b. School of Life Sciences, Guizhou Normal University, Guiyang 550001, Guizhou, China;
c. State Key Laboratory of Agricultural Genomics, BGI-Shenzhen, Shenzhen 518083, Guangdong, China
RNA editing is a post-transcriptional process that alters the genetic information of mRNAs, so that the translated proteins deviate from those predicted by the genomic DNA templates. RNA editing occurs in a wide range of eukaryotes, including protists, multicellular animals, and land plants (Gray, 2012; Ichinose and Sugita, 2016). RNA editing in animals, dominated by A-to-I synonymous editing with low editing efficiency (20–30%), might play important roles in the regulation of gene expression (Wang et al., 2013). In contrast, RNA editing in plant organelles is usually non-synonymous, affecting mostly the 2nd and 1st codon positions with very high editing efficiency (~80%). In plants, RNA editing is thought to affect plant phenotype and growth by restoring evolutionarily conserved amino acids and ensuring the correct folding of trans-membrane proteins on the respiratory chain complex (Sloan, 2017).
RNA editing occurs in the organellar genomes of all land plant lineages except marchantioid liverworts (Rudinger et al., 2008). Generally, mitochondrial genes have more RNA editing sites than their plastid counterparts. For example, in angiosperms, mitochondrial genomes contain between 300 and 500 RNA editing sites (Edera et al., 2018), whereas plastid genomes contain only between 30 and 50 RNA editing sites (Oldenkott et al., 2014). In addition, the number of RNA editing sites varies in different plant lineages. Lycophytes, which contain the largest numbers of RNA editing sites, possess 2139 RNA editing sites in 18 mitochondrial genes in Selaginella moellendorffii (Hecht et al., 2011), and 400–3100 in plastid genes in Selaginella (Du et al., 2020). Gymnosperms contain 11–418 (average: 245) RNA editing sites in mitochondrial genes, and 3–255 (average: 78) in plastid genes (Liu et al., 2022). Finally, early diverging lineages tend to have more RNA sites, e.g., note the high numbers of RNA editing sites in liverwort Haplomitrium (Rüdinger et al., 2012), lycophyte Selaginella (Hecht et al., 2011), ferns Ophioglossum and Psilotum, gymnosperm Ginkgo (Guo et al., 2016), angiosperms Amborella (Rice et al., 2013) and Liriodendron (Richardson et al., 2013).
RNA editing site abundance is correlated with the diversity of PPR PLS protein genes encoded by the nuclear genome (Rüdinger et al., 2012; Schallenberg-Rüdinger et al., 2014), the heterogeneity of GC content (Guo et al., 2017; Hecht et al., 2011; Malek et al., 1996; Smith, 2009), and substitution rates of genes (Cuenca et al., 2010). It has been well established that among-species heterogeneity of GC content and substitution rate result in systematic errors and affect phylogenetic analyses (Liu et al., 2014). Phylogenetic analyses of plant taxa are likely to be greatly affected as hundreds of RNA editing sites in plant organelles undergo biased conversions between C and T (Edera et al., 2018), introducing GC heterogeneity (Dong et al., 2022). Moreover, because there are 5–10 times more RNA editing sites in mitochondrial than in plastid genomes, the impact of RNA editing sites on phylogenetic analyses based on mitochondrial sequence data should be evaluated with more caution. However, studies on this issue have failed to yield a consensus (as reviewed in Dong et al., 2022). The prevailing opinions regarding the impact of RNA editing on phylogenetic reconstructions are that (1) RNA editing sites do not affect phylogenetic reconstructions (Boweand dePamphilis, 1996) and are reliable sources of phylogenetic information (Schmidt et al., 2002); (2) RNA editing sites in DNA sequences provide additional phylogenetic information, and phylogenomic inferences should be based on DNA sequences rather than on cDNA sequences (Petersen et al., 2006; Picardi and Quagliariello, 2008); and (3) RNA editing sites contain homoplasious signals (Groth-Malonek et al., 2005) and should be modified based on cDNA sequences (Du et al., 2020; Guo et al., 2016; Richardson et al., 2013) or be completely excluded in phylogenetic analyses (Bell et al., 2020).
Gymnosperms are mostly tall and woody trees, comprising about 1000 species in 12 families and 83 genera (Wang and Ran, 2014). Extant gymnosperms are distributed in all continents except Antarctica, and two-thirds of gymnosperms are conifers, constituting over 39% of the world's forests (Armenise et al., 2012). Gymnosperms represent the largest land-based biome, and accordingly are of high ecological and economic significance. Conservation and utilization of gymnosperm germplasm resources requires a well-resolved gymnosperm phylogeny. However, gymnosperm phylogenetic reconstruction has long been ambiguous, especially regarding the unstable placement of gnetophytes (Wang and Ran, 2014; Yang et al., 2022). The gnetophyte lineage has been proposed to be the sister to all seed plants (Soltis et al., 2002), sister to all other gymnosperms (Burleighand Mathews, 2004), sister to cupressophytes (Ran et al., 2010), and sister to Pinaceae (Zhong et al., 2010; Wu et al., 2011; Ran et al., 2018). Recent developments in sequencing techniques and genomic resources have greatly advanced our understanding of gymnosperm phylogeny (Gitzendanner et al., 2018; Ran et al., 2018; Leebens-Mack et al., 2019; Liu et al., 2022) and promise to resolve phylogenetic uncertainties. Indeed, phylogenomic studies have led to two hypotheses regarding the phylogenetic position of gnetophytes: nuclear phylogenomic analyses have indicated that gnetophytes are sister to Pinaceae (Ran et al., 2018), whereas organellar phylogenomic analyses have indicated that gnetophytes are sister to cupressophytes (Zhong et al., 2010). This cyto-nuclear incongruence remains unresolved (Liu et al., 2022).
In our recent study of the cycad genome (Liu et al., 2022), we generated empirical RNA editing data sets (by comparing the DNA and RNA sequences) for gymnosperm representatives, and analyzed both the original DNA data set and a data set with all RNA editing sites modified (based on the RNA sequences). The mitochondrial RNA editing-modified tree recovered the same topology as the original tree, only with slightly lower support for the sister relationship of gnetophytes and cupressophytes, i.e., BPP (Bootstrap Percentage Supports) and GCF (Gene Concordance Factor) decreased from 100% to 41% in original tree to 97% and 33% in the RNA editing-modified tree (Liu et al., 2022). The two data sets yielded the same topology, which might suggest that the impact of RNA editing was not completely mitigated. Here we revisit this empirical editing site data and further examine the impact of RNA editing on phylogenetic analysis. Specifically, we aim to re-evaluate the impact of mitochondrial RNA editing on the phylogenetic reconstruction of gymnosperms and construct a more reliable and robust mitochondrial genomic tree for gymnosperms. For this purpose, we completely excluded all RNA editing sites in our phylogenetic analysis of gymnosperms. Mitochondrial RNA editing data (Table S1), including all the annotated mitochondrial genomes for 46 gymnosperms, were retrieved from the data repository of the cycad genome study (Liu et al., 2022). The mitochondrial genome sequence of Liriodendron tulipifera was downloaded from NCBI organellar database.
Phylogenomic analyses of 46 gymnosperms were based on five mitochondrial genome data sets. The first data set consisted of original protein-coding genes these data included RNA editing sites. The second data set excluded RNA editing sites. The third data set consisted of sequences with degenerated codons (Regier et al., 2010). The fourth data set, called NT12, excluded 3rd codon positions. The final data set consisted of only RNA editing sites. For each of these data sets, we included 40 conserved protein-coding genes: atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFC, ccmFN, cob, cox1, cox2, cox3, matR, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpL2, rpL5, rpL16, rps1, rps2, rps3, rps4, rps7, rps10, rps11, rps12, rps13, rps14, rps19, sdh3, sdh4, and tatC.
Each protein-coding gene was extracted and aligned with MAFFT v5.0, with the '-localpair –maxiterate 1000' parameter, implemented in Geneious v.10.0.2 (http://www.geneious.com). The individual gene alignments and the corresponding RNA editing site annotation table were exported. Then an in-house python script was used to remove and retrieve all columns containing at least one RNA editing site annotation from the original data set. FASconCAT-G (Kück and Longo, 2014) was used to produce the codon degenerated and the NT12 data set, as well as to concatenate all the individual matrices into the concatenated super matrices for each data set.
Each concatenated data matrix was analyzed separately using PartitionFinder (Lanfear et al., 2012) for the optimal partitioning scheme and corresponding best-fit models, and subjected to maximum likelihood (ML) phylogenetic reconstruction using RAxML v.8.2.12 (Stamatakis, 2014) with 100 bootstrap replicates. The phylogenetic trees were visualized and modified with FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
In total, 11, 486 RNA editing sites were identified in the 40 conserved mitochondrial protein-coding genes of the 46 gymnosperms (Table S1). The number of mitochondrial RNA editing sites of gymnosperms is abundant, ranging from 11 to 418 sites, and averaged 245 sites per species. The abundance of RNA editing sites appears to be related to the function of genes, ranging from 7 sites in sdh4 to 147 sites in nad4, with an average of 63 sites per gene. Genes encoding the mitochondrial respiratory chain complex have the highest number of RNA editing sites, whereas genes encoding ribosomal proteins have the fewest RNA editing sites (Fig. 1), indicating that RNA editing may be important for plant respiration and energy metabolism.
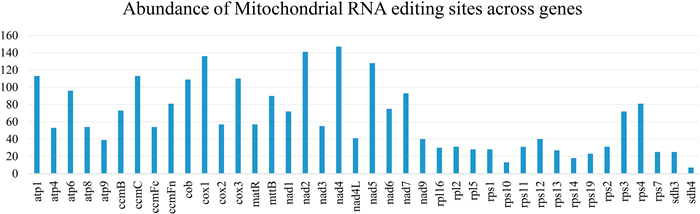
|
| Fig. 1 The abundance of mitochondrial editing sites across 40 protein-coding genes used for phylogenetic reconstruction. |
To create a data set that excluded RNA editing sites, we removed 2537 sites (or 6.49% of the total length of the protein-coding sequence alignment) that contained at least one RNA editing site from the original protein-coding gene matrix of the 40 conserved protein-coding gymnosperm genes (Fig. 1). This alignment without RNA editing sites consisted of 36, 531 sites. The alignment of degenerated codons consisted of 39, 068 sites; the alignment that excluded third codon positions consisted of 26, 059 sites; and the alignment of RNA editing sites contained 2537 sites. GC content was highest in the data set that consisted only of RNA editing site sequences (41.29%), followed by the data set of original protein-coding gene sequences (28.97%). The ratio of parsimony-informative sites was highest in the RNA editing site sequences (77.57%), followed again by the original protein-coding gene sequences (23.78%). The data set with degenerated codons contained the lowest ratio of parsimony informative sites with only 13.34%.
The ML trees inferred from three data sets (i.e., the original sequence data set, NT12 data set, and codon degenerated data set) collectively supported the sister relationship of gnetophytes and cupressophytes with maximum supports (Fig. 2a, c, and d). This relationship was also strongly supported by the ML tree recovered from a data set that consisted of only RNA editing site sequences (Fig. S1). However, after RNA editing sites were excluded from the matrix, the mitochondrial ML tree (Fig. 2b) strongly supported the sister relationship of gnetophytes and Pinaceae with high bootstrap percentages (BPP = 90%). This result is consistent with recent nuclear phylogenomic results (Ran et al., 2018; Liu et al., 2022). Thus, the removal of RNA editing sites provide evidence from the mitochondrial genome for the sister relationship of the gnetophytes and Pinaceae. Nevertheless, excluding RNA editing sites did not change the sister relationship of cycads to the rest of gymnosperms, even though the sister relationship of cycads and Ginkgo has been recovered by nuclear and plastid data sets (Liu et al., 2022). This finding suggests that RNA editing may not be the sole cause of cyto-nuclear incongruences in gymnosperm phylogentic analyses.
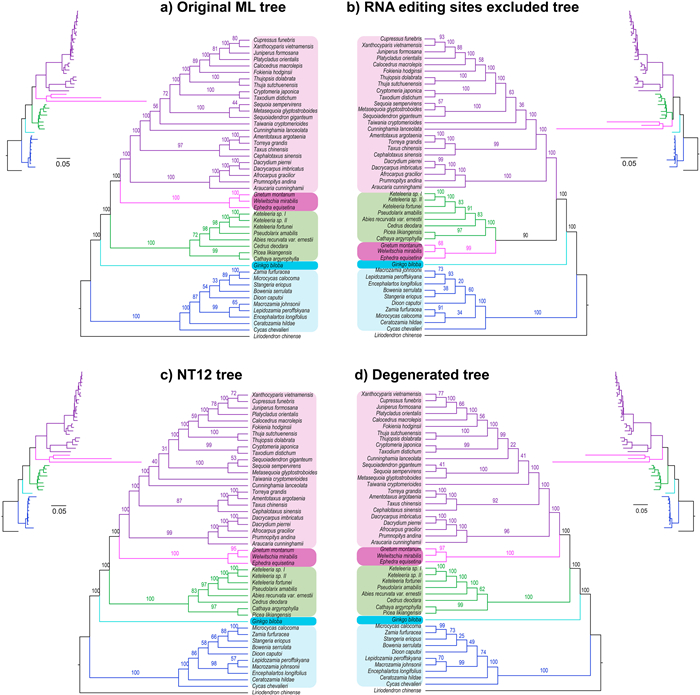
|
| Fig. 2 Maximum likelihood trees inferred from the concatenated mitochondrial sequences of a) the original data set, b) the RNA editing sites excluded data set, c) the 3rd codon positions excluded data set, d) the degenerated codon data set. Bootstrap support values are shown above the branches. The branches leading to five major groups of gymnosperms are highlighted in different colors: purple, magenta, green, blue and cyan for cupressophytes, gnetophytes, Pinaceae, cycads, and Ginkgo, respectively. |
Cyto-nuclear incongruences on the backbone phylogeny of gymnosperms mainly pertain to the placement of gnetophytes and cycads (Liu et al., 2022). Several approaches have been proposed to reconcile the incongruences related to the position of gnetophytes, including the curation of data biases (Burleighand Mathews, 2004; Wu et al., 2011; Zhong et al., 2010), the tree inference method (Huelsenbeck, 1995) and substitution models (Cox et al., 2014). However, the incongruence of the position of cycads might reflect ancient hybridization events. In extant cycads, Ginkgo, and gnetophytes both plastids and mitochondria are maternally inherited, whereas conifers have unstable maternal and paternal inheritance systems for mitochondria despite a consistent paternal inheritance system for plastids (Owens and Wilson, 1999). The complex cytoplasmic inheritance system in gymnosperms provides the possibility of cycads' ancestor hosting maternal plastid and paternal mitochondria, which may explain the mitochondrion-plastid conflict that obscures the phylogenetic position of cycads.
Nuclear and organellar RNA editing systems have opposite effects on protein diversity (Sloan, 2017). In animals and fungi, nuclear RNA editing increases protein diversity and creates novel protein types (Sloan, 2017). In plants, organellar RNA editing, which is functionally important, restores conserved amino acid sequences (Knoop, 2011; Bentolila et al., 2012; Sloan, 2017; Edera et al., 2018; Dong et al., 2019). Substitution of these edited sites, which is biased toward frequent changes from C and T (Edera et al., 2018), strongly deviate from neutral evolution and likely mislead phylogenetic inferences (Dong et al., 2022). An additional way in which RNA editing sites may affect phylogenetic reconstructions is that the abundance of these sites is positively correlated with GC content (Dong et al., 2019) and negatively correlated to substitution rates (Cuenca et al., 2010). Thus, RNA editing sites should be carefully considered in phylogenetic reconstructions.
In our study, GC content and the percentage of parsimony informative sites were both high in a data set that consisted of only RNA editing site sequences. The phylogenetic tree inferred from this data set recovered significantly long branches and aberrant relationships, indicating that RNA editing sites skew phylogenetic inferences. Although some studies have proposed revision of RNA editing sites rather than exclusion prior to phylogenetic reconstruction, this treatment might be too mild to thoroughly mitigate the impact of RNA editing, especially in plant groups containing large number of editing sites like gymnosperms and lycophytes (Richardson et al., 2013; Du et al., 2020; Dong et al., 2022; Liu et al., 2022). For example, in our study, revision of RNA editing sites generated a phylogenetic tree with the same topology as that based on RNA editing sites only. In fact, the tree topologies of phylogenies generated from several data sets (i.e., original sequences, NT12 data set, and sequences with degenerated codons), including revised RNA editing site sequences (Liu et al., 2022), did not differ. However, excluding RNA editing sites from the mitochondrial protein-coding gene data set restored the sister relationship of gnetophytes and Pinaceae.
Our findings are a reminder of the impact of RNA editing on plant phylogenetic reconstructions. Specifically, our work highlights how organellar genome sequences used as molecular markers in phylogenetic analyses must be carefully evaluated, taking into consideration the effect of RNA editing sites, especially in lineages with many RNA editing sites, like gymnosperms, lycophytes, and hornworts. We recommend that efforts to fully mitigate the effects of RNA editing in phylogenetic analysis exclude, instead of revise, RNA editing sites from sequence data.
AcknowledgementsThis work is supported by the Shenzhen Urban Management Bureau Fund (No. 202106 and 202302) and Fairy Lake Science Fund (No. FLSF-2021-02).
Data availability
The mitochondrial genes and RNA editing site annotation for 46 gymnosperms were downloaded from the figshare repository, the matrices and trees newly generated for current study were deposited in figshare repository https://doi.org/10.6084/m9.figshare.20485698. Other supporting results are included within the article.
Author contributions
S.D. and Y.L. designed the study. S.D. and X.Z. carried out bioinformatics and phylogenetic analyses. S.D. drafted the manuscript. Y.L. and T.P. modified the final manuscript, and all authors reviewed it.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.02.004.
Armenise, L., Simeone, M., Piredda, R., et al., 2012. Validation of DNA barcoding as an efficient tool for taxon identification and detection of species diversity in Italian conifers. Eur. J. For. Res., 131: 1337-1353. DOI:10.1007/s10342-012-0602-0 |
Bell, D., Lin, Q., Gerelle, W.K., et al., 2020. Organellomic data sets confirm a cryptic consensus on (unrooted) land-plant relationships and provide new insights into bryophyte molecular evolution. Am. J. Bot., 107: 91-115. DOI:10.1002/ajb2.1397 |
Bentolila, S., Heller, W.P., Sun, T., et al., 2012.
Rip1, a member of an arabidopsis protein family, interacts with the protein rare1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A., 109: 8372-8373. |
Bowe, L.M., dePamphilis, C.W.W., 1996. Effects of RNA editing and gene processing on phylogenetic reconstruction. Mol. Biol. Evol., 13: 1159-1166. DOI:10.1093/oxfordjournals.molbev.a025680 |
Burleigh, J.G., Mathews, S., 2004. Phylogenetic signal in nucleotide data from seed plants: Implications for resolving the seed plant tree of life. Am. J. Bot., 91: 1599-1613. DOI:10.3732/ajb.91.10.1599 |
Cox, C.J., Li, B., Foster, P.G., et al., 2014. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst. Biol., 63: 272-279. DOI:10.1093/sysbio/syt109 |
Cuenca, A., Petersen, G., Seberg, O., et al., 2010. Are substitution rates and RNA editing correlated?. BMC Evol. Biol., 10: 1-15. |
Dong, S., Zhao, C., Zhang, S., et al., 2019. The amount of RNA editing sites in liverwort organellar genes is correlated with GC content and nuclear PPR protein diversity. Genome Biol. Evol., 11: 3233-3239. DOI:10.1093/gbe/evz232 |
Dong, S.S., Li, H.L., Goffinet, B., et al., 2022. Exploring the impact of RNA editing on mitochondrial phylogenetic analyses in liverworts, an early land plant lineage. J. Syst. Evol., 60: 16-22. DOI:10.1111/jse.12706 |
Du, X.Y., Lu, J.M., Li, D.Z., 2020. Extreme plastid RNA editing may confound phylogenetic reconstruction: A case study of Selaginella (lycophytes). Plant Divers., 42: 356-361. |
Edera, A.A., Gandini, C.L., Sanchez-Puerta, M.V., 2018. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol., 97: 215-231. DOI:10.1007/s11103-018-0734-9 |
Gitzendanner, M.A., Soltis, P.S., Wong, G.K.S., et al., 2018. Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. Am. J. Bot., 105: 291-301. DOI:10.1002/ajb2.1048 |
Gray, M.W., 2012. Evolutionary origin of RNA editing. Biochemistry, 51: 5235-5242. DOI:10.1021/bi300419r |
Groth-Malonek, M., Pruchner, D., Grewe, F., et al., 2005. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol. Biol. Evol., 22: 117-125. |
Guo, W., Grewe, F., Fan, W., et al., 2016.
Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol., 33: 1448-1460. DOI:10.1093/molbev/msw024 |
Guo, W., Zhu, A., Fan, W., et al., 2017. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol., 213: 391-403. DOI:10.1111/nph.14135 |
Hecht, J., Grewe, F., Knoop, V., 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol., 3: 344-358. DOI:10.1093/gbe/evr027 |
Huelsenbeck, J.P., 1995. Performance of phylogenetic methods in simulation. Syst. Biol., 44: 17-48. |
Ichinose, M., Sugita, M., 2016. RNA editing and its molecular mechanism in plant organelles. Genes, 8: 5. DOI:10.3390/genes8010005 |
Knoop, V., 2011. When you can't trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci., 68: 567-586. DOI:10.1007/s00018-010-0538-9 |
Kück, P., Longo, G.C., 2014. Fasconcat-g: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool., 11: 1-8. |
Lanfear, R., Calcott, B., Ho, S.Y.W., et al., 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol., 29: 1695-1701. DOI:10.1093/molbev/mss020 |
Leebens-Mack, J.H., Barker, M.S., Carpenter, E.J., et al., 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574: 679-685. |
Liu, Y., Cox, C.J., Wang, W., et al., 2014. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst. Biol., 63: 862-878. DOI:10.1093/sysbio/syu049 |
Liu, Y., Wang, S., Li, L., et al., 2022. The Cycas genome and the early evolution of seed plants. Nat. Plants, 8: 389-401. DOI:10.1038/s41477-022-01129-7 |
Malek, O., Lättig, K., Hiesel, R., et al., 1996. RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J., 15: 1403-1411. DOI:10.1002/j.1460-2075.1996.tb00482.x |
Oldenkott, B., Yamaguchi, K., Tsuji-Tsukinoki, S., et al., 2014. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata
. RNA, 20: 1499-1506. DOI:10.1261/rna.045575.114 |
Owens, J.N., Wilson, V.R., 1999. Cytoplasmic inheritance in Podocarpus totara (Podocarpaceae). In: Ⅳ International Conifer Conference, vol. 615, pp. 171–172.
|
Petersen, G., Seberg, O., Davis, J.I., et al., 2006. RNA editing and phylogenetic reconstruction in two monocot mitochondrial genes. Taxon, 55: 871-886. DOI:10.2307/25065682 |
Picardi, E., Quagliariello, C., 2008. Is plant mitochondrial RNA editing a source of phylogenetic incongruence? An answer from in silico and in vivo data sets. BMC Bioinformatics, 9: S14. DOI:10.1186/1471-2105-9-S2-S14 |
Ran, J.-H., Gao, H., Wang, X.-Q., 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer Ⅱ and further evidence for the phylogeny of gymnosperms. Mol. Phylogenet. Evol., 54: 136-149. |
Ran, J.H., Shen, T.T., Wang, M.M., et al., 2018. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. Roy. Soc. B-Biol. Sci., 285: 20181012. DOI:10.1098/rspb.2018.1012 |
Regier, J.C., Shultz, J.W., Zwick, A., et al., 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature, 463: 1079-1083. DOI:10.1038/nature08742 |
Rice, D.W., Alverson, A.J., Richardson, A.O., et al., 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella
. Science, 342: 1468-1473. DOI:10.1126/science.1246275 |
Richardson, A.O., Rice, D.W., Young, G.J., et al., 2013. The "fossilized" mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology, 11: 29. |
Rudinger, M., Polsakiewicz, M., Knoop, V., 2008. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol. Biol. Evol., 25: 1405-1414. DOI:10.1093/molbev/msn084 |
Rüdinger, M., Volkmar, U., Lenz, H., et al., 2012. Nuclear dyw-type ppr gene families diversify with increasing rna editing frequencies in liverwort and moss mitochondria. J. Mol. Evol., 7: 37-51. DOI:10.1007/s00239-012-9486-3 |
Schallenberg-Rüdinger, M., Lenz, H., Polsakiewicz, M., et al., 2014. A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol., 10: 1549-1556. |
Schmidt, H.A., Strimmer, K., Vingron, M., et al., 2002. Tree-puzzle: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics, 18: 502-504. |
Sloan, D.B., 2017. Nuclear and mitochondrial RNA editing systems have opposite effects on protein diversity. Biol. Lett., 13: 20170314. DOI:10.1098/rsbl.2017.0314 |
Smith, D.R., 2009. Unparalleled GC content in the plastid DNA of Selaginella
. Plant Mol. Biol., 71: 627. DOI:10.1007/s11103-009-9545-3 |
Soltis, D.E., Soltis, P.S., Zanis, M.J., 2002. Phylogeny of seed plants based on evidence from eight genes. Am. J. Bot., 89: 1670-1681. DOI:10.3732/ajb.89.10.1670 |
Stamatakis, A., 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 9: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Wang, I.X., So, E., Devlin, J.L., et al., 2013.
ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep., 5: 849-860. |
Wang, X.Q., Ran, J.H., 2014. Evolution and biogeography of gymnosperms. Mol. Phylogenet. Evol., 75: 24-40. |
Wu, C.S., Wang, Y.N., Hsu, C.Y., et al., 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol., 3: 1284-1295. DOI:10.1093/gbe/evr095 |
Yang, Y., Ferguson, D.K., Liu, B., et al., 2022. Recent advances on phylogenomics of gymnosperms and a new classification. Plant Divers., 44: 340-350. |
Zhong, B., Yonezawa, T., Zhong, Y., et al., 2010. The position of Gnetales among seed plants: Overcoming pitfalls of chloroplast phylogenomics. Mol. Biol. Evol., 27: 2855-2863. DOI:10.1093/molbev/msq170 |



