b. Research and Collections Center, Illinois State Museum, 1011 East Ash Street, Springfield, IL 62703, USA
Phylogenetic trees have been commonly used in studies addressing evolutionary, ecological and biogeographical questions (e.g. Webb, 2000; Cavender-Bares et al., 2009; Qian and Jin, 2016; Jin and Qian, 2019, 2022; Yue and Li, 2021; Zhang et al., 2021; Qian, 2023; Zhou et al., 2023). Because gene-based, well resolved phylogenies including all species of a study area are rare, researchers have commonly used megatree-based (supertree) methods to generate phylogenetic trees for their studies. Specifically, researchers use existing megatrees as backbones to graft species in researchers' species lists to the backbones based on taxonomic information (e.g. genus, family, and order), and then prune the resulting phylogenetic trees to include only those species present in the researchers' species lists.
The software Phylomatic developed by Webb and Donoghue (2005) perhaps was the first tool to generate megatree-based phylogenies. However, it has a number of serious limitations, which have been discussed in previous studies (e.g. Qian and Jin, 2016). Qian and Jin (2016) and Jin and Qian (2019, 2022) developed a series of tools (S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2, which are collectively called the PhyloMaker series) to generate megatree-based phylogenies. The total number of studies using these tools has increased continuously since they were made available to researchers, and the total number of studies using these tools per year exceeds the number of studies using Phylomatic since 2020 (Fig. 1), based on Thompson Reuters ISI Web of Science. For example, the total number of studies citing the publications of the three PhyloMaker packages was 186 in 2021 whereas the number of the publications citing the publication of Phylomatic in the same year was only 45 (Fig. 1), indicating that the three PhyloMaker packages are useful tools.
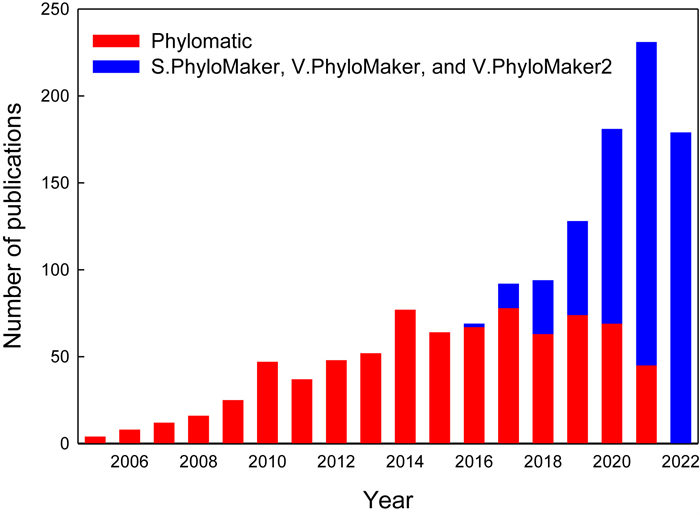
|
| Fig. 1 The number of the publications that used either Phylomatic (Webb and Donoghue, 2005) or one or more packages of the PhyloMaker series (S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2; Qian and Jin, 2016; Jin and Qian, 2019, 2022) in each year, based on Thompson Reuters ISI Web of Science (access on October 29, 2022). |
The three PhyloMaker packages were developed with a focus on plants and based on the megatrees embedded in the packages, but they can be used to generate megatree-based phylogenies for any groups of organisms, including animals, as long as megatree backbones and taxonomic information for them are provided. This was made clear by Jin and Qian (2019), and previous studies have used these packages to generate phylogenetic trees based on megatrees other than those embedded in the packages. For example, Qian et al. (2021) used V.PhyloMaker and a megatree published by Testo and Sundue (2016) for pteridophytes to generate a phylogenetic tree. In order for these PhyloMaker tools to be able to generate phylogenies based on megatree backbones other than those provided by the three published PhyloMaker packages, users need to modify R codes in some functions of the PhyloMaker packages. Although this is doable with little effort for those who are familiar with R language, doing so may be difficult for other users. Here, we add a new tool, which is called 'U.PhyloMaker' ('U' stands for 'universal', meaning that the tool can be used to generate phylogenetic trees for both plants and animals), to the PhyloMaker series. This new tool, which was developed based on the functions in S.Phylomaker (Qian and Jin, 2016) and V.PhyloMaker (Jin and Qian, 2019), allows users to easily generate megatree-based phylogenies for any groups of organisms based on megatrees available at present and in future.
In this article, in addition to introducing the package U.PhyloMaker, we provide information about how to use this package and some of the existing megatrees for plants (vascular plants) and vertebrate animals (amphibians, birds, fishes, mammals, and reptiles) to generate phylogenetic trees for users' species lists. We provide a simple R script that can be used to generate any phylogenetic trees when some minor changes (e.g. file names) are made by users (see Box 1).
| # load the package |
| library("U.PhyloMaker") |
| # input the sample species list, the megatree and genus-family relationship files |
| sp.list < - read.csv("amphibian_sample_species_list.csv") |
| megatree < - read.tree("amphibian_megatree.tre") |
| gen.list < - read.csv("amphibian_genus_list.csv") |
| # generate a phylogeny for the sample species list |
| result < - phylo.maker(sp.list, megatree, gen.list, nodes.type = 1, scenario = 3) |
| write.tree(result$phylo, "output_tree.tre") |
| write.csv(result$sp.list, "output_splist.csv") |
The package 'U.PhyloMaker' uses the functions reported in S.PhyloMaker and V.PhyloMaker with some modifications. Details about the package are described in the document entitled "Introduction to the 'U.PhyloMaker' package", which is provided along with the package (see below for website address), and the document entitled "Descriptions of scenarios and functions.doc", which is one of the supplementary files of this article. As in other packages of the PhyloMaker series (i.e. S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2), U.PhyloMaker allows the user to determine how the user wants U.PhyloMaker to graft species in user's species list to the specified megatree by making two selections (i.e. select one from the two node-adding options for non-monophyletic families or genera; select one of the three species-adding scenarios; see the supplementary information for details).
Unlike the other packages of the PhyloMaker series (i.e. S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2) in which data files (e.g. megatrees and taxonomic information) supporting the functions of the packages were embedded within the packages, in order to make U.PhyloMaker to be able to generate phylogenetic trees based on megatrees available at the present time and in future, we separated data files from the package U.PhyloMaker. Data files for some groups of organisms that can be directly used by the package are available from the megatree website at https://github.com/megatrees (see below for more information).
The U.PhyloMaker package was written in RStudio (RStudio Team, 2015), is for use in the R language (R Core Team, 2016), and requires a standard installation of R and the 'ape' package (Paradis et al., 2004). When the user loads U.PhyloMaker, the 'ape' package will be automatically loaded.
U.PhyloMaker is an open-source package (published under GPL-2). The R package of U.PhyloMaker, together with documentation, is available from GitHub (https://github.com/jinyizju/U.PhyloMaker).
3. Data filesTo run U.PhyloMaker, the user needs to specify three data files in the R console of U.PhyloMaker or the R script provided by us (Box 1): a file with user's species list, a file with the megatree that the user wants to use as a backbone, and a file with the genus–family relationship for the megatree. Files for megatrees and genus–family relationships for some groups of plants and animals are available from the website at https://github.com/megatrees/.
3.1. Species listA species list to be used by U.PhyloMaker should be in the format of comma-separated values (csv) with five columns (i.e. species, genus, family, species.relative, and genus.relative) as shown in the file 'sample_species_list.csv' in Supplementary data. The first column is mandatory and the other four columns are optional. However, if the user does not provide the family name of a species and if the genus–family relationship of the species is not available in the file with the genus–family relationship for the megatree, the species cannot be grafted to the megatree backbone (see below for details). Because the spelling and nomenclature of species names in many megatrees were standardized according to standard taxonomic databases (e.g. the plant megatrees used by V.PhyloMaker2, which are available at https://github.com/megatrees/; Jin and Qian, 2022), to maximize matching in species names between the user's species list and a megatree chosen by the user, we suggest the user standardize spelling and nomenclature of species using appropriate taxonomic databases. All names in the user's species list that are considered as synonyms should be replaced with their accepted names. Because taxa at terminal branches in most megatrees are species-level taxa (i.e. binomials), infraspecific taxa (e.g. subspecies, variety, and forma) in the user's species list should be combined with their parental species. Duplicate names should be removed. Zhang and Qian (2023) recently developed a tool (i.e. 'U.Taxonstand') for standardizing scientific names of any groups of organisms, and prepared databases for standardizing scientific names of bryophytes, vascular plants, amphibians, birds, fishes, mammals, and reptiles in the world, using U.Taxonstand to standardize scientific names for these groups of organisms is fairly easy.
Because a hybrid sign in front of the genus name of a species may cause a problem when adding the species to the megatree specified by the user, we suggest the user removes the hybrid sign in front of the genus name from both the megatree and user's species list. Hybrid species may be indicated with "×", "x" or "X" between the genus name and species epithet in a species name. However, for the same hybrid species, if different hybrid signs are used in the megatree and user's species list, the species in the user's species list cannot be matched to the species in the megatree. To maximize the matching of species names in the user's species list and species names in the megatree, the user may change the hybrid sign either in the megatree file or in the user's species list so that the same sign is used to indicate hybrid species in both the user's species list and the megatree.
Some species or genera that are absent from the megatree in use may be sister to or closely related with some species or genera in the megatree. When the information of their closely related species or genera is given in the user's species list (columns 'species.relative' and 'genus.relative'), these species or genera will be attached to their closely related species or genera as sister species or genera in a phylogeny generated by U.PhyloMaker (see fig. 1 of Jin and Qian (2019) for details).
3.2. MegatreeIn order for U.PhyloMaker to generate a phylogenetic tree for the species list provided by the user, the user must specify the megatree that the user wants to use as a phylogenetic backbone to generate a phylogenetic tree for the species list. U.PhyloMaker can work with a megatree file in the Newick or Nexus format, once the file is successfully imported in R as a 'phylo' object. Fortunately, megatrees for vascular plants and all major groups of vertebrates in the world are available online, although some of them do not include all families. We selected some of them as examples to show how they can be used by U.PhyloMaker. These megatrees are available from their original sources (see below) and from the website at https://github.com/megatrees/. New megatrees may be found at the website in future. When the users of U.PhyloMaker use the megatrees available from the website at https://github.com/megatrees/, the users should cite the original references of the megatrees in their publications, some of which are indicated below.
plant_megatree.tre: This megatree is the 'GBOTB.extended.WP.tre' megatree implemented in V.PhyloMaker2 (Jin and Qian, 2022), which was based on the botanical nomenclature of the World Plants (WP) database (https://www.worldplants.de). In addition to the 'GBOTB.extended.WP.tre' megatree, Jin and Qian (2022) also presented the magatrees 'GBOTB.extended.TPL.tre' and 'GBOTB.extended.LCVP.tre', which were based on the botanical nomenclature of The Plant List (TPL; http://www.theplantlist.org) and the LCVP database (Freiberg et al., 2020), respectively. Subsequently, the 'GBOTB.extended.WCVP.tre' megatree, which was based on the botanical nomenclature of the World Checklist of Vascular Plants (WCVP; http://sftp.kew.org/pub/data-repositories/WCVP/; Govaerts et al., 2021), was generated. All these plant megatrees are available from the website at https://github.com/megatrees/. When a phylogenetic tree generated based on any one of these megatrees is used in a publication, the user should cite Jin and Qian (2022) as the source of the megatree and should point out which of the original megatrees (e.g. GBOTB.extended.WP.tre) is used. Each of the megatrees includes all families of extant seed plants in the world and some families of pteridophytes. Each of the megatrees included the megatree of seed plants reported by Smith and Brown (2018) and the pteridophyte part of the megatree reported by Zanne et al. (2014). When the user uses a phylogenetic tree generated based on the 'plant_megatree.tre' megatree in a publication, the user should point out in the publication that the megatree is 'GBOTB.extended.WP.tre' reported by Jin and Qian (2022).
fern_megatree.tre: This megatree is the "ftol_sanger_con_dated.tre" megatree reported by Nitta et al. (2022). It includes all families of the extant ferns in the world.
amphibian_megatree.tre: This megatree is the same as the megatree "amph_shl_new_Consensus_7238.tre" available from https://data.vertlife.org/, which was originally reported by Jetz and Pyron (2018). This megatree includes all families of the extant amphibians in the world.
bird_megatree.tre: This megatree is one of the 10, 000 trees for birds provided by https://data.vertlife.org/, which were originally reported by Jetz et al. (2012).
fish_megatree.tre: This megatree is one of the 100 megatrees for fishes reported by Rabosky et al. (2018).
mammal_megatree.tre: This megatree is one of the 10, 000 trees (each with 5911 species) for mammals provided by https://data.vertlife.org/, which were originally reported by Upham et al. (2019).
reptile_megatree.tre: This megatree is one of the 10, 000 trees for species in the order Squamata of reptiles provided by https://data.vertlife.org/, which were originally reported by Tonini et al. (2016).
As shown in the VertLife website (https://data.vertlife.org/), many vertebrate groups each have multiple posterior phylogenies (e.g. 10, 000 trees for birds; Jetz et al., 2012). If a user would like to generate two or more phylogenetic trees based on different posterior phylogenies to account for uncertainty (variation) among different posterior phylogenies, the user may download two or more megatrees from https://data.vertlife.org/ or the original sources of the megatrees, and use U.PhyloMaker to generate more than one phylogenetic tree for the user's species list. Alternatively, the user may use the R script in the "R_codes_for_generating_trees_based_on_multiple_megatrees.r" file, which is available from the file "R codes and an example to use U.PhyloMaker to generate multiple trees.zip" located at https://github.com/jinyizju/U.PhyloMaker or https://github.com/ISMRCC/U.PhyloMaker, to automatically generate multiple trees from a single execution of U.PhyloMaker. We would like to point out that for many studies that are based on phylogenetic metrics such as phylogenetic diversity (PD), mean pairwise distance (MPD), mean nearest taxon distance (MNTD), net relatedness index (NRI), and nearest taxon index (NTI) (Faith, 1992; Webb, 2000; Webb et al., 2002), using the average value of a phylogenetic metric derived from two or more phylogenetic trees for an assemblage of species may not be necessary, because average values derived from multiple phylogenetic trees are often strongly and positively correlated with those derived from a singly phylogenetic tree (e.g. r = 0.999 for freshwater fish assemblages in North America; Qian et al., 2020).
3.3. The genus–family relationshipNearly all megatrees do not have information about the genus–family relationship. Without such information, species in those genera that are not present in the specified megatree (see above) cannot be grafted to the megatree. In the other packages of the PhyloMaker series (i.e. S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2), because megatrees with them were included in the packages, the genus–family relationship files for the megatrees were also included in the packages. Furthermore, a file with such relations for all genera of vascular plants is available online (https://github.com/jinyizju/genus.family.relationship). Because megatrees are not part of the package U.PhyloMaker, a file with the genus–family relationships for the megatree to be used by the user must be specified by the user (see below and Box 1 for details). We generated the genus–family relationship files for all the megatrees described above based on various data sources, and placed the files at https://github.com/megatrees/. When new megatrees are available from the website, we will upload the genus–family relationship files for them to the website. Because the genus–family relationship files are not part of U.PhyloMaker, we take no responsibility for the accuracy of the data in these files. We suggest users check and update the data if needed. Users can expand these files by adding those genera that are absent from the current versions of them, or modify the genus–family relationships. For example, the genus–family relationships for angiosperms in the current version of the plant genus list for the aforementioned four plant megatrees (i.e. GBOTB.extended.WP.tre, GBOTB.extended.TPL.tre, GBOTB.extended.LCVP.tre, and GBOTB.extended.WCVP.tre) were based on APG IV (Angiosperm Phylogeny Group, 2016). Thus, the genus–family relationships in the genus list may differ from those in the respective botanical databases based on which the botanical nomenclature in the megatrees was standardized. If the user would like to use the genus–family relationships in a particular botanical database (e.g. WP), the user needs to generate a genus list with genus–family relationships based on the botanical database, or modify the genus list provided by us (https://github.com/megatrees/).
4. Installation of U.PhyloMakerThe package U.PhyloMaker can be installed in R using the install_github function in the 'devtools' package (Wickham et al., 2018), as follows:
devtools: : install_github("jinyizju/U.PhyloMaker")
A stand-alone version of the package can be downloaded from the website at https://github.com/jinyizju/U.PhyloMaker or https://github.com/ISMRCC/U.PhyloMaker.
5. How to use U.PhyloMakerWe suggest the user puts the megatree and the genus–family relationship files in the folder where the user's species list is located. As stated above, megatrees for plants and vertebrates and their respective genus–family relationship files are available from the megatree website at https://github.com/megatrees/. We provided an R script in Box 1, as well as in Supplementary data, to run the package on a species list for amphibians as an example (the files specified in the R script of Box 1 are available from the website at https://github.com/megatrees/). If the user uses the file names and the default settings as shown in Box 1, the user does not need to make any change in the R script. Otherwise, the user only needs to specify (modify) five places at maximum in the five command lines in the R script as shown below.
amphibian_sample_species_list.csv: This is the name of a file with species list for which U.PhyloMaker will generate a phylogenetic tree for the user. The user may use a different file name.
amphibian_megatree.tre: This is the file name of the megatree for amphibians (see above for details).
amphibian_genus_list.csv: This is the name of the file with the genus–family relationship for amphibians (see above for details), and it works with the megatree "amphibian_megatree.tre".
nodes.type = 1: This is the default option, which means that U.PhyloMaker uses the "build.nodes.1" function to graft species to the megatree. If the user prefers to use the "build.nodes.2" function to graft species to the megatree, the user needs to change "nodes.type = 1" to "nodes.type = 2". Difference between these two node-finding functions was described in the Supplementary data. The previous users of S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2 used "nodes.type = 1" more frequently than "nodes.type = 2".
scenario = 3: This is the default option, which is more commonly used by the previous users of S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2, compared to the other two scenarios (i.e. scenarios 1 and 2). Differences among these three scenarios were described in the Supplementary data. If the user prefers to use scenario 1 or scenario 2 to generate a phylogenetic tree for the provided species list, the user needs to change "scenario = 3" to "scenario = 1" or "scenario = 2", respectively.
Note that due to potential changes during type-setting by the publisher of this article, the R script in Box 1 may not work if the user copies the command lines in Box 1 to a file or to the R console. If this happens, we suggest the user uses the R script in the Supplementary data, which are also available from the website at https://github.com/jinyizju/U.PhyloMaker or https://github.com/ISMRCC/U.PhyloMaker.
Each execution of U.PhyloMaker results in two output files. One file, which is called "output_tree.tre", as shown in Box 1, includes the phylogenetic tree generated by U.PhyloMaker for the species list provided by the user; and the other file, which is called "output_splist.csv", as shown in Box 1, includes the information in the species list provided by the user plus two additional columns, which are called "family.in.genus.list" and "output.note". The column "output.note" includes the information about if and how a particular species is grafted to the megatree: the term "present in megatree" means that the species is present in the megatree; the term "insertion based on genus" means that the species is absent from the megatree but its genus is present and the species is grafted to the genus; the term "insertion based on family" means that the genus of the species is absent from the megatree but its family is present and the species is grafted to the family; the term "no insertion" means that the family of the species is absent from the megatree and thus the species is not grafted to the megatree (note that if the genus is not present in the genus–family relationship file and the column family in the species list provided by the user is blank for the species, the species will not be grafted to the megatree even if the family of the species is present in the megatree). For a given species, if the family name in the species list provided by the user differs from that in the genus–family relationship file, U.PhyloMaker uses the family name in the latter when grafting the species to the megatree, and put the family name in the column "family.in.genus.list" of the "output_splist.csv" file. If the user prefers to use the genus–family relationship of the species in the species list provided by the user, the user needs to modify the genus–family relationship for its genus in the genus–family relationship file, and then re-runs U.PhyloMaker on the user's species list.
Box 1 provided an example to show how to use U.PhyloMaker to generate a phylogenetic tree for a species list of amphibians. If the user would like to use the script in Box 1 to generate a phylogenetic tree for a species list of a different group of organisms, the user needs to specify the file name of megatree and the file name of the genus list for the group of organisms in the script. For example, if the user's species list is for mammals, the user can use the script in Box 1 to generate a phylogenetic tree for the species list after replacing "amphibian_megatree.tre" and "amphibian_genus_list.csv" with "mammal_megatree.tre" and "mammal_genus_list.csv", respectively. As noted above, megatrees and genus lists for many major groups of organisms, including those mentioned above, are available from the website at https://github.com/megatrees/.
Warning or note messages may appear during the execution of the functions in the package. The user may ignore the messages if the execution can successfully generate a phylogenetic tree for the species list of the user.
6. DiscussionU.PhyloMaker is a new package of the PhyloMaker series. Although the main functions used in this package are the same as those used by the other packages of the PhyloMaker series (i.e. S.PhyloMaker, V.PhyloMaker, and V.PhyloMaker2; Qian and Jin, 2016; Jin and Qian, 2019, 2022), U.PhyloMaker makes it much easier to generate megatree-based phylogenetic trees for different groups of organisms, including both plants and animals. Jin and Qian (2019, 2022) showed that these packages can generate very large phylogenetic trees at a relatively fast speed. For example, Jin and Qian (2022) tested the functions used by the packages of the PhyloMaker series against a list of 365, 198 species, which included all angiosperm species in the world, and a phylogenetic tree was generated within about 42 h. We have tested U.PhyloMaker on multiple species lists, each of which included 20, 000 to 50, 000 species, and found that each test completed with success within less than 1 h in most cases, and the speed of U.PhyloMaker is faster than the other packages of the PhyloMaker series, even in the case that the nodes.info files were already embedded in the other packages of the PhyloMaker series and thus would save some execution time for them to execute the PhyloMaker functions. For example, in our tests on the species list with all 29, 158 seed plant species in China, 61% of which are absent from the "plant_megatree.tre", generating a phylogenetic tree for this species list took 55 min when using V.PhyloMaker2 and 49 min when using U.PhyloMaker (the same computer was used for the two tests). Because most of species lists at continental, regional, and local scales each include only thousands of species or fewer, and the running time of U.PhyloMaker on these species lists is usually much less than 1 h, thus, the speed of generating phylogenetic trees with the functions of U.PhyloMaker is relatively fast, although the packages may run faster when the functions within it are further optimized.
Several studies have shown that phylogenetic trees generated based on megatrees are robust for studies on community phylogenetics (Qian and Zhang, 2016; Li et al., 2019; Qian and Jin, 2021). For example, Qian and Zhang (2016) showed that values of phylogenetic metrics derived from the phylogenetic tree resolved at the family level are strongly and positively correlated with those derived from the phylogenetic tree resolved at the species level. Qian and Jin (2021) showed that values of phylogenetic metrics derived from a fully-resolved species level tree are nearly identical to those from a tree resolved only at the genus level (Pearson's correlation coefficient ranges from 0.966 to 1.000). Thus, megatree-based phylogenetic trees are appropriate for studies on community phylogenetics, particularly in the case that the vast majority of genera in a species list are resolved in the megatree. However, megatree-based phylogentic trees may not be appropriate for those studies that require a high resolution of a phylogenetic tree resolved at the species level.
In order for the users of U.PhyloMaker to be able to use the package easily, we make a number of megatrees available at https://github.com/megatrees/. We may place more megatrees on the website in future. We would like to make it clear that all megatrees at the website should not be considered as part of either the package U.PhyloMaker or this article. We recommend the users of U.PhyloMaker cite the original sources of the megatrees [e.g. citing Jin and Qian (2022) when using the 'plant_megatree.tre' megatree or one of the 'GBOTB.extended.WP.tre', 'GBOTB.extended.TPL.tre', 'GBOTB.extended.LCVP.tre', and 'GBOTB.extended.WCVP.tre' megatrees available at https://github.com/megatrees/ to generate a phylogenetic tree for plants, in addition to citing Smith and Brown (2018) and Zanne et al. (2014)].
AcknowledgementsWe are grateful to Shenhua Qian, Jian Zhang, and Yadong Zhou for testing U.PhyloMaker and anonymous reviewers for their comments. This work was supported by the Natural Science and Technology Foundation of Guizhou Province [[2020]1Z013] (to Y.J.); and the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou Province [U1812401] (to Y.J.).
Author contribution
H.Q. developed the idea of the software and worked on the lists for the genus–family relationships; Y.J. produced the package and wrote all R codes; H.Q. and Y.J. wrote the paper.
Declaration of competing interest
The authors declare no conflict of interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.12.007.
Angiosperm Phylogeny Group, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 181: 1-20. DOI:10.1111/boj.12385 |
Cavender-Bares, J., Kozak, K.H., Fine, P.V.A., et al., 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett., 12: 693-715. DOI:10.1111/j.1461-0248.2009.01314.x |
Faith, D.P., 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv., 61: 1-10. DOI:10.1016/0006-3207(92)91201-3 |
Freiberg, M., Winter, M., Gentile, A., et al., 2020. LCVP, the Leipzig catalogue of vascular plants, a new taxonomic reference list for all known vascular plants. Sci. Data, 7: 416. DOI:10.1038/s41597-020-00702-z |
Govaerts, R., Nic Lughadha, E., Black, N., et al., 2021. The World Checklist of Vascular Plants, a continuously updated resource for exploring global plant diversity. Sci. Data, 8: 215. DOI:10.1038/s41597-021-00997-6 |
Jetz, W., Pyron, R.A., 2018. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nat. Ecol. Evol., 2: 850-858. DOI:10.1038/s41559-018-0515-5 |
Jetz, W., Thomas, G.H., Joy, J.B., et al., 2012. The global diversity of birds in space and time. Nature, 491: 444-448. DOI:10.1038/nature11631 |
Jin, Y., Qian, H., 2019. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography, 42: 1353-1359. DOI:10.1111/ecog.04434 |
Jin, Y., Qian, H., 2022. V.PhyloMaker2: an updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Divers., 44: 335-339. DOI:10.1016/j.pld.2022.05.005 |
Li, D., Trotta, L., Marx, H.E., et al., 2019. For common community phylogenetic analyses, go ahead and use synthesis phylogenies. Ecology, 100: e02788. |
Nitta, J.H., Schuettpelz, E., Ramírez-Barahona, S., et al., 2022. An open and continuously updated fern tree of life. Front. Plant Sci., 13: 909768. DOI:10.3389/fpls.2022.909768 |
Paradis, E., Claude, J., Strimmer, K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20: 289-290. DOI:10.1093/bioinformatics/btg412 |
Qian, H., 2023. Patterns of phylogenetic relatedness of non-native plants across the introduction–naturalization–invasion continuum in China. Plant Divers., 45: 169-176. DOI:10.1016/j.pld.2022.12.005 |
Qian, H., Jin, Y., 2016. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J. Plant Ecol., 9: 233-239. DOI:10.1093/jpe/rtv047 |
Qian, H., Jin, Y., 2021. Are phylogenies resolved at the genus level appropriate for studies on phylogenetic structure of species assemblages?. Plant Divers., 43: 255-263. DOI:10.1016/j.pld.2020.11.005 |
Qian, H., Cao, Y., Li, D., et al., 2020. Geographic patterns and environmental correlates of phylogenetic relatedness and diversity for freshwater fish assemblages in North America. Ecography, 43: 1814-1824. DOI:10.1111/ecog.05280 |
Qian, H., Kessler, M., Deng, T., et al., 2021. Patterns and drivers of phylogenetic structure of pteridophytes in China. Global Ecol. Biogeogr., 30: 1835-1846. DOI:10.1111/geb.13349 |
Qian, H., Zhang, J., 2016. Are phylogenies derived from family-level supertrees robust for studies on macroecological patterns along environmental gradients?. J. Syst. Evol., 54: 29-36. DOI:10.1111/jse.12161 |
R Core Team, 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. URL. http://www.Rproject.org/.
|
Rabosky, D.L., Chang, J., Title, P.O., et al., 2018. An inverse latitudinal gradient in speciation rate for marine fishes. Nature, 559: 392-395. DOI:10.1038/s41586-018-0273-1 |
RStudio Team, 2015. RStudio. Integrated Development for R. RStudio, Inc., Boston, MA, USA. URL. http://www.rstudio.com/.
|
Smith, S.A., Brown, J.W., 2018. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot., 105: 302-314. DOI:10.1002/ajb2.1019 |
Testo, W., Sundue, M., 2016. A 4000-species dataset provides new insight into the evolution of ferns. Mol. Phylogenet. Evol., 105: 200-211. DOI:10.1016/j.ympev.2016.09.003 |
Tonini, J.F.R., Beard, K.H., Ferreira, R.B., et al., 2016. Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biol. Conserv., 204: 23-31. DOI:10.1016/j.biocon.2016.03.039 |
Upham, N.S., Esselstyn, J.A., Jetz, W., 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biology, 17: e3000494. DOI:10.1371/journal.pbio.3000494 |
Webb, C.O., 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat., 156: 145-155. DOI:10.1086/303378 |
Webb, C.O., Ackerly, D.D., McPeek, M.A., et al., 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst., 33: 475-505. DOI:10.1146/annurev.ecolsys.33.010802.150448 |
Webb, C.O., Donoghue, M.J., 2005. Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Notes, 5: 181-183. DOI:10.1111/j.1471-8286.2004.00829.x |
Wickham, H., Hester, J., Chang, W., 2018. Devtools: Tools to Make Developing R Packages Easier. R package version 1.13.6. https://CRAN.R-project.org/package=devtools.
|
Yue, J., Li, R., 2021. Phylogenetic relatedness of woody angiosperm assemblages and its environmental determinants along a subtropical elevational gradient in China. Plant Divers., 43: 111-116. DOI:10.1016/j.pld.2020.08.003 |
Zanne, A.E., Tank, D.C., Cornwell, W.K., et al., 2014. Three keys to the radiation of angiosperms into freezing environments. Nature, 506: 89-92. DOI:10.1038/nature12872 |
Zhang, J., Qian, H., 2023. U.Taxonstand: An R package for standardizing scientific names of plants and animals. Plant Divers., 45: 1-5. DOI:10.1016/j.pld.2022.09.001 |
Zhang, Y. -Z., Qian, L. -S., Spalink, D., et al., 2021. Spatial phylogenetics of two topographic extremes of the Hengduan Mountains in southwestern China and its implications for biodiversity conservation. Plant Divers., 43: 181-191. DOI:10.1016/j.pld.2020.09.001 |
Zhou, Y.-D., Qian, H., Jin, Y., et al., 2023. Geographic patterns of taxonomic and phylogenetic β-diversity of aquatic angiosperms in China. Plant Divers., 45: 177-184. DOI:10.1016/j.pld.2022.12.006 |



