Cancer at 23.5°N. |
Brandisia was originally described by Hooker and Thomson (1864), and was "approximately determined" as a member of Scrophulariaceae sensu lato due to the numerous ovules. However, Li (1947) proposed that transferring Brandisia (together with close-related taxa Paulownia Siebold & Zucc. and Wightia Wall.) into Bignoniaceae was more appropriate than retaining it in Scrophulariaceae. Currently, there are 13 taxa (11 species and two varieties) in Brandisia. Eight species and two varieties can be found in China, while three are endemic to Vietnam (Bonati, 1924; Tsoong and Lu, 1979; Hong et al., 1998; Pham, 2000). Over the last two decades, phylogenetic studies have shed new light on the position of Brandisia. Studies based on nuclear loci, plastid loci and plastid genomes have revealed that Brandisia is a member of Orobanchaceae (Oxelman et al., 2005; Zhou et al., 2014; Xia et al., 2019; Li et al., 2021a). However, the exact placement of Brandisia within Orobanchaceae varies among different molecular data. Studies based on nuclear coding gene phytochrome A (PHYA; Bennett and Mathews, 2006), combined loci (nuclear ribosomal internal transcribed spacer/ITS, phytochrome A/PHYA and phytochrome B/PHYB; plastid matK and rps2; McNeal et al., 2013), and ITS data (Yu et al., 2018) supported that Brandisia is sister to the major clade of hemiparasites (Rhinantheae, Buchnereae, Pedicularideae, and Pterygiella Oliv.). However, the results of plastid data showed incongruence, as phylogenies derived from plastome data supported Rhinantheae is sister to Brandisia (Xia et al., 2019; Li et al., 2021a).
In this study, we intended to investigate the evolution of Brandisia and its potential relationship with East Asian EBLFs. We first conducted phylogenetic analyses based on plastome and nuclear sequences, exploring the position of Brandisia and clarifying the species relationships within this genus. We then inferred divergence times and the biogeographic origin of Brandisia, which was used to explore its evolution and dispersal history.
2. Materials and methods 2.1. Taxa sampling, DNA extraction and sequencingEleven Brandisia taxa were used in this study (with 57 accessions; Table S1), including a potential hybrid population between B. discolor and B. hancei. Three of 13 Brandisia taxa (Brandisia rosea var. rosea, B. glabrescens var. hypochrysa and B. annamitica) were unavailable due to unclear locality records or habitat destruction. For phylogenetic analyses, 40 taxa of major lineages in Orobanchaceae were used. Paulownia tomentosa (Thunb.) Steud. was chosen as the outgroup based on previous studies (McNeal et al., 2013; Xia et al., 2019; Li et al., 2021a). All the DNA sequences but Brandisia were obtained from GenBank (Table S2).
For Brandisia, total genomic DNA was extracted from silica gel-dried leaves using the Plant Genomic DNA Kit DP305 (Tiangen Biotech, Beijing, China). DNA was fragmented with a focused ultrasonicator (Covaris) to construct a library. Paired-end reads of 150 bp were sequenced on the Illumina HiSeq 2000 platform at Novogene Co. (Beijing, China), generating 1.02–3.95 Gb of clean data (Table S1). Three samples of Brandisia kwangsiensis (MLP, XC and XY; Fig. 2, Table S1) were derived from herbarium specimens; the library construction followed a strategy customized for specimen materials (Zeng et al., 2018).
2.2. Sequence assembly and annotationClean sequencing data was analysed using the GetOrganelle pipeline (Jin et al., 2020) to assemble and select plastid genome and nuclear loci (nuclear ribosomal DNA (nrDNA), PHYA and PHYB). For the plastomes and nrDNA, the defaulted seed sequences of embryophyta plastome and plant nuclear ribosomal RNA were used, respectively. For PHYA and PHYB, sequences of Orobanchaceae from GenBank were used to build the seed files manually. The assembled plastomes were first annotated by PGA software (Qu et al., 2019) using the plastome of Paulownia tomentosa (NC_031436) as the reference and then manually checked and adjusted in Geneious 9.0.2. Contigs of nuclear loci were annotated in Geneious 9.0.2 using ribosomal DNA of P. tomentosa (KP718625), PHYA of Solanum lycopersicum L. (NM_001247561), and PHYB of S. lycopersicum (NM_001306202) as references.
2.3. Phylogenetic datasets construction and alignmentBased on the plastid genome and nuclear loci, seven datasets were constructed for phylogenetic analyses: 1) complete plastome sequences with the removal of inverted repeat B (IRb); 2) the concatenation of 81 plastid protein-coding genes (CDSs); 3) the nuclear ribosomal DNA sequences (nrDNA); 4) PHYA DNA sequences; 5) and 6) two copies of PHYB DNA sequences (PHYB_1 and PHYB_2); and 7) the combined sequences of nuclear loci (nrDNA, PHYA, PHYB_1 and PHYB_2). Datasets were aligned using MAFFT v.7.308 in Geneious 9.0.2 or MAFFT v.7 online. Because the plastomes of most taxa in Orobanchaceae have undergone serious rearrangement, we adjusted these plastomes prior to alignment using the online software Mulan (https://mulan.dcode.org/). Plastomes of Brandisia, Paulownia tomentosa, Triaenophora shennongjiaensis X.D. Li, Y.Y. Zan & J.Q. Li, Rehmannia Libosch. ex Fisch. & C.A. Mey and Lindenbergia philippensis (Cham. & Schltdl.) Benth. showed little rearrangement, thus, they were not processed in Mulan.
2.4. Phylogenetic analysesFor each dataset above, maximum likelihood (ML) analysis was conducted using RAxML-NG (1.0.0-master; Kozlov et al., 2019). Support values for nodes and clades were estimated through 1000 bootstrap replicates. For complete plastome sequences (dataset 1) and combined nuclear loci sequences (dataset 7), Bayesian inference (BI) analysis was also conducted using MrBayes (3.2.7a; Ronquist et al., 2012). Markov chain Monte Carlo (MCMC) analysis was conducted using two independent runs, each having four incrementally heated chains. A total of 5, 000, 000 and 10, 000, 000 generations of MCMC chains were run for dataset 1 and dataset 7, respectively. The first 25% of tree samples were discarded as burn-in, and the remaining samples were used to generate a majority-rule consensus tree. To find the best substitution model for nucleotide sequences, ModelTest-NG (0.1.6; Darriba et al., 2020), with the corrected Akaike information criterion (AICc), was used. For dataset 7, nuclear loci were treated as different partitions; the GTR + I + G model was the best model for each nuclear locus. RAxML-NG, MrBayes and ModelTest-NG analyses mentioned above were carried out on the CIPRES platform (Miller et al., 2010). Trees were visualized using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
2.5. Molecular datingBecause we believe that the combined sequences of nuclear loci (dataset 7) generated the true species tree of Brandisia species (detailed in the discussion), we used this dataset to estimate divergence times. BEAST (v.2.6.3; Bouckaert et al., 2014) was used at CIPRES. Because no reliable fossils have been found in Orobanchaceae, a secondary calibration strategy was adopted. We used two secondary calibration points derived from previous work involving Lamiales, which incorporated four fossils (Yu et al., 2018). The stem and crown ages of Orobanchaceae were set as 56.23 ± 10 and 54.56 ± 10 Mya with normal models, respectively. We set the nucleotide substitution model to GTR + I + Γ (selected by ModelTest-NG). We used an uncorrelated lognormal relaxed molecular clock model and the Yule speciation model for tree priors. The MCMC ran 100, 000, 000 generations and sampled every 1000 generations. The first 3000 generations were removed as "Pre-burn-in". The output log file was then checked for convergence of the chains using Tracer v.1.7 to confirm that the ESSs of all parameters were more than 200. TreeAnnotator v.1.7.5 was used to obtain the maximum clade credibility (MCC) tree with the posterior probability limit set to 0.5. The initial 10% of the trees were discarded as burn-in.
2.6. Estimation of ancestral areasTo infer ancestral area, we used the S-DIVA (Statistical dispersal-vicariance analysis) and BBM (Bayesian binary MCMC) methods implemented in RASP 4.2 (Reconstruct Ancestral State in Phylogenies; Yu et al., 2015, 2020). The 90, 001 trees (the first 10, 000 trees were discarded) from BEAST analysis above were used as input, and only Brandisia species were selected for analysis. For S-DIVA analysis, 1000 trees were sampled randomly, the number of maximum areas was set as 2, and only dispersals between adjacent areas were allowed. For BBM analysis, the consensus tree derived from the 90, 001 tree inputs was used. MCMC chains were run simultaneously for 500, 000 generations and sampled every 100 generations. Fixed JC + G (Jukes-Cantor + Gamma) was used for BBM analysis with null root distribution. The number of maximum areas was set to 1. Six distribution areas were set based on the distribution of Brandisia, the floristic regions of China proposed by Wu and Wu (1996; based on the floral composition and vegetation types), topography and the distribution of karst in China and SE Asia (Sweeting, 1995; Hollingsworth, 2009). They were A, Eastern Himalayas; B, Yunnan Plateau (and adjacent areas of SW Sichuan and W Guangxi); C, Core Karst in SW China (SE Yunnan, N Guizhou, W Guangxi; also including a part of N Vietnam); D, Southern China; E, Central China; F, Indochina (the wide area south of the Red River, including SW Yunnan, Myanmar, Thailand, Laos, and Vietnam).
3. Results 3.1. Assembly of plastomes and nuclear sequence fractionsComplete plastomes were assembled for most of the Brandisia samples, except for three samples, each with one gap located in the small single copy region (Table S1). The plastomes of Brandisia showed a typical quadripartite structure (Fig. S1), with large and small single copy (LSC and SSC, respectively) regions separated by two large inverted repeats (IRs). Three of the five B. kwangsiensis samples (MLP, XC and XY) showed plastomes much longer (159, 382–159, 469 bp) than the other Brandisia samples due to the expansions of their IRs (Fig. S2). For the remaining Brandisia samples, total length ranged from 154, 580 (B. hancei NP) to 155, 391 bp (B. swinglei). Gene contents included 115 unique genes for each sample, including 81 protein-coding genes (CDSs), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. Except for B. rosea var. flava, the slight expansions of the IRs toward the LSC led to the duplication of the rps19 gene in most samples and the duplication of the rpl22 gene further in B. hancei from BZ (Bazhong, Sichuan; Fig. S2). No other rearrangements or pseudogenes were found.
The nuclear ribosomal DNA (nrDNA) sequences were assembled for all the samples, which covered 18S, ITS1 (internal transcribed spacer), 5.8S, ITS2, 28S, and partial IGS (intergenic spacer). For IGSs, regions with good alignment (mainly the ETS (external transcribed spacer) regions) were used for phylogenetic analysis. Nuclear genes of PHYA and PHYB were assembled in some of our samples and often contained several contigs that composed partial sequences. Our results showed that PHYA was probably a single-copy gene in Brandisia. However, PHYB was probably a double-copy gene, as our assembled contigs showed different similarities to the two PHYB copies cloned in Pedicularideae in a previous study (McNeal et al., 2013). In our study, the two PHYB copies were named PHYB_1 and PHYB_2. The lengths of nrDNA, PHYA, PHYB_1 and PHYB_2 used are provided in Table S1.
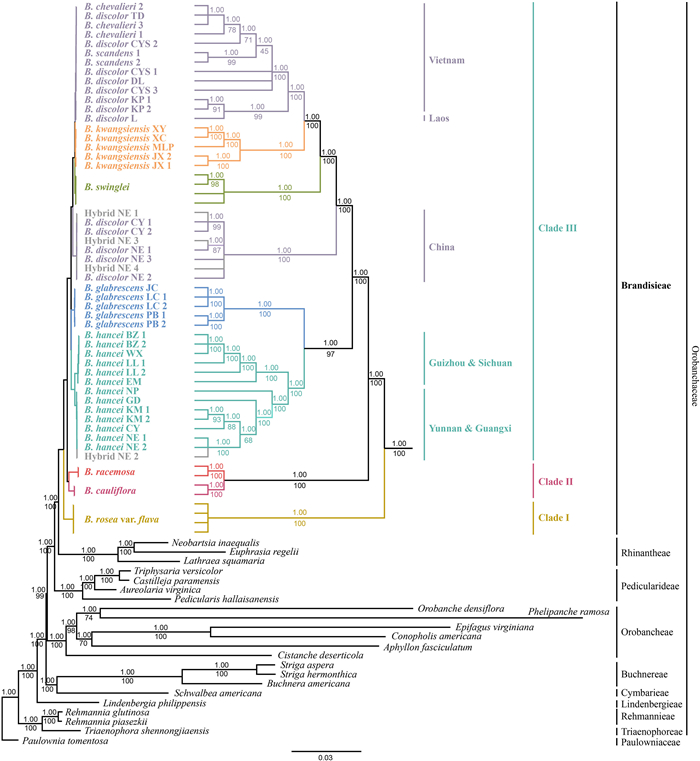
3.2. Phylogeny constructionInformation on sequence length, parsimony-informative (PI) sites and model selection of the seven aligned datasets are shown in Table S3. Phylogenies based on whole plastomes (with IRb removed; dataset 1) and CDS datasets (dataset 2) were consistent with each other (Figs. 3 and S3). The monophyly of Brandisia was well supported, and it was sister to Rhinantheae. Brandisia was divided into three clades. Clade Ⅰ contained only B. rosea var. flava. Clade Ⅱ contained B. racemosa and B. cauliflora. The remaining species were placed in Clade Ⅲ, within which B. glabrescens and B. hancei clustered in a clade that was sister to all other taxa.
|
| Fig. 3 Strict consensus tree derived from Bayesian inference of complete plastome. The values above and under the branches show posterior probability (PP) and bootstrap support (BS), respectively. The topology of Brandisia with short branch lengths is shown on the right. The bottom bar shows the number of substitutions per site. |
In addition, Brandisia discolor was divided into two clades, one from China and the other from Laos and Vietnam. Brandisia scandens and B. chevalieri were embedded in B. discolor from Laos and Vietnam. For the four potential hybrid samples between B. discolor and B. hancei, three were embedded within B. discolor from China, and the other one was nested within B. hancei.
The phylogenies derived from the individual dataset of the four nuclear loci (nrDNA, PHYA, PHYB_1 and PHYB_2; dataset 3–6) could hardly resolve the position of Brandisia in Orobanchaceae, and the relationships within Brandisia were generally not strongly supported (Figs. S4–S7). However, the topologies of the major clades within Brandisia were congruent with each other, except that B. swinglei and B. kwangsiensis formed a sister group with great support (BS = 100) based on PHYB_1 (Fig. S6).
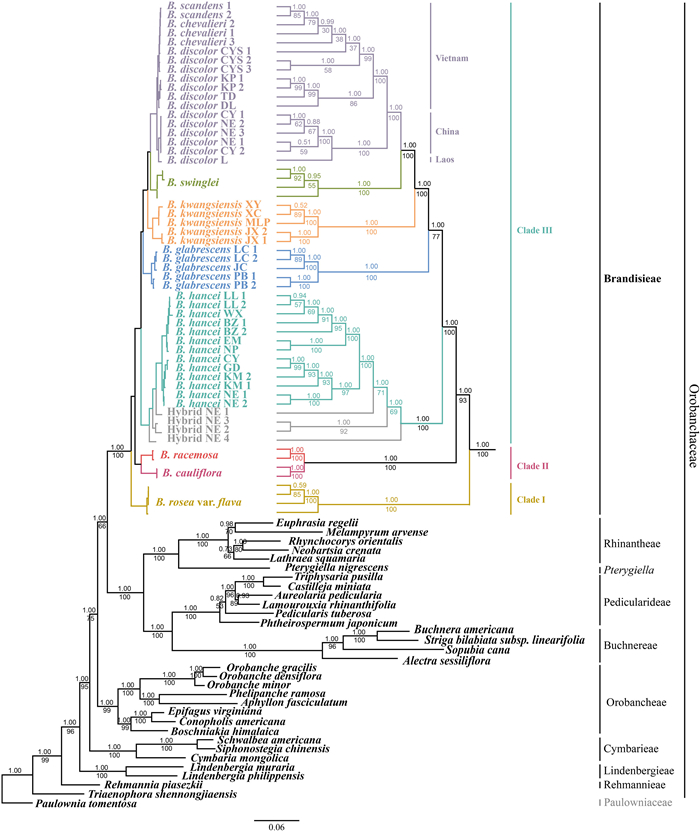
The combined dataset of four nuclear loci (dataset 7) generated a phylogeny with better resolution (Fig. 4). The monophyly of Brandisia was well supported, and was sister to a clade containing Rhinantheae, Buchnereae, Pedicularideae and Pterygiella (BS/PP = 66/1). Brandisia was divided into three clades, among which Clades Ⅰ and Ⅱ were identical to the results from the plastid datasets, while several major differences occurred in Clade Ⅲ. First, B. hancei was sister to the other taxa in Clade Ⅲ but not only B. glabrescens. Second, samples of B. discolor clustered together (with B. scandens and B. chevalieri embedded in), and samples of B. discolor from Vietnam were separate from those from China and Laos. Third, all the potential hybrid individuals (between B. discolor and B. hancei) were nested within B. hancei.
|
| Fig. 4 Strict consensus tree derived from Bayesian inference of combined datasets of four nuclear loci (3S/ITS, PHYA, PHYB_1 and PHYB_2). The values above and under the branches show posterior probability (PP) and bootstrap support (BS), respectively. |
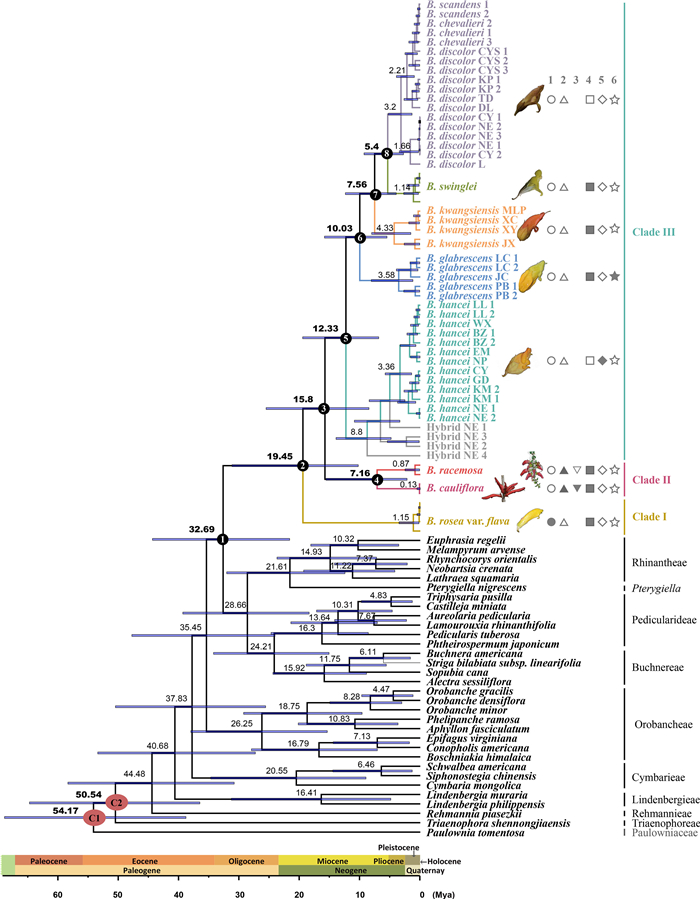
BEAST analysis showed that Brandisia and its sister diverged in the early Oligocene (32.69 Mya, 95% HPD = 21.66–44.39), and the diversification of Brandisia was dated to the early Miocene (19.45 Mya, 95% HPD = 10.28–31.22). Clades Ⅱ and Ⅲ diverged 15.8 Mya (95% HPD = 8.51–25.55), and their diversifications were dated back to 7.16 (95% HPD = 2.19–15.62) and 12.33 Mya (95% HPD = 6.89–19.5), respectively (Fig. 5). The exact values for the two calibrations and 8 annotated nodes are detailed in Table 1.
|
| Fig. 5 The maximum clade credibility (MCC) tree derived from BEAST based on combined datasets of four nuclear markers (3S/ITS, PHYA, PHYB_1 and PHYB_2) for Brandisia and other related taxa. The blue bars show 95% higher posterior densities (HPD). Two calibrated (red) and 8 key stem/crown nodes (black) are annotated by letters and/or numbers. Six diagnostic characteristics are provided. Trait 1, calyx bilabiate with two lobes (solid circles); calyx with five lobes (circles). Trait 2, raceme (solid triangles); single flower (triangles). Trait 3, raceme borne on main stem, compact (solid inverted triangles); raceme terminal, long (inverted triangles). Trait 4, flower in summer (-autumn) (solid squares); flower in winter (-spring) (squares). Trait 5, blade base subcordate, margin revolute (solid diamonds); blade base cuneate to subrounded, margin not revolute (diamonds). Trait 6, calyx big, vivid yellow (solid stars); calyx small to medium, gray or brown (stars). |
| Nodes | Calibration ages (Mya) | Estimation | ||
| Mean (Mya) | 95% HPD (Mya) | PP | ||
| C1: Orobanchaceae stem | 56.23 | 54.17 | 38.85–68.92 | 1 |
| C2: Orobanchaceae crown | 54.56 | 50.54 | 36.5–64.79 | 1 |
| 1: Brandisia stem | 32.69 | 21.66–44.39 | 1 | |
| 2: Brandisia crown | 19.45 | 10.28–31.22 | 1 | |
| 3: all Brandisia - B. rosea crown | 15.8 | 8.51–25.55 | 1 | |
| 4: B. racemosa + B. cauliflora crown | 7.16 | 2.19–15.62 | 1 | |
| 5: B. hancei + hybrid + B. glabrescens + B. kwangsiensis + B. swinglei + B. discolor crown | 12.33 | 6.89–19.5 | 1 | |
| 6: B. glabrescens + B. kwangsiensis + B. swinglei + B. discolor crown | 10.03 | 5.52–15.88 | 1 | |
| 7: B. kwangsiensis + B. swinglei + B. discolor crown | 7.56 | 3.98–12.42 | 1 | |
| 8: B. swinglei + B. discolor crown | 5.4 | 2.78–9.32 | 1 | |
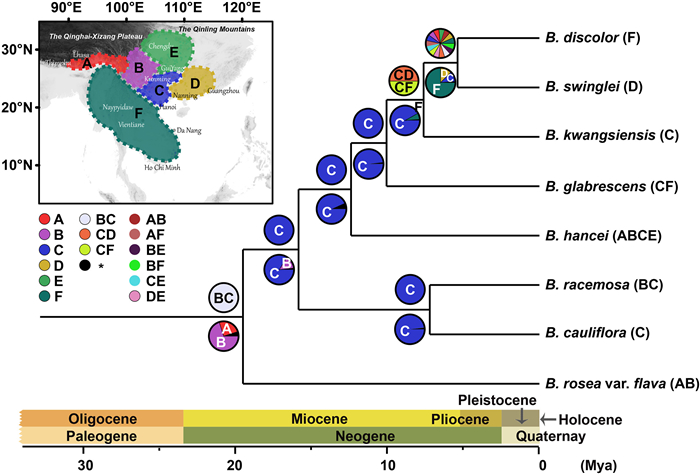
Brandisia is mainly distributed in southern China, Vietnam, Laos, Thailand, and India (Assam), with a current distribution center around the karst regions of Yunnan-Guizhou-Guangxi (Fig. 2). S-DIVA and BBM analyses inferred that Brandisia originated in the Yunnan Plateau and Eastern Himalayas (BA) and the Yunnan Plateau and Core Karst in SW China (BC), respectively (Fig. 6). These results indicate this genus probably originated in the Eastern Himalayas–SW China, formed distribution center in the karst region, and dispersed eastwards into the evergreen broadleaved forests in South China (Fig. 6). In addition, B. hancei and B. discolor are the two most widely distributed species in this genus. B. hancei extends northwards up to the southern margin of the Qinling Mountains (ca. 33°N), whereas B. discolor reaches southern Vietnam (Da Lat city, Lam Dong province, ca. 12°N; Fig. 2).
|
| Fig. 6 Ancestral area reconstruction of Brandisia. Statistical dispersal-vicariance analysis (S-DIVA) and Bayesian binary MCMC (BBM) were carried out for ancestral range reconstruction. The results are shown above and below the branches, respectively. The inserted picture shows the geographic regions. A is the Eastern Himalayas; B, Yunnan Plateau (and adjacent areas of SW Sichuan and W Guangxi); C, Core Karst in SW China (SE Yunnan, N Guizhou, W Guangxi; also including a part of N Vietnam); D, southern China; E, central China; F, Indochina (the wide area south of the Red River, including SW Yunnan, Myanmar, Thailand, Laos, and Vietnam). |
Orobanchaceae is a diverse family where parasitic herbs are predominant. Except for several autotrophic taxa (Triaenophora (Hook. f.) Soler., Rehmannia and Lindenbergia Lehm.), plastome rearrangements (including IR loss/expansion/contraction, gene order change, pseudogenization and loss of genes) are common across the whole family, which reflects the loss of pressure on the plastome in their evolutionary progress (Krause, 2008; Wicke et al., 2013; Cusimano and Wicke, 2016; Frailey et al., 2018). It is believed that there is a single origin of parasitism in Orobanchaceae, followed by several independent evolutions of holoparasitism (Young et al., 1999; Bennett and Mathews, 2006; McNeal et al., 2013). Brandisia is a relatively later-derived genus, and is sister to the major hemiparasites in Orobanchaceae (detailed below). In addition, although certain hosts are unknown, haustoria has been discovered in the roots of Brandisia (Personal communication with Dr. Wen-Bin Yu). Therefore, it is reasonable to expect hemiparasitic habits in Brandisia. However, the plastomes of Brandisia are complete and conventional, with full gene contents and merely a slight expansion of IRs (except for three samples of B. kwangsiensis), which are highly similar to the nonparasitic taxa in Orobanchaceae. Brandisia is one of the few shrubby genera (such as Alectra Thunb. and Xylocalyx Balf. f.) in Orobanchaceae and has the largest size (B. discolor can reach 3 m high; field observation). This bushing habit, which requires high photosynthetic productivity for development, probably explains the constant selection on plastomes and consequently their conservatism. Brandisia presumably relies less on its host, perhaps only in its early stage (such as seedling), which requires further study in the future.
4.2. Phylogenies of Orobanchaceae and BrandisiaBrandisia is a unique genus whose taxonomic placement has been arguable since its first description. Morphologically, it shows affinity to several families (such as Scrophulariaceae and Bignoniaceae; Hooker and Thomson, 1864; Li, 1947). The woody genera Paulownia and Wightia were deemed close to Brandisia and showed similarities in capsules and seeds (Hooker and Thomson, 1864). However, phylogenetic evidence supports the placement of Brandisia in Orobanchaceae, and Paulownia as an outgroup of Orobanchaceae (Wightia, however, is relatively more distant; Olmstead et al., 2001; Xia et al., 2019; Li et al., 2021a). The exact position of Brandisia in Orobanchaceae has been unstable because phylogenetic research based on different (nuclear vs. plastid) sequence data has reached discordant conclusions (McNeal et al., 2013; Yu et al., 2018; Xia et al., 2019). Our study shows a similar incongruence between nuclear-plastid data. Our plastome phylogeny placed Rhinantheae as sister to Brandisia (Fig. 4), which agrees with previous studies by Xia et al. (2019) and Li et al. (2021a); in contrast, our nuclear phylogeny placed the major clade of hemiparasites (Rhinantheae, Buchnereae, Pedicularideae, and Pterygiella) sister to Brandisia (Fig. 5), which agrees with previous studies by Bennett and Mathews (2006), McNeal et al. (2013), and Yu et al. (2018). In addition, Pterygiella (Xia et al., 2019) or the Pterygiella group (Pterygiella, the Phtheirospermum Bunge ex Fisch. & C.A. Mey. complex and Xizangia D.Y. Hong; Yu et al., 2018) were resolved as sister to Brandisia further based on plastid locus; and morphological affinity between Brandisia and Pterygiella have been noted by Xia et al. (2019). Currently, the position of Brandisia in Orobanchaceae is still unresolved and requires further study. This could be essential to understanding other important matters in this highly diverse family, such as the evolution of parasitism habits.
The three clades of Brandisia in both plastome and nuclear phylogenies (Figs. 3 and 4) correspond to the three subgenera proposed by Li (1947) based on morphological traits. Clade Ⅰ (B. rosea only) corresponds to subg. Rhodobrandisia, which has two-lobed calyces. Clade Ⅱ (B. cauliflora and B. racemosa) corresponds to subg. Coccineabotrys, which has inflorescences with red corollas. Clade Ⅲ (B. hancei, B. glabrescens, B. kwangsiensis, B. swinglei and B. discolor (including B. chevalieri and B. scandens)) corresponds to subg. Eubrandisia (should be revised as subg. Brandisia; autonym; Turland et al., 2018), which has solitary or paired axillary flowers and five-lobed calyces. B. cauliflora was described later and thus not included in Li's study (1947). Owing to its red inflorescence, B. cauliflora should be a member of subg. Coccineabotrys. B. laetevirens was accepted as a distinct species and was placed into B. subg. Brandisia by Li (1947). However, it was treated as a synonym of B. hancei in Flora Reipublicae Popularis Sinicae (FRPS; Tsoong and Lu, 1979) and Flora of China (FOC; Hong et al., 1998) later. After examining the type specimens (A00056851, A; MO-503773, MO; K000961210, K; and US00122408, US) and the protologue of B. laetevirens, we found this "species" showed high similarity to the potential hybrid individuals we found, as it showed intermediate features between B. hancei and B. discolor. B. laetevirens differs from B. hancei by larger leaf with longer petiole and less cordate base, and a narrower calyx; and it differs from B. discolor by its shorter petiole and larger calyces. Therefore, B. laetevirens is presumably a hybrid between B. discolor and B. hancei. The potential hybrid individuals we found (between B. discolor and B. hancei) clustered with B. discolor or B. hancei in the plastid phylogeny (Fig. 4), whereas in the nuclear phylogeny they only clustered with B. hancei, which possibly indicates different directions of hybridization further, if the plastome is maternally inherited. In the contact community of B. hancei and B. discolor we found, their flowering phenologies overlap highly (in Jan.–Feb.) and both provide copious nectar to avian pollinators. In addition, birds seemed not to differentiate between B. hancei, B. discolor and their hybrids in the field (for their similar reward; Chen et al., unpublished data). These factors make hybridization and backcross possible.
Phylogenetically, the positions and compositions of Clade Ⅰ and Clade Ⅱ within Brandisia were robust, whereas nuclear-plastid incongruences occurred in Clade Ⅲ (Figs. 3 and 4). These results indicate different evolutionary trajectories of plastome and nuclear loci, which may be attributed to incomplete lineage sorting and hybridization (Rieseberg and Soltis, 1991; Jakob and Blattner, 2006; Pelser et al., 2010). The nuclear phylogeny possibly revealed the true species tree of Brandisia for two reasons. First, the plastome phylogeny separated B. discolor into two clades that were not sisters to each other (Fig. 3). This molecular result conflicts with morphological evidence. In contrast, in the nuclear phylogeny, all the samples of B. discolor formed a monophyletic clade (Fig. 4). Second, plastomes are primarily maternally inherited, which illuminates only half of the parentage. Therefore, plastome phylogenies may not reveal hybrid history correctly (Small et al., 2004). Consequently, the phylogeny based on nuclear loci (biparentally inherited) in our study may be better at illuminating species relationships, if hybridization occurred in Brandisia.
Brandisia chevalieri and B. scandens were nested within B. discolor clade, and B. chevalieri was not even monophyletic (Figs. 3 and 4). Morphologically, B. discolor, B. chevalieri and B. scandens showed great similarity and can hardly be distinguished (Fig. 1). They all have ovate-lanceolate to narrowly lanceolate blades with irregularly undulate margins, which are glabrescent adaxially but densely gray to tawny tomentose abaxially and turn brown-black when dry (Fig. 1K–L). They all flower in winter–spring with brown-red corollas and small calyces. Therefore, both molecular and morphological evidence support the treatment of B. chevalieri, B. scandens and B. discolor as a single species. As B. discolor was validly published earlier than B. chevalieri and B. scandens, the latter two should be synonyms of B. discolor.
4.3. Origin and coevolution of Brandisia with karst evergreen broadleaved forestBrandisia is a characteristic component of karst evergreen broadleaved forests (EBLFs) in E Asia for its distribution, and hemiparasitic and liana characteristics. Most species in this genus inhabit East Asian EBLF communities (ca. 23–39°N), especially in the karst regions in southern China and northern Vietnam nearby (Fig. 2). They usually grow under forests, along forest edges, mountain slopes, trails and riverbanks. In addition, the hemiparasitic habit of Brandisia means certain host plants in their communities are essential for their lifecycles (Nickrent, 2020), indicating that Brandisia species are highly dependent on the EBLFs.
We estimate that Brandisia originated in the early Oligocene (32.69 Mya) in SW China (from the Eastern Himalayas through Yunnan Plateau to the core karst areas in SW China, ABC; Figs. 5 and 6). The date of Brandisia origin is consistent with that of the establishment of EBLFs in these regions (Xiang et al., 2016; Tian et al., 2021). However, the diversification of Brandisia started only after the early Miocene (19.45 Mya). Specifically, it was after the Oligocene–Miocene (O–M) boundary (ca. 23 Mya), when modern East Asian flora arose (Axelrod et al., 1996; Chen et al., 2018). In concert with the development of the Asian monsoon and the accompanying copious precipitation (especially the winter precipitation; Li et al., 2021b) in this region (Sun and Wang, 2005; Lu and Guo, 2014; But see e.g. Spicer, 2017), many essential elements (Fagaceae, Lauraceae, Theaceae and Magnoliaceae) and characteristic components (such as Mahonia and Oreocharis) of East Asian evergreen broadleaved forests originated or colonized around the O–M boundary and diversified from this time onwards (Yu et al., 2017; Chen et al., 2018, 2020; Deng et al., 2018; Hai et al., 2022; Kong et al., 2022; Xiao et al., 2022). The establishment of E Asian EBLFs probably provided suitable habits and hosts for Brandisia. After its origin, Brandisia did not spread widely, but concentrated in the core karst areas in SW China and nearby northern Vietnam, which may have been facilitated by the establishment of the karst flora and EBLFs in the early Miocene (Deng et al., 2018; Li et al., 2022a, 2022b).
The karst area in subtropical East Asia is a biodiversity hotspot that harbours high plant diversity with numerous endemic species (Davis et al., 1995; Zhu, 2007). Previous studies have suggested that Quercus sect. Ilex (Fagaceae) and the Old World gesneriads (Gesneriaceae) colonized the karst regions in southwestern China in the early Miocene (ca. 20 and 22 Mya, respectively; Deng et al., 2018; Li et al., 2022b). A study on caves of karst areas in southern China also illustrates that species colonization occurred mainly after the O-M boundary, which was in concert with the establishment of EBLFs (Li et al., 2022a). Precipitation from the East Asia monsoon likely accelerated the dissolution of the limestone substrate and deeply influenced the development of the karst region (Zhang, 1980; Liu, 1997; Li et al., 2007), which possibly created novel habitats for species diversification (Kong et al., 2017; Li et al., 2022a). In this process, newly disturbed and open habitats may have appeared, which may have contributed to the expansion and divergence of Brandisia, since Brandisia likes relatively open habitats with more solar energy to afford the development of their large bodies. In addition, southwestern China and northern Vietnam possessed a long-term stable climate, which may have preserved species during the ice ages (Tang et al., 2018; Huang et al., 2022). This may partly explain why the karst area of southern China and northern Vietnam nearby are distribution centers of Brandisia. There are three Brandisia species (B. swinglei, B. hancei and B. discolor) whose distributions are no longer restricted to limestone habitats (Fig. 2). B. swinglei spread eastwards into the EBLFs in southern China. B. hancei and B. discolor have extended northwards and southwards into Central China and tropical Indochina, respectively. Their distribution patterns may indicate re-adaptation and occupation of novel habitats.
Considering the relatively early origin of Brandisia, its species diversity is rather low (ca. eight species), and its distribution is relatively limited. This low diversity and narrow distribution may have several explanations. First, the long initialization period of Brandisia may be an overestimation caused by the possible extinction of its sister group. Brandisia is morphologically unique (Li, 1947) and uncertain in phylogenetic placement (such as McNeal et al., 2013; Li et al., 2021a; and results in our study), and thus seems rather isolated (Nickrent, 2020). This may be the result of the extinction of its close relatives, which consequently adds to the evolutionary time of their common ancestors to the origin of Brandisia. Second, the early origin and lack of very close taxa indicate that Brandisia is a relict genus. If so, the current distribution of Brandisia may be characteristic of a relic distribution. This scenario suggests that the possible original widespread distribution of Brandisia during the Oligocene–Miocene retreated to the current distribution surrounding Sino-Himalayan regions. This may have coincided with the contraction of the distribution of EBLFs caused by historical climate change (Wolfe, 1975; Kubitzki and Krutzsch, 1996; Milne and Abbott, 2002; Wang and Shu, 2013). Third, the ecosystems of the karst EBLFs in southwestern China and northern Vietnam have been relatively stable since the Tertiary (Tang et al., 2018; Huang et al., 2022). Being restricted to stable and favourable environments may lead to low levels of speciation and dispersal because of limited resources and niches (Malohlava and Bocak, 2010; Schluter, 2016; Igea and Tanentzap, 2020; Sun et al., 2020). This may explain why the karst regions possess fewer taxa with rapid radiations and diversity centers than do the Himalaya–Hengduan Mountains regions (Chen et al., 2018), and may be responsible for the observed low species diversity in Brandisia. Fourth, plants with woody habit are associated with lower diversification rates, possibly caused by greater longevity and longer generation time (Eriksson and Bremer, 1992; Dodd et al., 1999). This may partly explain why Brandisia has lower diversity than that of other taxa in Orobanchaceae. In addition, the unitary growth form (shrub), hemiparasitic habit (may not afford their expansion to much different habitats with no proper host plants) and seeds with tiny wings (cannot support dispersal over long distances) possibly lead to niche conservatism and limited competition, which hinder the diversification and dispersal of Brandisia further (Wiens and Donoghue, 2004). Fifth, pollination is important for plant propagation, and pollinators can not only affect plant establishment and persistence (Sargent and Ackerly, 2008), but also flower evolution and diversification (Dodd et al., 1999; Kay and Sargent, 2009; Van der Niet et al., 2014). Brandisia is an ornithophilous genus (Chen et al., unpublished data) whose distribution is consistent with its main avian pollinators (Nectariniidae, Dicaeidae, and Zosteropidae), which mainly occur at low latitudes where high productivity and stability of flower nectar resources can afford bird pollination (Proctor et al., 1996; Sekercioglu, 2006). In addition, avian pollination is less developed in mainland East Asia (compared with the Americas, where hummingbirds and related flowers flourish; Fleming and Muchhala, 2008; Funamoto, 2019). Therefore, limited avian pollination niches in this region may not support a great diversification of Brandisia.
5. ConclusionsIn this study, we used both plastome and nuclear sequences to construct the most comprehensive phylogenies of Brandisia to date. These results have updated our understanding of intrageneric relationships within Brandisia. We found that Brandisia, a typical hemiparasitic component of evergreen broadleaved forests (EBLFs), originated in the early Oligocene in the Eastern Himalayas–SW China and diversified in the early Miocene (19.45 Mya) mainly in E Asia karst EBLFs. Its evolution and distribution were shaped by the establishment of E Asia EBLFs, especially the historical development of karst forest ecosystems in southwestern China. In addition, some characteristics, such as hemiparasitism and pollination, are important factors in impacting plant evolution and dispersal, which deserve more attention in biogeography in the further.
Several fundamental issues regarding Brandisia have not been fully resolved and require further study. First, the phylogenetic position of Brandisia in Orobanchaceae was not ultimately determined. We recommend that future studies combine phylogenetic and morphological methods to resolve phylogenetic uncertainties within Brandisia. Second, B. annamitica, which is endemic to Vietnam, was not examined in this study. This species is morphologically unique, having two staminodes. Further study is needed to clarify its phylogenetic and taxonomic position in Brandisia. Third, the extent of parasitism in Brandisia is currently unclear. Specific host plants have yet to be identified and our understanding of the lifecycle of Brandisia is rather limited. Considering that Brandisia is a unique (both morphologically and phylogenetically) genus in Orobanchaceae, further studies on this genus should provide us with a better understanding of the evolution of parasitism in Orobanchaceae and the relationship between parasitic and host plants.
AcknowledgementsWe thank the Herbarium of Kunming Institute of Botany (KUN) for providing specimen samples and the Molecular Biology Experiment Center, Germplasm Bank of Wild Species in Southwest China (KIB) for sequencing these samples. We thank the Vietnam National Museum of Nature, Vietnam Academy of Science and Technology and the staff at National Parks and Natural Reserves for helping us in our fieldwork in Vietnam. This work was funded by the Key Projects of the Joint Fund of the National Natural Science Foundation of China (U1802232), the national youth talent support program, CAS "Light of West China" Program, Yunnan youth talent support program (YNWR-QNBJ-2018-183 to Y.N.), and Vietnam Academy of Science and Technology (UQĐTCB.06/22-23).
Author contributions
H.S. and Y.N. conceived the study; Y.N., Z.C., T.V.D, Z.-M.G. and Z.Z. performed the fieldwork; Z.C. and Z.-M.G. analyzed the data; Z.C. drafted the manuscript; H.S., Y.N. and Z.Z. revised the manuscript; all authors reviewed the manuscript and gave final approval for publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.03.005.
Axelrod, D.I., Al-Shehbaz, I., Raven, P.H., 1996. History of the modern flora of China. In: Zhang, A.-L., Wu, S.-G. (Eds.), Floristic Characteristics and Diversity of East Asian Plants, Proceedings of the First International Symposium on Floristic Characteristics and Diversity of East Asian Plants (IFCD). Higher Education Press, Beijing, China, pp. 43–55.
|
Bennett, J.R., Mathews, S., 2006. Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot., 93: 1039-1051. DOI:10.3732/ajb.93.7.1039 |
Bonati, G., 1924. Scrofulariacées nouvelles de l'Indo-Chine. Bull. Soc. Bot. France, 71: 1091-1100. DOI:10.1080/00378941.1924.10837015 |
Bouckaert, R., Heled, J., Kühnert, D., et al., 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol., 10: e1003537. DOI:10.1371/journal.pcbi.1003537 |
Chen, X.-H., Xiang, K.-L., Lian, L., et al., 2020. Biogeographic diversification of Mahonia (Berberidaceae): implications for the origin and evolution of East Asian subtropical evergreen broadleaved forests. Mol. Phylogenet. Evol., 151: 106910. DOI:10.1016/j.ympev.2020.106910 |
Chen, Y.-S., Deng, T., Zhou, Z., et al., 2018. Is the East Asian flora ancient or not?. Natl. Sci. Rev., 5: 920-932. DOI:10.1093/nsr/nwx156 |
Cusimano, N., Wicke, S., 2016. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol., 210: 680-693. DOI:10.1111/nph.13784 |
Darriba, D., Posada, D., Kozlov, A.M., et al., 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol., 37: 291-294. DOI:10.1093/molbev/msz189 |
Davis, S.D., Heywood, V.H., Hamilton, A.C., 1995. Centres of Plant Diversity. Vol. 2: Asia, Australasia and the Pacific. WWF/IUCN, Gland, Switzerland.
|
Deng, M., Jiang, X.-L., Hipp, A.L., et al., 2018. Phylogeny and biogeography of East Asian evergreen oaks (Quercus section Cyclobalanopsis; Fagaceae): insights into the Cenozoic history of evergreen broad-leaved forests in subtropical Asia. Mol. Phylogenet. Evol., 119: 170-181. DOI:10.1016/j.ympev.2017.11.003 |
Dodd, M.E., Silvertown, J., Chase, M.W., 1999. Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. Evolution, 53: 732-744. DOI:10.2307/2640713 |
Eriksson, O., Bremer, B., 1992. Pollination systems, dispersal modes, life forms, and diversification rates in angiosperm families. Evolution, 46: 258-266. DOI:10.2307/2409820 |
Fleming, T.H., Muchhala, N., 2008. Nectar-feeding bird and bat niches in two worlds: pantropical comparisons of vertebrate pollination systems. J. Biogeogr., 35: 764-780. DOI:10.1111/j.1365-2699.2007.01833.x |
Ford, D.C., Williams, P.W., 2007. Karst Hydrogeology and Geomorphology. Wiley, London, UK.
|
Frailey, D.C., Chaluvadi, S.R., Vaughn, J.N., et al., 2018. Gene loss and genome rearrangement in the plastids of five hemiparasites in the family Orobanchaceae. BMC Plant Biol., 18: 30. DOI:10.1186/s12870-018-1249-x |
Funamoto, D., 2019. Plant-pollinator interactions in East Asia: a review. J. Pollinat. Ecol., 25: 46-68. |
Hai, L., Li, X.-Q., Zhang, J.-B., et al., 2022. Assembly dynamics of East Asian subtropical evergreen broadleaved forests: new insights from the dominant Fagaceae trees. J. Integr. Plant Biol., 64: 2126-2134. DOI:10.1111/jipb.13361 |
Hollingsworth, E., 2009. Karst Regions of the World (KROW)–Populating Global Karst Datasets and Generating Maps to Advance the Understanding of Karst Occurrence and Protection of Karst Species and Habitats Worldwide. University of Arkansas, Fayetteville, US.
|
Hong, D.-Y., Yang, H.-B., Jin, C.-L., et al., 1998. Scrophulariaceae. In: Wu, Z.-Y., Raven, P.H. (Eds.), Flora of China. Science Press, Beijing.
|
Hooker, J.D., Thomson, T., 1864. Description of a new genus of Scrophularineæ from martaban. Bot. J. Linn. Soc., 8: 11-12. DOI:10.1111/j.1095-8312.1864.tb01070.x |
Huang, J., Spicer, R.A., Li, S.-F., et al., 2022. Long-term floristic and climatic stability of northern Indochina: evidence from the oligocene ha long flora. Vietnam. Palaeogeogr. Palaeoclimatol. Palaeoecol., 593: 110930. DOI:10.1016/j.palaeo.2022.110930 |
Huang, J., Su, T., Jia, L.-B., et al., 2018. A fossil fig from the Miocene of southwestern China: indication of persistent deep time karst vegetation. Rev. Palaeobot. Palynol., 258: 133-145. DOI:10.1016/j.revpalbo.2018.07.005 |
Igea, J., Tanentzap, A.J., 2020. Angiosperm speciation cools down in the tropics. Ecol. Lett., 23: 692-700. DOI:10.1111/ele.13476 |
Jakob, S.S., Blattner, F.R., 2006. A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol., 23: 1602-1612. DOI:10.1093/molbev/msl018 |
Jacques, F.M.B., Shi, G., Wang, W., 2011. Reconstruction of neogene zonal vegetation in South China using the integrated plant record (IPR) analysis. Palaeogeogr. Palaeoclimatol. Palaeoecol., 307: 272-284. DOI:10.1016/j.palaeo.2011.05.025 |
Jin, J.-J., Yu, W.-B., Yang, J.-B., et al., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 241. DOI:10.1186/s13059-020-02154-5 |
Kay, K.M., Sargent, R.D., 2009. The role of animal pollination in plant speciation: integrating ecology, geography, and genetics. Annu. Rev. Ecol. Evol. Syst., 40: 637-656. DOI:10.1146/annurev.ecolsys.110308.120310 |
Kong, H.-H., Condamine, F.L., Harris, A., et al., 2017. Both temperature fluctuations and East Asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol. Ecol., 26: 6414-6429. DOI:10.1111/mec.14367 |
Kong, H.-H., Condamine, F.L., Yang, L.-H., et al., 2022. Phylogenomic and macroevolutionary evidence for an explosive radiation of a plant genus in the Miocene. Syst. Biol., 71: 589-609. DOI:10.1093/sysbio/syab068 |
Kozlov, A.M., Darriba, D., Flouri, T., et al., 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics, 35: 4453-4455. DOI:10.1093/bioinformatics/btz305 |
Krause, K., 2008. From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants. Curr. Genet., 54: 111. DOI:10.1007/s00294-008-0208-8 |
Kubitzki, K., Krutzsch, W., 1996. Origins of East and south asian plant diversity. In: Zhang, A.-L., Wu, S.-G. (Eds.), Floristic Characteristics and Diversity of East Asian Plants, Proceedings of the First International Symposium on Floristic Characteristics and Diversity of East Asian Plants (IFCD). Higher Education Press, Beijing, China, pp. 56–70.
|
Li, H.-L., 1947. Relationship and taxonomy of the genus Brandisia. J. Arnold Arbor., 28: 127-136. DOI:10.5962/p.185569 |
Li, H.-T., Luo, Y., Gan, L., et al., 2021a. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biology, 19: 232. DOI:10.1186/s12915-021-01166-2 |
Li, J.-W., Vasconcelos, P., Duzgoren-Aydin, N., et al., 2007. Neogene weathering and supergene manganese enrichment in subtropical South China: an 40Ar/39Ar approach and paleoclimatic significance. Earth Planet. Sci., 256: 389-402. DOI:10.1016/j.epsl.2007.01.021 |
Li, S.-F., Valdes, P., Farnsworth, A., et al., 2021b. Orographic evolution of northern Tibet shaped vegetation and plant diversity in eastern Asia. Sci. Adv., 7: eabc7741. DOI:10.1126/sciadv.abc7741 |
Li, X.-Q., Xiang, X.-G., Jabbour, F., et al., 2022a. Biotic colonization of subtropical East Asian caves through time. Proc. Natl. Acad. Sci. U.S.A., 119: e2207199119. DOI:10.1073/pnas.2207199119 |
Li, X.-Q., Xiang, X.-G., Zhang, Q., et al., 2022b. Immigration dynamics of tropical and subtropical Southeast Asian limestone karst floras. Proc. R. Soc. B: Biol. Sci., 289: 20211308. DOI:10.1098/rspb.2021.1308 |
Liu, J.-R., 1997. The development history of the Guangxi tropical karst geomorphology and its sequences. Carsol. Sin., 16: 332-345. |
Lu, H.-Y., Guo, Z.-T., 2014. Evolution of the monsoon and dry climate in East Asia during late Cenozoic: a review. Sci. China Earth Sci., 57: 70-79. DOI:10.1007/s11430-013-4790-3 |
Malohlava, V., Bocak, L., 2010. Evidence of extreme habitat stability in a Southeast Asian biodiversity hotspot based on the evolutionary analysis of neotenic net-winged beetles. Mol. Ecol., 19: 4800-4811. DOI:10.1111/j.1365-294X.2010.04850.x |
McNeal, J.R., Bennett, J.R., Wolfe, A.D., et al., 2013. Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot., 100: 971-983. DOI:10.3732/ajb.1200448 |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE): 1-8. |
Milne, R., Abbott, R., 2002. The origin and evolution of Tertiary relict flora. Adv. Bot. Res., 38: 281-314. |
Nickrent, D.L., 2020. Parasitic angiosperms: how often and how many?. Taxon, 69: 5-27. DOI:10.1002/tax.12195 |
Olmstead, R.G., Depamphilis, C.W., Wolfe, A.D., et al., 2001. Disintegration of the Scrophulariaceae. Am. J. Bot., 88: 348-361. DOI:10.2307/2657024 |
Oxelman, B., Kornhall, P., Olmstead, R.C., et al., 2005. Further disintegration of Scrophulariaceae. Taxon, 54: 411-425. DOI:10.2307/25065369 |
Pelser, P.B., Kennedy, A.H., Tepe, E.J., et al., 2010. Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies. Am. J. Bot., 97: 856-873. DOI:10.3732/ajb.0900287 |
Pham, H.H., 2000. Brandisia. In: An Illustrated Flora of Vietnam, vol. 3. Young Publishing House, Ho Chi Minh, Vietnam, p. 901.
|
Proctor, M., Yeo, P., Lack, A., 1996. The Natural History of Pollination. Harper Collins, London, UK.
|
Qu, X.-J., Moore, M.J., Li, D.-Z., et al., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. DOI:10.1186/s13007-019-0435-7 |
Rieseberg, L.H., Soltis, D., 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants, 5: 65-84. |
Ronquist, F., Teslenko, M., van der Mark, P., et al., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Sargent, R.D., Ackerly, D.D., 2008. Plant–pollinator interactions and the assembly of plant communities. Trends Ecol. Evol., 23: 123-130. DOI:10.1016/j.tree.2007.11.003 |
Schluter, D., 2016. Speciation, ecological opportunity, and latitude. Am. Nat., 187: 1-18. DOI:10.1086/684193 |
Sekercioglu, C.H., 2006. Increasing awareness of avian ecological function. Trends Ecol. Evol., 21: 464-471. DOI:10.1016/j.tree.2006.05.007 |
Small, R., Cronn, R., Wendel, J., 2004. Use of nuclear genes for phylogeny reconstruction in plants. Aust. Syst. Bot., 17: 145-170. DOI:10.1071/SB03015 |
Song, Y.-C., Da, L.-J., 2016. Evergreen broad-leaved forest of East Asia. In: Box, E.O. (Ed.), Vegetation Structure and Function at Multiple Spatial, Temporal and Conceptual Scales. Springer International Publishing, Cham, pp. 101–128.
|
Spicer, R.A., 2017. Tibet, the Himalaya, Asian monsoons and biodiversity – in what ways are they related?. Plant Divers., 39: 233-244. DOI:10.1016/j.pld.2017.09.001 |
Sun, X.-J., Wang, P.-X., 2005. How old is the Asian monsoon system?—palaeobotanical records from China. Palaeogeogr. Palaeoclimatol. Palaeoecol., 222: 181-222. DOI:10.1016/j.palaeo.2005.03.005 |
Sun, B.-N., Wu, J.-Y., Liu, Y.-S., et al., 2011. Reconstructing Neogene vegetation and climates to infer tectonic uplift in western Yunnan, China. Palaeogeogr. Palaeoclimatol. Palaeoecol., 304: 328-336. DOI:10.1016/j.palaeo.2010.09.023 |
Sun, M., Folk, R.A., Gitzendanner, M.A., et al., 2020. Recent accelerated diversification in rosids occurred outside the tropics. Nat. Commun., 11: 3333. DOI:10.1109/bigdata50022.2020.9378247 |
Sweeting, M.M., 1995. Karst in China: its Geomorphology and Environment. Springer, Berlin, Germany.
|
Takhtajan, A.L., 1969. Flowering Plants: Origin and Dispersal. Oliver & Boyd, Edinburgh, UK.
|
Tang, C.Q., 2015. Evergreen broad-leaved forests. In: Tang, C.Q. (Ed.), The Subtropical Vegetation of Southwestern China: Plant Distribution, Diversity and Ecology. Springer Netherlands, Dordrecht, pp. 49–112.
|
Tang, C.Q., Matsui, T., Ohashi, H., et al., 2018. Identifying long-term stable refugia for relict plant species in East Asia. Nat. Commun., 9: 4488. DOI:10.1038/s41467-018-06837-3 |
Tian, Y.-M., Spicer, R.A., Huang, J., et al., 2021. New early oligocene zircon U-Pb dates for the 'Miocene' Wenshan basin, Yunnan, China: Biodiversity and paleoenvironment. Earth Planet. Sci., 565: 116929. DOI:10.1016/j.epsl.2021.116929 |
Tsoong, P.-C., Lu, L.-D., 1979. Scrophulariaceae. In: Flora Reipublicae Popularis Sinicae. Science Press, Beijing, China, pp. 1–242.
|
Turland, N.J., Wiersema, J.H., Barrie, F.R., et al., 2018. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017, vol. 159. Koeltz Botanical Books, Regnum Vegetabile, Glashütten.
|
Van der Niet, T., Peakall, R., Johnson, S.D., 2014. Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Ann. Bot., 113: 199-211. DOI:10.1093/aob/mct290 |
Wang, K., Zhang, C., Chen, H., et al., 2019. Karst landscapes of China: patterns, ecosystem processes and services. Landsc. Ecol., 34: 2743-2763. DOI:10.1007/s10980-019-00912-w |
Wang, W.-M., Shu, J.-W., 2013. Cenozoic xeromorphic vegetation in China and its spatial and temporal development in connection with global changes. Palaeoworld, 22: 86-92. DOI:10.1016/j.palwor.2013.08.001 |
Wicke, S., Müller, K.F., de Pamphilis, C.W., et al., 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell, 25: 3711. DOI:10.1105/tpc.113.113373 |
Wiens, J.J., Donoghue, M.J., 2004. Historical biogeography, ecology and species richness. Trends Ecol. Evol., 19: 639-644. DOI:10.1016/j.tree.2004.09.011 |
Wolfe, J.A., 1975. Some aspects of plant geography of the northern hemisphere during the Late Cretaceous and Tertiary. Ann. Mo. Bot. Gard., 62: 264-279. DOI:10.2307/2395198 |
Wu, Z.-Y., 1980. Vegetation of China. Science Press, Beijing, China.
|
Wu, Z.-Y., Wu, S.-G., 1996. A proposal for a new floristic kingdom (realm): the E. Asiatic Kingdom, its delineation and characteristics. In: Zhang, A.-L., Wu, S.-G. (Eds.), Floristic Characteristics and Diversity of East Asian Plants, Proceedings of the First International Symposium on Floristic Characteristics and Diversity of East Asian Plants (IFCD). Higher Education Press, Beijing, China, pp. 3–42.
|
Xia, Z., Wen, J., Gao, Z., 2019. Does the enigmatic Wightia belong to Paulowniaceae (Lamiales)?. Front. Plant Sci., 10: 528. DOI:10.3389/fpls.2019.00528 |
Xiang, X.-G., Mi, X.-C., Zhou, H.-L., et al., 2016. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr., 43: 1310-1323. DOI:10.1111/jbi.12726 |
Xiao, T.-W., Yan, H.-F., Ge, X.-J., 2022. Plastid phylogenomics of tribe Perseeae (Lauraceae) yields insights into the evolution of East Asian subtropical evergreen broad-leaved forests. BMC Plant Biol., 22: 32. DOI:10.1186/s12870-021-03413-8 |
Young, N.D., Steiner, K.E., Depamphilis, C.W., 1999. The evolution of parasitism in Scrophulariaceae/Orobanchaceae: plastid gene sequences refute an evolutionary transition series. Ann. Mo. Bot. Gard., 86: 876-893. DOI:10.2307/2666173 |
Yu, W.-B., Randle, C.P., Lu, L., et al., 2018. The hemiparasitic plant Phtheirospermum (Orobanchaceae) is polyphyletic and contains cryptic species in the Hengduan Mountains of Southwest China. Front. Plant Sci., 9: 142. DOI:10.3389/fpls.2018.00142 |
Yu, X.-Q., Gao, L.-M., Soltis, D.E., et al., 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol., 215: 1235-1248. DOI:10.1111/nph.14683 |
Yu, Y., Blair, C., He, X., 2020. RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol., 37: 604-606. DOI:10.1093/molbev/msz257 |
Yu, Y., Harris, A.J., Blair, C., et al., 2015. RASP (reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol., 87: 46-49. DOI:10.1016/j.ympev.2015.03.008 |
Zeng, C.-X., Hollingsworth, P.M., Yang, J., et al., 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods, 14: 43. DOI:10.1186/s13007-018-0300-0 |
Zhang, Z.-G., 1980. Karst types in China. Geojournal, 4: 541-570. DOI:10.1007/BF00214219 |
Zhao, L.-C., Wang, Y.-F., Liu, C.-J., et al., 2004. Climatic implications of fruit and seed assemblage from Miocene of Yunnan, southwestern China. Quat. Int., 117: 81-89. DOI:10.1016/S1040-6182(03)00118-6 |
Zhou, Q.-M., Jensen, S.R., Liu, G.-L., et al., 2014. Familial placement of Wightia (Lamiales). Plant Syst. Evol., 300: 2009-2017. DOI:10.1007/s00606-014-1029-5 |
Zhu, H., 2007. The karst ecosystem of southern China and its biodiversity. Trop. For., 35: 44-47. |



