b. Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China;
c. Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China (Ministry of Education), Southwest Forestry University, Kunming 650224, China;
d. College of Life Sciences, Shaanxi Normal University, Xi'an 710119, China;
e. University of Chinese Academy of Sciences, Beijing 100101, China
Vegetable storage lipids (mainly triacylglycerol, TAG) accumulated mostly in specific oil-producing tissues not only give essential supply for human diets, but also provide critical raw materials in industry (Baud and Lepiniec, 2010; Cahoon et al., 2007; Slocombe et al., 2009). Generally, TAG biosynthesis can be simplified into two main steps: fatty acid biosynthesis in the plastids and TAG assembly in the endoplasmic reticulum (Chapman and Ohlrogge, 2012; Li-Beisson et al., 2013). After fatty acids are synthesized and exported from plastids, fatty acids are converted into acyl-CoA and assembled into TAG via many key enzymes (Baud and Lepiniec, 2010; Li et al., 2016; Ohlrogge and Chapman, 2011). Although the pathway of oil biosynthesis is conserved in plants and many studies on dissecting the molecular mechanism of oil accumulation, the regulation of TAG biosynthesis in oil-producing tissues not completely understood in plants, involving in diverse regulatory pathways. The identification of gene regulators and dissecting their regulatory mechanisms are critical for both understanding the oil accumulation and designing genetic strategies to efficiently increase plant oil production (Izadi-Darbandi et al., 2020; Wang et al., 2007; 2015; 2018; Weselake et al., 2009).
The WRINKLED1 (WRI1) transcription factor has been relatively well known as a key regulator of oil biosynthesis in oilseeds (Cernac and Benning, 2004; Chapman and Ohlrogge, 2012; Chen et al., 2018). The seed oil content in loss-of-function mutant of AtWRI1 (wri1) in Arabidopsis was reduced by 80% compared to the wild-type, reflecting its importance in regulation of oil biosynthesis in Arabidopsis seeds (Focks and Benning, 1998). WRI1 and its function in directly regulating many genes of glycolysis and fatty acid biosynthesis (such as ketoacyl-acyl carrier protein synthase/KAS1, acyl carrier protein/ACP1, enoyl-ACP reductase/EAR1 and biotin carboxyl carrier protein/BCCP2) by binding the AW-box cis-elements [CnTnG(n)7CG] within up-stream promoter regions of these lipid genes in oil biosynthesis pathway has broadly been identified in many plants (Bates et al., 2013; Baud et al., 2007; Maeo et al., 2009). Furthermore, WRI1 was often considered as a key master regulator that could be applied towards enhancing seed oil content by transgenic engineering techniques in plants (Cernac and Benning, 2004; Ohlrogge and Jaworski, 1997; Vanhercke et al., 2019). Although studies have revealed that WRI transcription factors, as members of the APETALA2 (AP2) family comprised of AP2, ERF, DREB, RAV and Soloist subfamilies, are distinct in sequence structure from other members because they have two AP2 domains comprising 60–70 conserved amino acid residues (other members have only single AP2 domain) in AP2 family (Magnani et al., 2004; Riechmann and Meyerowitz, 1998; Wessler, 2005), how WRI members can regulate the expression of many lipid genes whereas other members of AP2 family do not, remains unknown. Besides, homologous genes of WRI1 often varied in different plants and whether their functions are convergent or divergent remain uncertain in different plants. It is important to investigate how WRI members function in regulating oil biosynthesis toward understanding the molecular mechanism of oil accumulation in developing seeds of different oilseed crops.
Castor bean (Ricinus communis L., 2n = 20) is an important non-edible oilseed crop, widely cultivated in many countries and regions (in particular India, China and Brazil). Castor oil is unique among all the vegetable oils because they are mainly composed of ricinoleic acid (over 85%), a fatty acid consisting of 18 carbons, a hydroxyl group attached to C12 and a double bond between C9 and C10. Owing to the structural peculiarity of ricinoleic acid, castor oils with the highest and most stable viscosity index among all the vegetable oils are economically important in industry, because they are often used as raw materials for producing lubricants, aviation oil, nylon, inks, soaps, and adhesive (da Silva Romas et al., 1984; da Silva et al., 2006). In particular, castor oils are soluble with ethanol at the any proportion and have been considered as ideal feedstock for making biodiesel (Goodrum et al., 2005; Ogunniyi, 2006; Scholz and Silva, 2008). Due to its growth property such as small gestation period, drought hardiness and wide adaptation to soil conditions, castor bean has created tremendous interest worldwide for the utilization of marginal lands to develop biodiesel productions. With increasing demands of castor oils in industry, there is an immediate need for genetic improvement and metabolic engineering of castor bean to develop or create new cultivars with high oil content (Ji et al., 2018). Investigating key regulatory factors controlling oil accumulation in the developing seeds is an essential prerequisite to understand the molecular mechanism of oil biosynthesis with seed development in castor bean. Although previous studies had found that RcWRI1 was critical in controlling oil accumulation during seed development (Haque et al., 2018; Yang et al., 2019), the regulatory mechanism of RcWRI1 largely remains uncertain. In this study, we have tried to identify RcWRIs and investigate their expression profiles with seed development and oil accumulation. In particular, we identified potential targeted DNA sequences bond with RcWRI1 by DAP-seq (DNA Affinity Purification sequencing) technique and confirmed both AP2 domains of RcWRI1 are required to function in activating the expression of lipid genes in castor bean. This study provides significant information in understanding the molecular mechanism of WRI1 in regulation of oil accumulation during seed development of castor bean.
2. Materials and methods 2.1. Plant materials and growth conditionsCastor bean seeds var. ZB107 elite (kindly provided by Zibo Academy of Agriculture Science, Shandong, China) were sterilized and germinated in an incubator at 30 ℃, grown in the greenhouse of Kunming Institute of Botany under normal conditions from 2017 to 2020 (Wang et al., 2018; Wang et al., 2022). To protect against cross-pollination, hand pollination was carried out, while the inflorescences were covered with paper bags and tagged to keep records from the days after pollination (DAP). Seeds were collected at five different developmental stages (stage1-stage5) from 10 to 50 days after pollination (Zhang et al., 2016). 0–10 days after artificial pollination was stage1. Similarly, 10–20 days, 20–30 days, 30–40 days and 40–50 days after artificial pollination are Stage2, Stage3, Stage4 and Stage5 respectively.
2.2. Identification of RcWRIs in castor beanIdentification of RcWRIs in Castor Bean according to the method of Han et al. (2020). Based on the castor bean genome (http://castorbean.jcvi.org/index.php), an extensive search was performed to identify all WRI genes in castor beans. First, BLAST search was conducted to identify candidate genes in castor bean, using WRI1 homologs AtWRI1(AT3G54320), AtWRI2(AT1G16060), and AtWRI3(AT1G79700) from Arabidopsis thaliana at phytozome v.13 in JGI (https://phytozome-next.jgi.doe.gov/) with E-value < 10−5 (Tajima et al., 2013). After that, the online programs SMART (http://smart.embl-heidelberg.de/) and pfam (http://pfam.sanger.ac.uk/) were used to check the predicted AP2 domains in these candidate proteins (Letunic and Bork, 2018). The genes which showed the most significant E-value with AP2 domains were considered as putative WRIs of castor beans (RcWRIs).
2.3. RNA extraction and cDNA synthesisTotal RNA was extracted from castor bean seeds of 35 DAP (Haque et al., 2018; Xu et al., 2013) using TRIzol (Invitrogen, Carlsbad, CA) followed by RNeasy Plant Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Then the concentration and quality of total RNA were checked by the NANODROP-2000 spectrophotometer (Thermo Scientific) followed by 1.5% agarose gel electrophoresis. Complementary DNA was synthesized from 1 μg of total RNA by EasyScript First-Strand cDNA Synthesis SuperMix (TIANGEN, Beijing, China) according to the manufacturer's instructions, and then the cDNA samples were stored at −20 ℃ until use.
2.4. Yeast one hybrid assayThree tandem repeats of AW-box and mutated AW-box with upstream and downstream flanking regions from the RcBCCP2 promoters (Table S1) along with two restriction enzyme (Kpn Ⅰ and Xho Ⅰ) sites at the 5′ and 3′ ends were synthesized by TSINGKE Biological Technology Co., Ltd (Beijing, China). For generating of mutant baits, the conserved bases were substituted in the AW-box. The synthesized fragments so called the bait sequences were cloned into Kpn Ⅰ-Xho Ⅰ site of pAbAi vector (Clontech, http://www.clontech.com). The designed bait vectors were digested with Bst BI to make them linearized and then transformed into YM4271 competent yeast cells (Clontech, http://www.clontech.com) using Frozen-EZ Yeast Transformation Ⅱ Kit (ZYMO RESEARCH, USA) for homologously binding to the ura3-52 locus of the yeast genome. Positive clones were selected on the SD/-Ura agar plate after 3 days of culture. The bait auto-activation was checked by culturing the positive yeast cells on the SD/-Ura/ABA (100–300 ng/mL) agar plates according to the Matchmaker® Gold Yeast One-Hybrid Library Screening System User Manual (http://www.clontech.com). For construction of prey vectors, the CDS sequences of RcWRI1 were amplified from cDNA libraries by RcWRI1-FP/RcWRI1-RP (Table S1). The structural designs of AP2 domains of WRI1 used for cloning in the yeast one hybrid assay and later for transgene expression in tobacco leaves are shown in Supplementary Table 1. After PCR amplifications, the amplified sequences were cloned into Eco RI-Bam HI site of pGADT7 AD vector (Clontech, http://www.clontech.com) by the Gibson assembly method (Gibson et al., 2009) using the Trelief−TM SoSoo Cloning Kit (Lot#TSV-S1, TSINGKE Biological Technology Co., Ltd) according to the manufacturer's instructions. The recombinant prey-vectors were named as RcWRI1-pGADT7AD, RcWRI1AP2D1-pGADT7AD, RcWRI1AP2D2-pGADT7AD. Then the constructed vectors were transformed into the competent YM4271 yeast cells containing the bait-pAbAi recombinant vectors using Frozen-EZ Yeast Transformation Ⅱ Kit (ZYMO RESEARCH, USA) and cultured on the SD/-Ura/-Leu agar plates followed by the SD/-Ura/-Leu/ABA (100–300 ng/mL) agar plates and the results were analyzed.
2.5. Transient expression in tobacco leavesFor transient expression of fusion protein in tobacco (Nicotiana benthamiana) leaf epidermal cells. RcBCCP2 (−1/-1022 bp), RcEAR1 (−1/-1603 bp) promoter or mutant promoter regions in pA7 was digested with Hind Ⅲ/Nco Ⅰ and cloned into 3301vacGFPMP vector. The WRI1, WRI1-AP2D1, and WRI1-AP2D2 of castor bean were cloned into Nco Ⅰ-Pml Ⅰ site of pCAMBIA3301 fused with GFP protein (Table S1). The various constructs of effector and reporter were transformed individually into Agrobacterium strain EHA105 and then co-infiltrated into the leaves of tobacco (Smith et al., 2003). After infiltration for 2 days, the fluorescence of GFP was examined with a confocal laser scanning microscope Olympus FV1000 (Japan).
2.6. Protein sequences retrieveThe Arabidopsis WRI1 protein sequence was downloaded from (http://www.arabidopsis.org). BLASTP searches were performed in the NCBI database (http://www.ncbi.nlm.nih.gov) with the Arabidopsis WRI1 as query (Marchler-Bauer et al., 2015). All corresponding protein sequences of the putative RcWRI1 and its isoform were retrieved with maximum query coverage and with lower threshold of E-value (< 1e-5).
2.7. Co-expression gene network analysisCo-expression analysis was conducted by repeated iteration of network topological parameters as a function of the Pearson correlation coefficient (PCC) threshold in order to select significant correlation values. The correlation matrix was visualized and analyzed by Short Time-series Expression Miner (STEM, http://www.sb.cs.cmu.edu/stem/) for co-expression network of genes.
2.8. DNA affinity purification sequencing experimentsDAP-Seq experiments were performed following the method described by O'Malley et al. (2016). First, a DAP-Seq genomic DNA (gDNA) was extracted from castor bean seeds (stage 3, rapid accumulation stage of oil accumulation), the enriched DNAs were fragmented into short fragments by ultrasonic. Next, the DNA fragments were end repaired, 3′A added, and ligated to Illumina sequencing adapters. Separately, RcWRI1 fused to HaloTag were expressed in an in vitro wheat germ system. RcWRI1-HALO were immobilized on Magne HALO-Tag beads, washed, and incubated with 500 ng of DNA library in 40 μL phosphate-buffered saline (PBS) with slow rotation in a cold room for 1.5 h. The beads were washed five times, DNA was eluted and amplified with indexed TruSeq primers. Sequencing was performed on an Illumina HiSeq 2500 with 100-bp SR reads. The correct DAP-Seq library concentration for a specific read count was calculated based on the library fragment size. Negative control mock DAP-Seq libraries were prepared as described above but without the addition of protein to the beads.
2.9. Determination of seed FA and oil content, and gas chromatography (GC) analysisSeed FA and oil contents were determined according to the method of Li et al. (2006). In brief, the seeds were ground to powder in a ball mill, 0.2 g powder was dissolved in 2 mL hexane and isopropanol (hexane: isopropanol = 3:2, v/v). After homogenization, total lipids were extracted using the hexane/isopropanol method (Hara and Radin, 1978). FA methyl esters were derivatized in 1 mL of 5% (v/v) sulfuric acid in methanol for 1.5–2 h at 85 ℃. FA methyl esters were quantified by GC with flame ionization detection on a wax column (EC wax; 30m × 0.53 mm i.d. × 1.20 μm; Alltech). GC parameters were as follows: from 155 ℃; heated to 220 ℃ (1.5 ℃/min), 10 min isotherm; injector 250 ℃, detector 250 ℃; carrier gas 36 cm/s hydrogen; split ratio 1:50; detector gas 30 mL/min hydrogen; 300 mL/min air and 30 mL/min nitrogen; manual injection volume less than 1 μL. The peak areas were computed by the integration software, and percentages of fatty acid methyl esters (FAME) were obtained as weight percent by direct internal normalization (Adhikari et al., 2016).
2.10. DAP-seq data analysisWe defined target genes as those that contain DAP-Seq peaks located within the transcribed regions of genes, in introns, 2 kb upstream of the transcription start site (TSS), or 2 kb downstream of the transcription termination site. Bowtie 2 (v.2.2.5) was used to align the clean reads from each sample against the reference genome. All reads from transcriptional start site (TSS) to transcriptional termination site (TES) interval and upstream and downstream 2 k interval were counted by deepTools (version: 3.2.0) software. MACS2 (v.2.1.2) software was designed to identify read-enriched regions from DAP-seq data. Dynamic Poisson Distribution was used to calculated p-value of the specific region based on the unique mapped reads. The region would be defined as a peak when q-value < 0.05 (Cao et al., 2020).
2.11. Protein motif analysisThe MEME-ChIP suite was used to identify de novo DNA sequence motifs enriched in the WRI1-bound regions. DNA sequences from the 600 most-enriched target genes, trimmed to 100 bp centered on the region maximum (peak summit), were submitted to MEME-ChIP using a custom first-order background Markov model and an E-value cutoff of 1e-2.The motifs identified by MEME or DREME from the MEME Suite were further screened for enrichment using HOMER (homer.ucsd.edu/homer/motif/index.html) on the complete set of bound regions from target genes (250 bp, centered on the summit) against a negative set of regions, equivalent in number, length, and distribution, relative to the TSS. The cumulative hypergeometric distribution was used to test for motif enrichment significance. Hypergeometric optimization of motif enrichment (HOMER) is a suite of tools for motif discovery and various next-generation sequencing analyses. It is a versatile application that can analyze most epigenomic and transcriptomic NGS data types, including RNA-seq, ChIP-seq, MeDIP-seq, ATAC-seq and DNase-seq (O'Malley et al., 2016).
3. Results 3.1. Seed-specific expression of WRI1 is tightly associated with oil accumulation with seed developmentBased on castor reference genome we used amino acids of AtWRIs (AT3G54320, AT1G16060 and AT1G79700) in Arabidopsis as a query respectively and searched the homologs, resulting in three homolog castor WRI genes (30069.m000440, 30114.m000507 and 30131.m006896). Based on the WRI nomenclatures (Haque et al., 2018; Tajima et al., 2013), the identified WRI genes were named as RcWRI1 (30069.m000440), RcWRI2 (30114.m000507) and RcWRI3 (30131.m006896), as shown in Table 1.
| Homologs in Arabidopsis | Genes | Castor Bean ID | No. of Amino Acid | Mw (kDa) | PI | E-value to Arabidopsis |
| AtWRI1:AT3G54320 | RcWRI1 | 30069.m000440 | 426 | 49.7 | 5.45 | 8.00e-133 |
| AtWRI2:AT1G16060 | RcWRI2 | 30114.m000507 | 327 | 36.81 | 5.87 | 4.07e-87 |
| AtWRI3:AT1G79700 | RcWRI3 | 30131.m006896 | 330 | 56.63 | 8.84 | 1.51e-81 |
While we inspected the oil accumulation with seed development, we found that the oil accumulation did not start at Stage 1 in early stage of seed development. As shown in Fig. 1A, oil accumulation started at Stage 2 (as initial stage of oil accumulation), fast accumulated from Stage 2 to Stage 4 (as rapid accumulation stage of oil accumulation), and mostly stopped from Stage 4 to Stage 5. This observation suggest that the oil accumulation might be finely regulated at the molecular level with seed development (Ma et al., 2013; To et al., 2012).
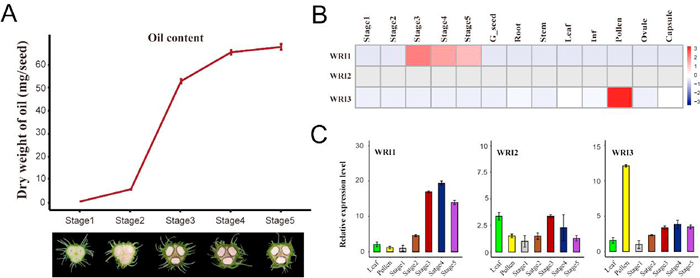
|
| Fig. 1 RcWRI1 regulates oil accumulation during seed development in castor bean. (A) Oil contents and seed morphology in development stages of the castor bean seeds. Bars = means ± SD from three biological replicates. (B) Transcriptional expression of RcWRI1, RcWRI2 and RcWRI3 in different tissues. The FPKM values used for heatmap were acquired from the castor bean genome database (https://woodyoilplants.iflora.cn/). (C) Relative expression of RcWRI1, RcWRI2 and RcWRI3 in different tissues. The data of histogram were derived from real-time PCR. |
To investigate the expressional profiles of identified RcWRIs among tissues we used the available transcriptome data that we have previously developed in diverse castor bean tissues (https://woodyoilplants.iflora.cn/). We inspected the expression levels of RcWRI1, RcWRI2 and RcWRI3 in 13 different tissues, covering root, stem, leaf, inflorescence, pollen, ovule, capsule, germinating seed, and developing seeds (including the five Stages 1–5 of seed development). As shown in Fig. 1B, we found that RcWRI2 was lowly expressed all tested tissues, RcWRI3 was highly expressed only in pollen, and RcWRI1 was highly expressed only in developing seeds, including Stages 2, 3 and 4. The RT-qPCR analysis in leaf, pollen, and developing seeds at the five Stages 1–5 further verified the expression profiles of RcWRI1, RcWRI2 and RcWRI3 (Fig. 1C; Table S3). In addition, the expression patterns of other transcription factors (LEC1, LEC2, ABI3 and FUS3) regulating oil synthesis in castor bean were inconsistent with the pattern of oil accumulation in castor seeds and the expression levels of these other factors in seeds were relatively low (Baud et al., 2007; Mu et al., 2008; Shen et al., 2010; Wang and Perry, 2013), only the expression pattern of WRI1 was highly consistent with the pattern of oil accumulation in castor seeds (Fig. S1; Table S2). Combined with the research progress of WRI1 function in A. thaliana, it suggests that WRI1 gene may be an important regulator of oil accumulation in castor bean seeds.
In order to verify whether RcWRI1 directly regulated oil accumulation, we heterologously transferred the RcWRI1 into tobacco leaf tissue, and sorted out five transgenic individuals. Semi-quantitative PCR technique confirmed that RcWRI1 had been significantly expressed in each transgenic individual in contrast to the control (Fig. S2). The oil content of leaves in the five independent transgenic individuals varied from 3.7304 to 5.1186 mg/g, significantly increased (Fig. 2A), about 1.2–1.7 folds comparing with the control (3.0234 mg/g). We further inspected the changes of fatty acid composition between transgenic leaves and the control and found that the changes of fatty acid composition mainly occurred in palmitic acid (C16:0) and linolenic acid (C18:3) (Fig. 2B). This result suggests that the heterologous expression of RcWRI1 in tobacco significant enhance oil accumulation in leaf.
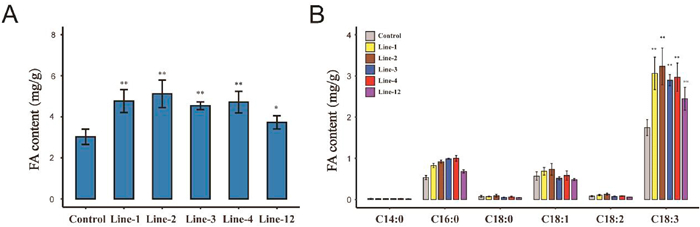
|
| Fig. 2 The content of total fatty acid (left) and different fatty acid components (right) in control and transgenic tobacco. Bars = means ± SD from three biological replicates. |
Although many studies have shown that WRI1 could regulate the expression of diverse lipid genes in plants, how WRI1 directly regulated the expression of lipid genes is still unclear in different plants. In order to globally detect the potential targeted genes bound by RcWRI1, we conducted the DAP-seq analysis. In total, 64112994 reads were generated, of which 94.76% reads were mapped into the castor bean genome with 36721298 unique mapped reads and 36317235 clean reads. After removing low-quality peaks, 29551 peaks were obtained (Table 2).
| DAP-seq | Total reads | Mapped Reads (ratio) | Unique Mapped Reads (ratio) | Clean reads (ratio) | Peak number |
| RcWRI1 | 64, 112, 994 | 60, 754, 018 (94.76%) |
36, 721, 298 (57.28%) |
36, 317, 235 (56.65%) |
29, 551 |
While we inspected the reads distribution on genome, we found that most of reads were fallen into the promoter regions (32.12% of reads within −2 kb regions) and intergenic regions (including 11.99% of reads within downstream of genes and 51.81% of reads within distal intergenic regions), according with the binding nature of transcription factors on genome (Fig. 3A; Table S4). In particular, it was clear that reads were enriched around TSS (transcription start site) regions (Fig. 3B). Furthermore, to investigate the putative functions of WRI1-binding candidate genes, we identified 7961 WRI1-binding candidate genes that contained promoters of WRI1-binding sites (Table S5). The GO (Gene Ontology) functional annotation for these identified WRI1-binding candidate genes revealed that these genes were mainly involved in multi metabolism and cell development processes. The top 20 functions of GO enrichment were concentrated on metabolism processes such as lipid metabolism, secondary metabolism and purine nucleotide biosynthesis (Fig. 3C). These results suggest that RcWRI1 be, as a master transcription factor, globally involved in metabolism processes by binding promoter regions of many genes (in particular lipid genes).
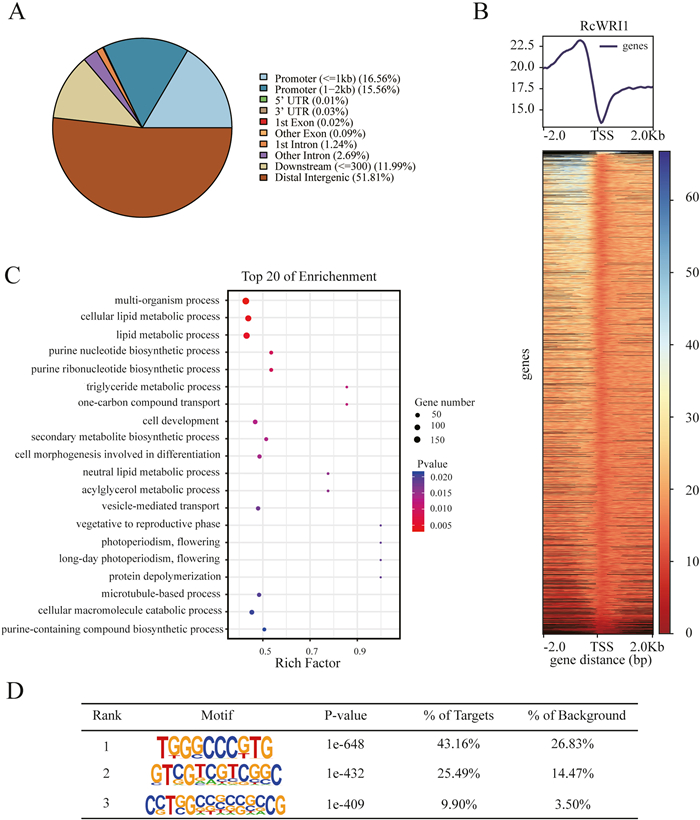
|
| Fig. 3 DAP-seq analysis of RcWRI1 in castor bean genome. (A) Relative proportions of RcWRI1 DAP-seq peaks in genic and intergenic regions. (B) The average enrichment abundance of RcWRI1 near TSS site. (C) GO analysis of genes that were marked by RcWRI1 on promoter regions. (D) The motif prediction of promoter regions bound by RcWRI1. |
To detect the specific cis-acting elements on promoter regions that binds with RcWRI1, we made a protein motif analysis to predict the cis-acting elements for WRI1 bound by MEME and HOMER (Fig. 3D). It was found that AW-box ([CnTnG](n)7[CG]) and two AW-boxes like ([GnAnC](n)6[GC]/[GnAnC](n)7[G]) were the main cis-acting element for WRI1 recognition. In particular, RcWRI1 is bound to its promoter and contains AW-box (Tables S5–S6), which indicates that WRI1 may have self-activation.
3.3. Co-expression network analysis of RcWRI1For a given transcription factor, its expression is usually associated with its targeted genes. To detect downstream the potential targeted genes of RcWRI1, we investigated the gene expression profiles of identified WRI1-binding candidate genes in developing seeds using RNA-seq data obtained from Stages 1–5. We determined expressed genes in developing seeds with a criterion that the expression level of a given gene has FPKM≥2 at least in each of the five stages. We sorted out 5405 genes (including 116 lipid genes) for subsequent expression cluster analysis. We identified six distinct profiles of gene expression (Profiles 1–6) in developing seeds (Fig. 4A). Profiles 1 and 6 exhibited that the stable down-expression or up-expression pattern with seed development, covering most of identified genes, Profile 2 and 3 exhibited the fast or slow down-expression with seed development, and Profile 5 exhibited the fast or slow up-expression with seed development. Profile 4 (including 344 genes) exhibited the no-expression at the early stage, fast up-expression after Stage 2, slow up-expression after Stage 3, and the no-change during seed maturation. In particular, we noticed that RcWRI1 and most genes related to lipid metabolism (including 25 lipid key-synthetase genes) were nested in Profile 4, highly consistent with oil accumulation with seed development (Tables S6–S7). While we further checked the organization of cis-elements in the promoter regions of 25 lipid key-synthetase genes, we found that 17 of the 25 lipid genes harbored AW-box or AW-box like domain, indicating that WRI1 may regulate the expression of lipid genes through AW-box or AW-box like (Fig. 4B; Tables S6–S8).
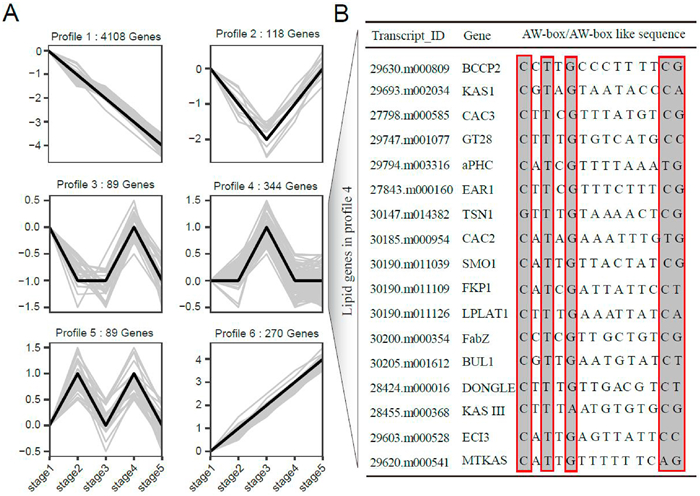
|
| Fig. 4 The co-expression network of RcWRI1 in stage1-stage5. (A) The expression trend diagrams of genes marked by RcWRI1 on promoter regions at different stages of seed development (stage1-stage5). (B) The AW-box or AW-boxlike of lipid genes in profile 4. |
The specific domain sequences within transcription factors usually determined the main function of a given transcription factor. The identified 19 members in the AP2 subfamily contained two AP2 domains comprising 60–70 conserved amino acid residues (Xu et al., 2013). Why only RcWRI1 (others do not) functions in regulating lipid genes remains unknown. While inspecting the sequences of two AP2 domains in AP2 family we found that amino acid sequence of AP2 domains were quite variable though the featured structure (Figs. S3–S4) present in most of (such as because of a 10-aa insertion in the R1 domain or a 1-aa insertion in the R2 domain, suggesting their functions divergence (see Xu et al., 2013 Additional file 3). To further investigate which domain of two domains within RcWRI1 is required in the regulation of downstream lipid genes, we conducted the transcription factor and DNA interaction, Yeast one-hybrid assay by constructing vectors containing domain 1 (AP2D1), domain 2 (AP2D2), and the whole RcWRI1 sequences, respectively. The known Biotin Carboxyl Carrier Protein 2 (BCCP2) gene and enoyl-ACP reductase (EAR1) gene (Maeo et al., 2009) were selected as candidate genes. As shown in Fig. 5, only the whole RcWRI1 sequences (containing two AP2 domains) could bind to AW-box (Fig. 5A), while the AW-box sequence was mutated, the whole RcWRI1 sequences could not bind with the mutant AW-box (MAW-box). The single AP2D1 or AP2D2 could all not bind with AW-box (Fig. 5A). Further, transcription factor activation experiments in tobacco leaf also verified that only complete RcWRI1 sequences had the binding ability to AW-box, while either AP2D1 or AP2D2 could not bind to AW-box (Fig. 5B). These results suggest that both AP2 domains within RcWRI1 be required to function in the regulation of downstream lipid genes.
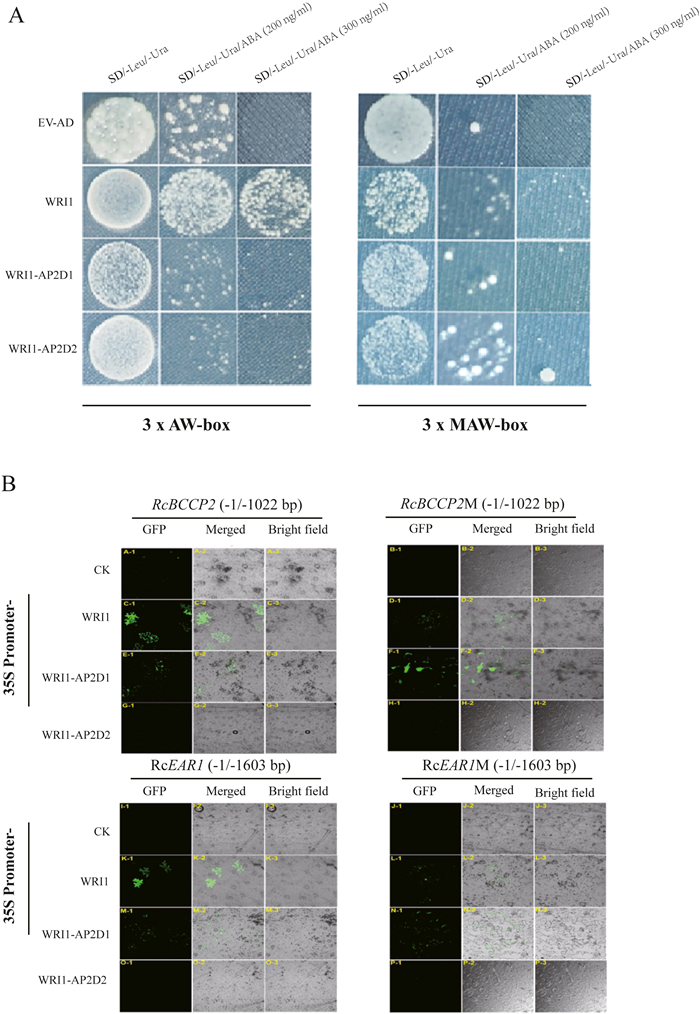
|
| Fig. 5 Both AP2 domains of RcWRI1 are necessary for transcriptional regulation. (A) Yeast one hybrid assay. Three replicates of AW-box and MAW-box with flanking regions was used as bait sequences and inserted into pAbAi vectors. WRI1, WRI1-AP2D1 and WRI1-AP2D2 used as prey sequences inserted into pGADT7 AD. EV-AD indicates empty AD vector was used for negative control. Yeast cells were grown on synthetic defined medium without Leu and Ura and with/without different concentrations of ABA. The yeast one hybrid assay was performed for RcWRI1. (B) Transactivation of GFP with promoter of BCCP2, BCCP2M, EAR1 and EAR1M by RcWRI1, WRI1-AP2D1 and WRI1-AP2D2 in tobacco leaves. Figure A, C, E and G indicate transactivation of RcBCCP2 derived by WRI1, WRI1-AP2D1 and WRI1-AP2D2. Figure B, D, F and H indicate transactivation of RcBCCP2M derived by WRI1, WRI1-AP2D1 and WRI1-AP2D2. Fig. I, K, M and O indicate transactivation of RcEAR1 derived by WRI1, WRI1-AP2D1 and WRI1-AP2D2. Figure J, L, N and P indicate transactivation of RcEAR1M derived by WRI1, WRI1-AP2D1 and WRI1-AP2D2. CK indicate negative control experiments by infiltrating of the tobacco leaves with Agrobacterium containing only promoter-vector constructs. Expressions were observed in GFP, Merged and Bright field. |
Oil biosynthesis in oilseeds is finely regulated, WRI1 has been well-known as a master regulator in several oilseed crops though its regulatory mechanism has not been clearly dissected in different plants. We here identified RcWRI1 as a master regulator in castor bean seeds and dissected the regulatory mechanism of RcWRI1 in regulating expressions of lipid genes in developing castor seeds.
According to WRI nomenclatures, three RcWRIs genes (RcWRI1, RcWR2 and RcWRI3) were identified. Though their structure similarity (containing two AP2 domain), the identified RcWRIs genes are functionally divergent based on their expression patterns in different tissues. It seems that only RcWRI1 is functionally involved in regulating oil biosynthesis of castor bean seeds. In particular, the heterologous transformation of RcWRI1 significantly increased the oil content of tobacco leaf tissues, suggesting that the regulatory mechanism of RcWRI1 in promoting oil accumulation is conserved.
Based on the DAP-seq analysis, 7961 WRI1-binding candidate genes were identified. Functionally, these identified genes were mainly involved in diverse metabolism pathways. Because of the false positive of the DAP-Seq method (Cao et al., 2020; O'Malley et al., 2016), that contained promoters of WRI1-binding sites were selected, and RcWRI1 be, as a master transcription factor, globally involved in metabolism processes by binding promoter regions of many genes. The GO functional annotation for these identified WRI1-binding candidate genes revealed that these genes were mainly involved in multi metabolism and cell development processes. The top 20 functions of GO enrichment were concentrated on metabolism processes such as lipid metabolism, secondary metabolism and purine nucleotide biosynthesis (Fig. 3C). These results suggest that RcWRI1 be, as a master transcription factor, globally involved in metabolism processes by binding promoter regions of many genes (in particular lipid genes). In addition, based on DAP-seq results we identified three AW-box and AW-boxes like bind sites of RcWRI1. Similarly, studies have been made on WRI1 for downstream regulation described that a 15 bp conserved binding site [CnTnG(n)7CG] called AW-box located at the upstream regions of the target genes, which was directly bound by WRI1 (Maeo et al., 2009). In addition, WRI1 was also bound with AW-box like [GnAnC](n)6[GC]/[GnAnC](n)7[G] in the upstream regions of PIN4 and PIN5 (Kong et al., 2017). Taken together, these studies clearly suggest that the specific binding of RcWRI1 with AW-box bind site though the sequences within AW-box bind sites may be not strictly conserved.
Co-expression network analysis of RcWRI1 identified 343 genes co-expressed with RcWRI1, including 25 genes in lipid metabolism pathway. Seemingly, most of co-expressed genes with RcWRI1 might not be directly involved in regulating the transcription of lipid genes. In fact, previous studies had shown that WRI1 also was functionally involved in glycolytic pathway of metabolism, widely participating the metabolism of other storage materials such as starch and protein in developing seeds (Baud et al., 2009; Pouvreau et al., 2011; Wu et al., 2014). Thus, except for regulating lipid gene expression RcWRI1 might also be important in regulating other genes involved in storage material accumulation in castor bean.
Indeed, the specific domain sequences within transcription factors usually determined the targeted genes bound with a given transcription factor (Riechmann and Meyerowitz, 1998; Magnani et al., 2004). The structural feature with two specific AP2 domains within RcWRI1s is distinct in AP2 family (Kumar et al., 2016; Xu et al., 2013; Figs. S3–S4). Here our results demonstrated that both AP2 domains, for RcWRI1, are required to activate downstream gene expression. The result suggests that the specific two AP2 domains co-determine the function of RcWRI1 in regulating transcription of lipid genes. This finding provides a new insight into understanding the function of WRI1 in specifically regulating lipid genes in oilseed crops.
AcknowledgementsThis work was supported by National Natural Science Foundation of China (grant number 31701465). We are grateful to the Service Center for Experimental Biotechnology at the Kunming Institute of Botany, CAS, for supporting plant cultivation.
Author contributions
A.L. conceived the research. Q.T., B.H., M.H, Y.W. and Y. L. performed the experiments. All authors analyzed the data. Q.T., B.H., and A.L. wrote the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Appendice A and B. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.09.003.
Adhikari, N.D., Bates, P.D., Browse, J., 2016. WRINKLED1 rescues feed back inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol., 171: 179-191. DOI:10.1104/pp.15.01906 |
Bates, P.D., Stymne, S., Ohlrogge, J., 2013. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol., 16: 358-364. DOI:10.1016/j.pbi.2013.02.015 |
Baud, S., Lepiniec, L., 2010. Physiological and developmental regulation of seed oil production. Prog. Lipid Res., 49: 235-249. DOI:10.1016/j.plipres.2010.01.001 |
Baud, S., Mendoza, M.S., To, A., et al., 2007. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J., 50: 825-838. DOI:10.1111/j.1365-313X.2007.03092.x |
Baud, S., Wuillème, S., To, A., et al., 2009. Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J., 60: 933-947. DOI:10.1111/j.1365-313X.2009.04011.x |
Cahoon, E.B., Shockey, J.M., Dietrich, C.R., et al., 2007. Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr. Opin. Plant Biol., 10: 236-244. DOI:10.1016/j.pbi.2007.04.005 |
Cao, Y., Zeng, H., Ku, L., et al., 2020. ZmIBH1-1 regulates plant architecture in maize. J. Exp. Bot., 71: 2943-2955. DOI:10.1093/jxb/eraa052 |
Cernac, A., Benning, C., 2004. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J., 40: 575-585. DOI:10.1111/j.1365-313X.2004.02235.x |
Chapman, K.D., Ohlrogge, J.B., 2012. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem., 287: 2288-2294. DOI:10.1074/jbc.R111.290072 |
Chen, L., Zheng, Y., Dong, Z., et al., 2018. Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genom., 293: 401-415. DOI:10.1007/s00438-017-1393-2 |
da Silva, N.L., Maciel, M.R., Batistella, C.B., et al., 2006. Optimization of biodiesel production from castor oil. Appl. Biochem. Biotechnol., 130: 405-414. DOI:10.1385/abab:130:1:405 |
da Silva Romas, L.C., Tango, S.J., Savi, A., et al., 1984. Variability for oil and fatty acid composition in castor bean varieties. J. Am. Oil Chem. Soc., 61: 1841-1843. DOI:10.1007/BF02540812 |
Focks, N., Benning, C., 1998. wrinkled 1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol., 118: 91-101. DOI:10.1104/pp.118.1.91 |
Gibson, D.G., Young, L., Chuang, R.Y., et al., 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods, 6: 343-345. DOI:10.1038/nmeth.1318 |
Goodrum, J.W., Geller, D.P., 2005. Influence of fatty acid methyl esters from hydroxylated vegetable oils on diesel fuel lubricity. Bioresour. Technol., 96: 851-855. DOI:10.1016/j.biortech.2004.07.006 |
Han, B., Xu, H., Feng, Y., et al., 2020. Genomic characterization and expressional profiles of autophagy-related genes (ATGs) in oilseed crop Castor bean (Ricinus communis L.). Int. J. Mol. Sci., 21. DOI:10.3390/ijms21020562 |
Haque, M.E., Han, B., Wang, B., et al., 2018. Development of an efficient chromatin immunoprecipitation method to investigate protein-DNA interaction in oleaginous castor bean seeds. PLoS One, 13: e0197126. DOI:10.1371/journal.pone.0197126 |
Hara, A., Radin, N.S., 1978. Lipid extraction of tissues with a low-toxicity solvent. Anal. Biochem., 90: 420-426. DOI:10.1016/0003-2697(78)90046-5 |
Izadi-Darbandi, A., Younessi-Hamzekhanlu, M., Sticklen, M., 2020. Metabolically engineered rice biomass and grain using genes associated with lipid pathway show high level of oil content. Mol. Biol. Rep., 47: 7917-7927. DOI:10.1007/s11033-020-05837-1 |
Ji, X.J., Mao, X., Hao, Q.T., et al., 2018. Splice variants of the Castor WRI1 gene upregulate fatty acid and oil biosynthesis when expressed in tobacco leaves. Int. J. Mol. Sci., 19: 146. DOI:10.3390/ijms19010146 |
Kong, Q., Ma, W., Yang, H.B., et al., 2017. The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J. Exp. Bot., 68: 4627-4634. DOI:10.1093/jxb/erx275 |
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Letunic, I., Bork, P., 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res., 46: 493-496. DOI:10.1093/nar/gkx922 |
Li, Y., Beisson, F., Pollard, M., et al., 2006. Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry, 67: 904-915. DOI:10.1016/j.phytochem.2006.02.015 |
Ma, W., Kong, Q., Arondel, V., et al., 2013. Wrinkled 1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS One, 8: e68887. DOI:10.1371/journal.pone.0068887 |
Maeo, K., Tokuda, T., Ayame, A., et al., 2009. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J., 60: 476-487. DOI:10.1111/j.1365-313X.2009.03967.x |
Magnani, E., Sjölander, K., Hake, S., 2004. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell, 16: 2265-2277. DOI:10.1105/tpc.104.023135 |
Li, N., Xu, C.C., Li-Beisson, Y.H., et al., 2016. Fatty acid and lipid transport in plant cells. Trends Plant Sci., 21: 145-158. DOI:10.1016/j.tplants.2015.10.011 |
Li-Beisson, Y., Shorrosh, B., Beisson, F., et al., 2013. Acyl-lipid metabolism. Arabidopsis Book 11, e0161. https://doi.org/10.1199/tab.0161. Epub 2013 Jan 29. PMID: 23505340; PMCID: PMC3563272.
|
Marchler-Bauer, A., Derbyshire, M.K., Gonzales, N.R., et al., 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res., 43: D222-D226. DOI:10.1093/nar/gku1221 |
Mu, J., Tan, H., Zheng, Q., et al., 2008. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol., 148: 1042-1054. DOI:10.1104/pp.108.126342 |
Ogunniyi, D.S., 2006. Castor oil: a vital industrial raw material. Bioresour. Technol., 97: 1086-1091. DOI:10.1016/j.biortech.2005.03.028 |
Ohlrogge, J.B., Chapman, K.D., 2011. The seeds of green energy: expanding the contribution of plant oils as biofuels. Biochemist, 33: 34-38. DOI:10.1042/BIO03302034 |
Ohlrogge, J.B., Jaworski, J.G., 1997. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol., 48: 109-136. DOI:10.1146/annurev.arplant.48.1.109 |
O'Malley, R.C., Huang, S.C., Song, L., et al., 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 166: 1598. DOI:10.1016/j.cell.2016.08.063 |
Pouvreau, B., Baud, S., Vernoud, V., et al., 2011. Duplicate maize Wrinkled 1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol., 156: 674-686. DOI:10.1104/pp.111.173641 |
Riechmann, J.L., Meyerowitz, E.M., 1998. The AP2/EREBP family of plant transcription factors. Biol. Chem., 379: 633-646. DOI:10.1515/bchm.1998.379.6.633 |
Scholz, V., da Silva, J.N., 2008. Prospects and risks of the use of castor oil as a fuel. Biomass Bioenerg., 32: 95-100. |
Shen, B., Allen, W.B., Zheng, P., et al., 2010. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol., 153: 980-987. DOI:10.1104/pp.110.157537 |
Slocombe, S.P., Cornah, J., Pinfield-Wells, H., et al., 2009. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J., 7: 694-703. DOI:10.1111/j.1467-7652.2009.00435.x |
Smith, M.A., Moon, H., Chowrira, G., et al., 2003. Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta, 217: 507-516. DOI:10.1007/s00425-003-1015-6 |
Tajima, D., Kaneko, A., Sakamoto, M., et al., 2013. Wrinkled 1 (WRI1) homologs, AP2-type transcription factors involving master regulation of seed storage oil synthesis in Castor bean (Ricinus communis L.). Am. J. Plant Sci., 4: 333-339. DOI:10.4236/ajps.2013.42044 |
To, A., Joubès, J., Barthole, G., et al., 2012. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell, 24: 5007-5023. DOI:10.1105/tpc.112.106120 |
Wang, X.D., Long, Y., Yin, Y.T., et al., 2015. New insights into the genetic networks affecting seed fatty acid concentrations in Brassica napus. BMC Plant Biol., 15: 91. DOI:10.1186/s12870-015-0475-8 |
Wang, F., Perry, S.E., 2013. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol., 161: 1251-1264. DOI:10.1104/pp.112.212282 |
Wang, W., Ao, T., Zhang, Y., et al., 2022. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis L.). Plant Divers., 44: 201-212. DOI:10.1016/j.pld.2021.06.008 |
Vanhercke, T., Dyer, J.M., Mullen, R.T., et al., 2019. Metabolic engineering for enhanced oil in biomass. Prog. Lipid Res., 74: 103-129. DOI:10.1016/j.plipres.2019.02.002 |
Wang, L., Du, X.L., Feng, Y.Z., et al., 2018. Ectopic expression of EuWRI1, encoding a transcription factor in E. ulmoides, changes the seeds oil content in transgenic tobacco. Biotechnol. Prog., 34: 337-346. DOI:10.1002/btpr.2606 |
Wang, H.Y., Guo, J.H., Lambert, K.N., et al., 2007. Developmental control of Arabidopsis seed oil biosynthesis. Planta, 226: 773-783. DOI:10.1007/s00425-007-0524-0 |
Weselake, R.J., Taylor, D.C., Rahman, M.H., et al., 2009. Increasing the flow of carbon into seed oil. Biotechnol. Adv., 27: 866-878. DOI:10.1016/j.biotechadv.2009.07.001 |
Wessler, S.R., 2005. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci., 10: 54-56. DOI:10.1016/j.tplants.2004.12.007 |
Wu, X.L., Liu, Z.H., Hu, Z.H., et al., 2014. BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J. Integr. Plant Biol., 56: 582-593. DOI:10.1111/jipb.12158 |
Xu, W., Li, F., Ling, L., et al., 2013. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genomics, 14: 785. DOI:10.1186/1471-2164-14-785 |
Yang, Z., Liu, X., Li, N., et al., 2019. WRINKLED1 homologs highly and functionally express in oil-rich endosperms of oat and castor. Plant Sci., 287. DOI:10.1016/j.plantsci.2019.110193 |
Zhang, Y., Mulpuri, S., Liu, A., 2016. High light exposure on seed coat increases lipid accumulation in seeds of castor bean (Ricinus communis L.), a nongreen oilseed crop. Photosynth. Res., 128: 125-140. DOI:10.1007/s11120-015-0206-x |



