b. Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Menglun, Mengla, Yunnan 666303, China;
c. University of Chinese Academy of Sciences, Beijing 100049, China;
d. School of Life Sciences, Henan University, Kaifeng 475004, China
Cotton (Gossypium hirsutum), an essential crop-producing natural textile fiber, is cultivated worldwide because of its high yield. However, the cotton growth, yield, and fiber quality were severely threatened by the destructive and soil-borne fungal pathogen Verticillium dahliae. It invades the roots of the cotton and then colonizes the vascular bundle in the soil. The microsclerotia of V. dahliae is extremely viable in the field, and it can resist abiotic stresses such as high temperature, low temperature, and high osmotic pressure. Few resistant varieties have been successfully bred through traditional crossbreeding in G. hirsutum (Zhang et al., 2012). Thus, the current method of preventing V. dahliae is still mainly using chemical methods. However, in order to decrease the usage of chemicals, genetic engineering using plant resistance genes has also been applied to improve cotton resistance to V. dahliae.
As sessile organisms, plants constantly face various biotic stresses which may greatly threaten their survival in the ever-changing natural circumstance. Fortunately, they have evolved intricate defense mechanisms for perception of stress signals and activation of specific resistance responses, contributing to their survivability under various adverse conditions. First, resistant plants use a group of plasma membrane anchored pattern-recognition receptors (PRRs) to perceive microbes or pathogen-associated molecular patterns (PAMP), leading to PAMP-triggered immunity (PTI). Second, plant hosts also have evolved disease resistance proteins to recognize specific effector molecules (Avr proteins) and activate effector-triggered immunity (ETI) (Jones and Dangl, 2006; Bacete et al., 2018).
Phytohormones play vital roles in both plant growth and response to various stresses. Many studies have shown that salicylic acid (SA), ethylene (ET), and jasmonic acid (JA) are closely related to plant defense response to diverse pathogens (Li et al., 2006). According to their lifestyles, pathogens can be classified as biotrophs and necrotrophs. The SA signaling pathway is often associated with resistance to biotrophs that usually obtain nutrition from living tissues, while the JA and ET signaling pathways plant important roles in plant defense against necrotrophs which often feed on dead tissues (Glazebrook, 2005). The wilting process of cotton after infection by V. dahliae includes two stages: living nutrient type and dead body nutrition type, indicating that both SA and JA signaling pathways are closely associated with cotton and V. dahliae interaction (Zhang et al., 2013). One recent study demonstrated that overexpression of Gbvdr6 increases plant tolerance to V. dahliae in both Arabidopsis and cotton through the modulation of both SA and JA/ET signaling pathways (Yang et al., 2018). Thus, cotton defense responses to V. dahliae involve both SA and JA/ET signals through their synergistic or antagonistic interactions. A better understanding of plant hormonal response to V. dahliae will contribute to the effective management of Verticillium wilt while maintaining both cotton yield and quality.
The phytohormone JA is a ubiquitous molecule and acts as an important regulatory signal to regulate diverse physiological processes (Kazan and Manners, 2012; Pieterse et al., 2012; Hu et al., 2017; Huang et al., 2017). It can be perceived by the F-box protein CORONATINE INSENSITIVE1 (COI1), which contributes to the ubiquitination and degradation of a group of downstream repressors of the JA signaling, namely JASMONATE ZIM-DOMAIN (JAZ) proteins (Zhang et al., 2017). Studies have revealed that JAZ proteins play their biological functions via physical interactions with various transcription factors; their degradation contributes to the release of downstream transcription factors, finally leading to the activation of jasmonate signaling. One recent study has shown that GhJAZ2 functions negatively to regulate cotton resistance to V. dahliae through physical interaction with GhbHLH171 and repression of its transcriptional activity (He et al., 2018). Identification of more JAZ direct targets during cotton-V. dahliae interaction will provide new insights into the roles of JA signaling in cotton defense response.
WRKY transcription factor superfamily has been shown to play critical roles in diverse plant physiological processes through forming integral parts of signaling webs. It can be divided into three major subfamilies based on both the number of WRKY domains and the type of their zinc fingerlike motifs at the C terminals. Interestingly, several WRKY members have been identified as positive or negative regulators in plant defense against various pathogens (Pandey and Somssich, 2009). For example, mutations of Arabidopsis WRKY8 or structurally related WRKY18, WRKY40, and WRKY60 increased basal resistance to the biotroph Pseudomonas syringae (Chen et al., 2010; Xu et al., 2006). Disruption of Arabidopsis WRKY33 or WRKY75 promoted susceptibility to the necrotrophs Botrytis cinerea and Alternaria brassicicola (Chen et al., 2020; Zheng et al., 2006). In cotton, GbWRKY1 positively regulates plant defense response to both V. dahliae and B. cinerea (Li et al., 2014). Furthermore, GhWRKY70 negatively regulates the resistance to V. dahliae in Arabidopsis through SA/JA signaling pathway (Xiong et al., 2019). Thus, WRKY transcription factors may function as key components to elaborately modulate the intricate transcriptional networks of plant defense.
In this work, we showed that a cotton transcription factor, GhWRKY33, is involved in Verticillium wilt resistance. Our results demonstrated that GhWRKY33 inhibits the expression of AtPDF1.2 by inhibiting the expression of AtERF1, thereby impairing the resistance to V. dahliae in Arabidopsis. Moreover, protein–protein interaction analysis revealed that GhJAZ3 physically interacts with GhWRKY33. Our results thus imply that GhWRKY33 and GhJAZ3 may function as negative regulators in plant defense response against V. dahliae.
2. Materials and methods 2.1. Growth of plant material and pathogen culturesThe Gossypium hirsutum cultivar Zhongzhimian 2 was cultured in a growth cabinet at 28 ℃ under LDs (16-h light/8-h dark cycle), and the Arabidopsis plants were cultured in a growth cabinet at 22 ℃ under SDs (8-h light/16-h dark cycle). The highly virulent V. dahliae strain Vd080 was cultivated on potato dextrose agar (PDA) for 7 days and then was transferred to liquid Czapek medium for another 7 days at 25 ℃ and 120 rpm to yield conidia. Conidia were harvested by filtration with funnel, and the final concentration was adjusted to 1 × 107conidia/mL prior to use.
2.2. Construction of transgenic overexpression linesTo generate the 35S: GhWRKY33 and 35S: GhJAZ3 constructs, their cDNA fragments were first cloned into the pOCA30 vector, and then the resulting vectors were transformed into WT Arabidopsis through floral dip method. Harvested seeds were selected by 1/2 Murashige & Skoog medium containing 50 mg/mL kanamycin. The selected plants were transferred to soil and used for further studies.
2.3. Induction treatmentsThe cotton seedlings at 3 leaf stage were sprayed with 100 μM methyl jasmonate (MeJA) and 10 mM amino cyclopropane-1-carboxylic acid (ACC). Water was used as a control. The aerial parts were collected at different time points after treatment and stored in a −80 ℃ refrigerator.
2.4. Pathogen inoculation and detection of Verticillium wilt resistanceThe cotton seedlings at the 3-leaf stage or three-week-old Arabidopsis seedlings were inoculated with a Verticillium dahliae conidial suspension as described previously (He et al., 2018; Zhang et al., 2011). Three weeks after inoculation, the symptoms of Verticillium wilt were observed, and the fungal biomass was counted. Fungal biomass was measured by an approach as depicted formerly (Xiong et al., 2019).
2.5. Gene expression analysisPlant material was broken up using a high-throughput tissue grinder (SCIENTZ-48, China) and total RNA was extracted from plant materials by TRIzol kit (Invitrogen, USA). RNA concentration was measured and RNA quality was assessed using NanoDrop ONE (Thermo Fisher Scientific, USA). The Superscript II (Invitrogen, USA) reverse transcription system was used to reverse-transcribe1μg total RNA in a 20-uL reaction mixture. Half-reactions (10 μl system with 1ul aliquots as template) were performed on a Light Cycler 480 real-time PCR machine (Roche, Germany) using the Light Cycler FastStart DNA Master SYBR Green I Kit. Quantitative RT–PCR was conducted using AtActin2 (At3G18780) and GhUBQ7 (DQ116441) as internal controls, and gene-specific primers are listed in Table S1.
2.6. Subcellular localization analysisTo research into the subcellular localization of GhWRKY33 and GhJAZ3 in plants, the CDS of GhWRKY33 and GhJAZ3 were cloned into the pOCA30-GFP vector respectively. Plasmids with GFP alone was used as control. These plasmids were transformed into EHA105 to prepare cell suspension. The cell suspension was infiltrated into Nicotiana benthamiana leaves, and then N. benthamiana were incubated at 22° for another 48 h. The GFP fluorescence signal was observed in the infected leaves under confocal laser microscopy (Leica, Germany).
2.7. Yeast one-hybrid assayThe full-length GhWRKY33 coding sequence was fused into the EcoRI and BamHI sites of pGADT7 vector to generate pGADT7-GhWRKY33. The W-box motifs within the different promoter regions of AtERF1 and GhERF2 and the motifs mutating off the core bases of the W-box (mW-box motif) were ligated to the pAbAi vector. The detailed procedure for the yeast one-hybrid was implemented as stated in the manufacturer's instructions (Clontech, USA).
2.8. Yeast two-hybrid screening and confirmationThe coding region of GhWRKY33 was fused to pGBKT7 after PCR amplification, while the sequence of the coding region of GhJAZ3 was fused to the pGADT7 vector and then co-transformed into the yeast strain Y2HGold (Clontech, USA). Protein–protein interactions were verified by colony growth of co-transformed yeast cells in SD/-Trp-Leu-Ade-His solid medium.
2.9. Luciferase complementation imaging assay (LCI)The full-length CDS of GhWRKY33 and GhJAZ3 were fused with the C-terminal (pCAMBIA1300-cLUC) and N-terminal (pCAMBIA1300-nLUC) portions of the luciferase reporter gene, respectively. Then the assays were performed according to Zhang et al. (2021).
2.10. Transient expression assayThe promoter regions of AtERF1 (~2500 bp) and GhERF2 (~2100 bp) were inserted into pGreenII 0800-LUC as reporters. The full-length CDS of GhWRKY33, GhJAZ3, and GFP were inserted into the pGreenII62-SK as effectors. Then the transient expression assay was performed according to Wang et al. (2016).
2.11. Accession numbersSequence data for the genes described in this article can be found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the following accession numbers: KC414677 for GhWRKY33, AY781117 for GhERF2, DQ116441 for GhUBQ7. Sequence data for the genes described in this article can be found in the COTTONGEN (https://www.cottongen.org/) following accession numbers: CotAD_41943 for GhJAZ3(Yu et al., 2021). Sequence data for the genes described in this article can be found in TAIR (https://www.arabidopsis.org/) with the following accession numbers: AT3G18780 for AtACTIN2, AT3G23240 for AtERF1, AT5G44420 for AtPDF1.2.
3. Results 3.1. Expression pattern and subcellular localization of GhWRKY33The WRKY family of transcription factors has been revealed to be widely associated with plant defense against various phytopathogens, however, little information is available about its role in defense against V. dahliae in G. hirsutum. Here, GhWRKY33 was found to be induced by V. dahliae strain Vd080 (Fig. 1A). Further expression analysis showed that GhWRKY33 also responded to ET and JA, with a stronger expression upon combined treatment of ET and JA (Fig. 1B). These results implied that GhWRKY33 may play a role in cotton defense against V. dahliae.
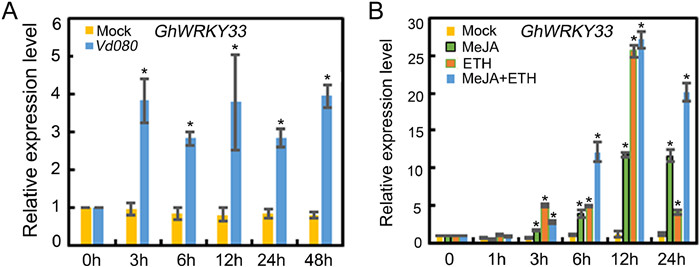
|
| Fig. 1 Expression of GhWRKY33. (A) Expression of GhWRKY33 upon infection by Verticillium dahliae in cotton. (B) Expression of GhWRKY33 in response to MeJA and ET treatment in cotton. For A and B, values are mean ± SD of three independent biological replicates (P < 0.05). |
To determine the subcellular localization of GhWRKY33, its full-length cDNA was fused with the green fluorescent protein (GFP) gene driven by CaMV35S promoter and subsequently expressed transiently in young leaves of Nicotiana benthamiana. The fluorescence of GhWRKY33-GFP was observed exclusively in the nucleus (Fig. S1). Contrarily, the fluorescence of free GFP was expressed in both nucleus and cytoplasm (Fig. S1). These results suggested that GhWRKY33 is localized in the nucleus and may perform its role as a transcription factor.
3.2. Overexpression of GhWRKY33 in Arabidopsis decreases plant tolerance to V. dahliaeTo confirm its role in defense against V. dahliae, we first generated transgenic Arabidopsis plants heterologously expressing GhWRKY33 under the control of the CaMV35S promoter (35S: GhWRKY33-L3 and 35S: GhWRKY33-L7). When homozygous GhWRKY33-transgenic lines (T3) were obtained, the wild-type, 35S: GhWRKY33-L3 and 35S: GhWRKY33-L7 transgenic Arabidopsis plants were simultaneously inoculated with V. dahliae. As shown in Fig. 2A, GhWRKY33-overexpressing plants displayed more serious disease symptoms than WT two weeks after inoculation (Fig. 2A). The ratio of disease leaves and the fungal biomass was also significantly higher in GhWRKY33-overexpressing plants (Fig. 2B and C).
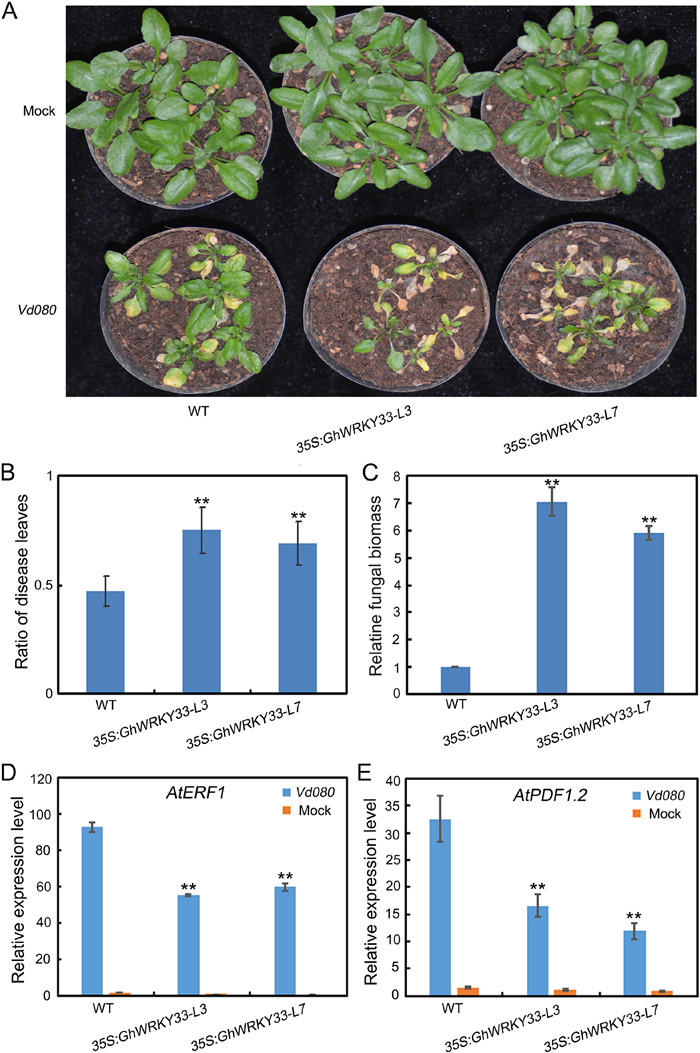
|
| Fig. 2 GhWRKY33 over-expressed Arabidopsis reduces resistance to V. dahliae. (A) Disease symptom development. The Verticillium wilt phenotypes were identified and photographed 21 days after inoculation. (B) Disease index for the indicated genotypes. (C) Fungal biomass upon inoculation with V. dahliae. (D and E) Expression of AtERF1 and AtPDF1.2 in GhWRKY33 over-expressed Arabidopsis plants and WT after inoculation with V. dahliae. For B-E, values are mean ± SD of three independent biological replicates (**P < 0.01). |
To understand the potential mechanisms of GhWRKY33-mediated Verticillium wilt resistance, we determined the expression levels of several JA signaling pathway-associated genes in these plants after inoculation with water of V. dahliae. These genes included Arabidopsis ETHYLENE RESPONSE FACTOR 1 (AtERF1) and Arabidopsis PLANT DEFENSIN gene (AtPDF1.2). As expected, the expression of both AtERF1and AtPDF1.2 was significantly up-regulated upon infection by V. dahliae in all plants, however, their expression in GhWRKY33-overexpressing lines was dramatically lower than that of WT (Fig. 2D and E). Thus, these results demonstrated that the JA signaling pathway-associated genes were down-regulated in GhWRKY33-overexpressing plants, implying that GhWRKY33 may negatively regulate plant defense against V. dahliae.
3.3. GhWRKY33 inhibits the expression of both AtERF1 and GhERF2WRKY proteins have been revealed to perform their distinct roles by binding directly to the W-box (T/CTGACC/T) in the promoters of their target genes (Ülker and Somssich, 2004; Chen et al., 2017). The above work indicated that GhWRKY33 might function as a critical regulator during plant–pathogen interactions by negatively modulating JA signaling pathway-associated genes. Thus, we speculated that GhWRKY33 may bind to the W-box elements in the promoter of AtERF1. To confirm the negative regulatory functions of GhWRKY33, the luciferase (LUC) reporter approach was first employed to analyze the effects of GhWRKY33 in Arabidopsis mesophyll protoplasts. Here we also analyze the possible regulation of Gossypium ETHYLENE RESPONSE FACTOR 2 (GhERF2), a homologous gene of AtERF1. Then both the promoters of AtERF1 and GhERF2were fused with LUC gene as reporters (AtERF1:LUC and GhERF2:LUC) (Fig. 3A). At the same time, the full-length CDS of GhWRKY33 was ligated with the CaMV35S promoter as an effector (Fig. 3A). Co-expression of reporter with effector plasmid in Arabidopsis mesophyll protoplasts led to the repression of LUC compared with the control (Fig. 3B and C). This result indicates that GhWRKY33 can inhibit the expression of both AtERF1and GhERF2.
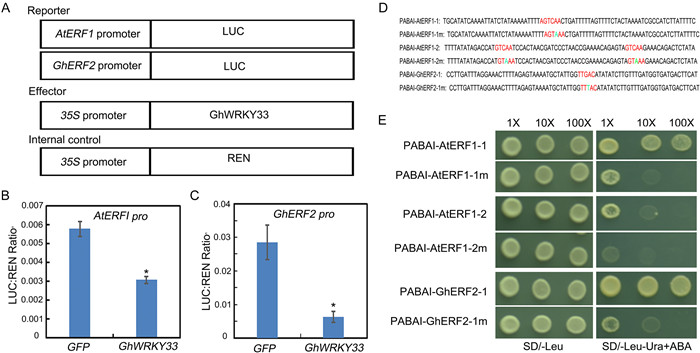
|
| Fig. 3 GhWRKY33 negatively regulates AtERF1 and GhERF2 expression. (A) Diagrammatic drawing of the reporters and effector utilized in the test. (B and C) Transient dual-LUC reporter assay shows inhibition of AtERF1 and GhERF2 expression by GhWRKY33. Values are mean ± SD of three independent biological replicates (*P < 0.05). (D) Schematic diagram of the AtERF1 and GhERF2 promoters for yeast one-hybrid hybridization experiments, showing the W-BOX(TTGAC) as well as the W-box base mutations (TTTAC) in the promoter. (E) GhWRKY33 binds to the TTGAC motif on the promoters of AtERF1 and GhERF2. The experiment was carried out three times and all yielded similar results, with representative results depicted in the figure. |
To examine whether GhWRKY33 can bind the W-boxes of AtERF1 and GhERF2, yeast one-hybrid approach was utilized. Three copies of the W-box motif and their mutant forms (TGAC mutated to TTAC) were synthesized and inserted into the pBait-AbAi plasmid (Fig. 3D) and the CDS of GhWRKY33 was fused with the pGADT7 vector. As shown in Fig. 3E, GhWRKY33 could bind to the W-box motifs in the promoters of AtERF1 and GhERF2, but could not bind to their mutant forms. These results indicate that GhWRKY33 may inhibit the expression of both AtERF1 and GhERF2 through targeting their promoters.
3.4. Physical interaction between GhWRKY33 and GhJAZ3To clarify how GhWRKY33 modulates V. dahliae resistance, we used the yeast two-hybrid system to determine its interaction partners. After screening and sequence analysis, GhJAZ3 was identified as potential interaction partner of GhWRKY33 (Fig. 4A). We further determine their interaction by Luciferase complementation imaging (LCI). For LCI analysis, cLUC/nLUC, cLUC/GhJAZ3-nLUC, GhWRKY33-cLUC/nLUC, and GhWRKY33-cLUC/GhJAZ3-nLUC were co-injected into young leaves of N. benthamiana and the LUC signals were observed 48 h after injection. As shown in Fig. 4B, LUC signal was only detected when GhWRKY33-cLUC/GhJAZ3-nLUC was co-expressed in N. benthamiana leaves. These results suggest that GhWRKY33 physically interacts with GhJAZ3.
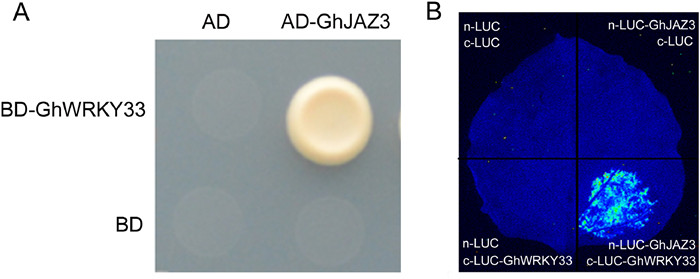
|
| Fig. 4 Physical interaction between GhWRKY33 and GhJAZ3. (A) Yeast two-hybrid assay. Co-transformed yeast cells with AD-GhJAZ3 and BD-GhWRKY33 were able to thrive on synthetic dropout media lacking Leu, Trp, His, and Ade. The activation domain (AD) of Gal4 was employed as a negative control. The experiment was carried out three times and all yielded similar results, with representative results depicted in figure. (B) Luciferase complementation imaging (LCI) assay. The fluorescence resulted from co-expressed GhWRKY33-cLUC/GhJAZ3-nLUC in Nicotiana benthamiana leaf shows that GhWRKY33 can bind to GhJAZ3. The experiment was carried out three times and all yielded similar results, with representative results depicted in the figure. |
The above results show that GhWRKY33 can interact with GhJAZ3, implying that GhJAZ3 may participate in plant disease resistance through the JA pathway. Then, we conduct expression analysis of GhJAZ3. As shown in Fig. 5A and Fig. 5B, GhJAZ3 was induced by V. dahliae infection, and also strongly induced by JA. Furthermore, GhJAZ3 is also localized in nucleus (Fig. 5C). These results imply that GhWRKY33 may function together with GhJAZ3 to participate in plant defense against V. dahliae.
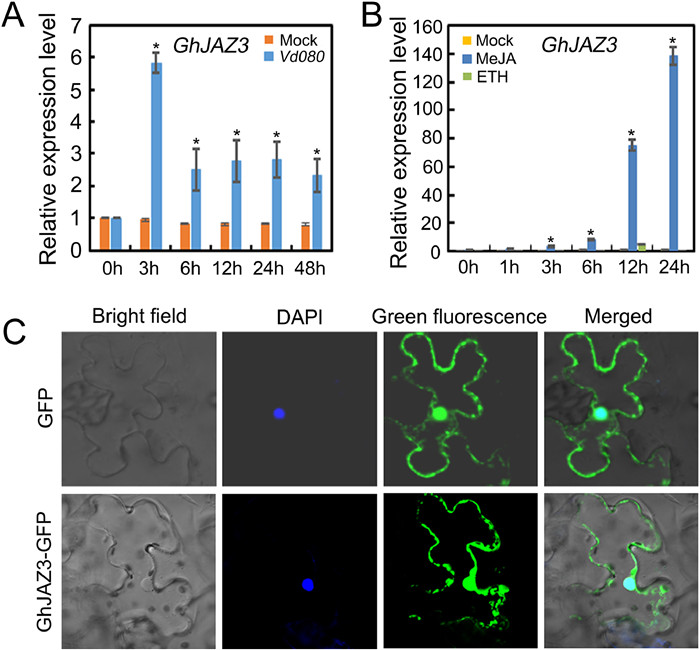
|
| Fig. 5 Expression and subcellular localization of GhJAZ3. (A) Expression of GhJAZ3 upon infection by V. dahliae in cotton. (B) Expression of GhJAZ3 in response to MeJA and ET treatment in cotton. For A and B, values are mean ± SD of three independent biological replicates (*P < 0.05). (C) Subcellular localization of GhJAZ3. Free GFP was found in both the nucleus and the cytoplasm, whereas GhJAZ3-GFP (GhJAZ3-green fluorescent protein) was only found in the nucleus. The nucleus is stained with 4′, 6-Diamidino-2-phenylindole (DAPI). |
To further confirm its role in defense against V. dahliae, we then generated transgenic Arabidopsis plants heterologously expressing GhJAZ3 driven by CaMV35S promoter (35S: GhJAZ3-L4 and 35S: GhJAZ3-L9) (Table S2). When homozygous GhJAZ3-transgenic lines (T3) were obtained, the wild-type, 35S: GhJAZ3-L4 and 35S: GhJAZ3-L9 transgenic Arabidopsis plants were simultaneously inoculated with V. dahliae. GhJAZ3-overexpressing plants displayed more serious disease symptoms than WT two weeks after inoculation (Fig. 6A); this was correlated with elevated ratio of disease leaves, higher fungal biomass, and lower expression of AtPDF1.2 in GhJAZ3-overexpressing plants (Fig. 6B–D). Thus, these results imply that GhJAZ3 may negatively regulate plant defense against V. dahliae.
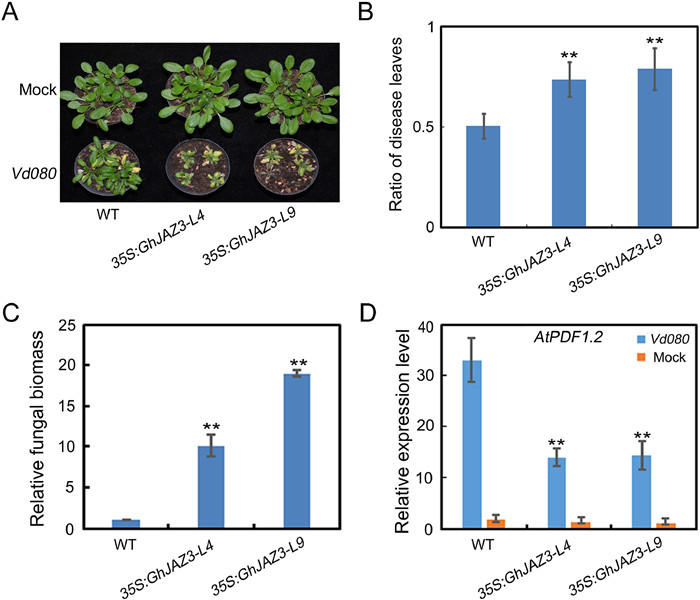
|
| Fig. 6 GhJAZ3 over-expressed Arabidopsis reduces resistance to V. dahliae. (A) Disease symptom development. The Verticillium wilt phenotypes were identified and photographed 21 days after inoculation. (B) Disease index for the indicated genotypes. (C) Fungal biomass upon inoculation with V. dahliae. (D) Expression of AtPDF1.2 in GhJAZ3 over-expressed Arabidopsis plants and WT after inoculation with V. dahliae. For C and D, values are mean ± SD of three independent biological replicates (**P < 0.01). |
Having demonstrated that GhJAZ3 physically interacts with GhWRKY33, we speculated that it might affect GhWRKY33's transcriptional function. We use LUC reporter approach to determine GhJAZ3's effect on the transcriptional activity of GhWRKY33. AtERF1:LUC and GhERF2:LUC were again used as reporters. We also generated effector constructs that contained either a GFP, GhWRKY33, or GhJAZ3 gene driven by the CaMV35S promoter (35S: GFP, 35S: GhWRKY33, and 35S: GhJAZ3) (Fig. 7A). As shown in Fig. 7B–E, the expression of GhWRKY33 or GhJAZ3 resulted in reduced LUC signals compared with the reporters alone. Interestingly, coexpression of GhWRKY33 with GhJAZ3 further reduced the LUC signals compared with the expression of GhWRKY33 or GhJAZ3 alone (Fig. 7B–E). These results support the hypothesis that GhJAZ3 acts synergistically with GhWRKY33 to suppress AtERF1 and GhERF2 expression.
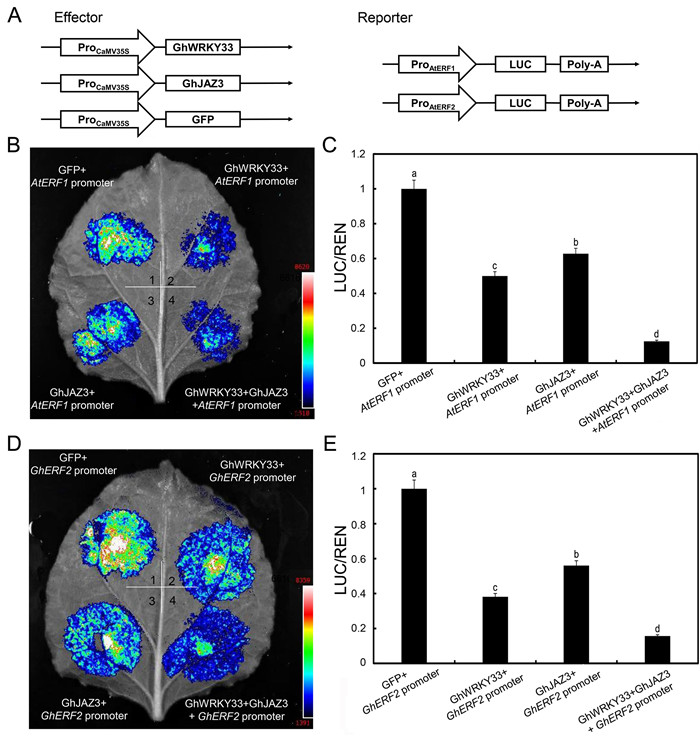
|
| Fig. 7 GhJAZ3 acts synergistically with GhWRKY33 to suppress AtERF1 and GhERF2 expression. (A) Diagrammatic drawing of the reporters and effector utilized in the test. (B–E). GhJAZ3 acts synergistically with GhWRKY33 to inhibit AtERF1 and GhERF2 transcription in Nicotiana benthamiana transient expression assays. Images of representative leaves are shown. Values are mean ± SD of three independent biological replicates (one-way ANOVA, P < 0.05). |
Verticillium dahliae, a destructive soil-borne vascular pathogen, causes a great threat to many crops worldwide, such as cotton, tomato, and sunflower (Klosterman et al., 2009). Typical V. dahliae symptoms, such as progressive leaf necrosis and severe growth stunting, are consistent with a switch from a biotrophic to a necrotrophic lifestyle (Dhar et al., 2020; Fradin et al., 2009). Although substantial progress in functional research on cotton and V. dahliae interaction has been achieved over the past several years, the underlying mechanisms related to their interaction are still poorly understood. Previous studies have suggested that certain transcription factors can incorporate with phytohormone signaling pathways to participate in cotton defense response to V. dahliae (Dhar et al., 2020).
Due to the sessile lifestyle of plants, plants have evolved various adaptive mechanisms to activate proper immune responses to ensure plant survival upon various pathogen attacks. Transcriptional regulatory networks play critical roles among these processes. Interestingly, the WRKY transcription factor family has been demonstrated to be widely involved in transcriptional reprogramming associated with both plant growth and stress responses (Chen et al., 2012, 2017; Rushton et al., 2010; Wang et al., 2019; Zhang et al., 2022). Although numerous studies have revealed that WRKY TFs play essential roles in various defense responses, their specific biological functions in these processes remain elucidated. Especially, the functional elucidation of WRKY TFs in cotton remains to be a major challenge. Several WRKY members in cotton, including GbWRKY1, GhWRKY22, and GhWRKY70, have been shown to play important roles in defense against the pathogen V. dahliae (Li et al., 2014; Xiong et al., 2019; Zhang et al., 2016). Recently, AtWRKY33 was also shown to participate in plant defense responses against V. dahliae toxins by directly binding to the NADPH oxidase gene RbohD and activateing RbohD-dependent hydrogen peroxide signaling(Zhao et al., 2020). Here, we found that GhWRKY33 is strongly induced by V. dahliae and MeJA (Fig. 2) and transgenic Arabidopsis plants overexpressing GhWRKY33 exhibit enhanced susceptibility to V. dahliae (Fig. 1). These results implied that GhWRKY33 may negatively regulate JA-mediated plant defense to the pathogen V. dahliae. Interestingly, contrary to our results, it was shown that virus-induced gene silencing of GhWRKY33 leaded to enhanced susceptibility of resistant cotton to V. dahliae (Zhang et al., 2016). We speculate that GhWRKY33 may have distinctive roles in different species, and it is still necessary to further determine its role in defense against V. dahliae using cotton Ghwrky33 mutants.
WRKY genes respond differentially to various environmental stresses and their proteins subsequently execute their transcriptional activating or repressing activity to enable their specific roles. Genetic and molecular biological studies have revealed that WRKY proteins function positively or negatively to regulate plant defense responses against various pathogens (Pandey and Somssich, 2009). As shown in Fig. 2, GhWRKY33 responded strongly to both JA and V. dahliae. Thus, its protein can accumulate and subsequently regulated potential target genes. Based on our Y1H results, GhWRKY33 may directly associate with the W-box elements in the promoters of both AtERF1 and GhERF2 (Fig. 3B), indicating that both AtERF1 and GhERF2 may be direct targets of GhWRKY33. The opposite expression patterns of GhWRKY33 and AtERF1 or GhERF2 in GhWRKY33 overexpression lines (Figs. 1 and 3), and the reduced expression of LUC as showed in the transient expression assays (Fig. 3), further suggest that GhWRKY33 is a negative regulator of both AtERF1 and GhERF2. These results further indicated that GhWRKY33 may negatively regulate JA-mediated defense responses to V. dahliae.
The phytohormone JA is required for the regulation of multiple physiological processes, such as anthocyanin accumulation, root growth, plant fertility, and defense responses (Pauwels and Goossens, 2011). It's well-known that JAZ proteins act as inhibitors in JA signaling. They physically interact with various transcription factors to inhibit the JA signaling pathway under favorable conditions. When plants are subjected to biotic and abiotic stresses, the JA content is increased. After COI1 senses the stimulation of JA, under the action of the SCFCOI1 complex, the JAZ protein is ubiquitinated and degraded by the 26S proteasome pathway. Their degradation contributes to the releasement of their downstream transcription factors, leading to the activation of JA signaling (Pauwels and Goossens, 2011). Several types of TFs have been reported as targets of JAZ proteins, such as MYB TFs, bHLH TFs, and EINs, to positively or negatively regulate diverse JA responses (Kazan and Manners, 2012). In cotton, GhJAZ2 can target GhbHLH171 to inhibit its transactivation activity and they antagonistically regulate cotton tolerance to V. dahliae (He et al., 2018). Recently, several WRKY proteins were also identified as targets of JAZs in Arabidopsis. For example, AtWRKY57 interacts with AtJAZ4/8 to negatively modulate JA-mediated Leaf Senescence (Jiang et al., 2014). Similarly, AtWRKY75 can also interact with AtJAZ4/8 to positively modulate JA-mediated defense to necrotrophs (Chen et al., 2020). However, little information is available about the possible interaction between GhWRKYs and GhJAZs and their involvement in cotton and V. dahliae interaction. In this study, we found that GhJAZ3 can interact with GhWRKY33 (Fig. 4). Similar toGhWRKY33, GhJAZ3 is induced by V. dahliae infection and is also induced by exogenous MeJA application. Furthermore, GhJAZ1 also inhibits the transcription of AtERF1, GhERF2, and GhPDF1.2 and functions as an inhibitor in cotton resistance to V. dahliae. Further analysis using LUC assay demonstrated that GhJAZ3 enhances the transrepression activity of GhWRKY33 and subsequently synergistically represses the expression of AtERF1 and GhERF2 and finally negatively modulates JA-mediated defense response (Fig. 7). Thus, our results indicated that JAZ-targeted GhWRKY33 may negatively modulate plant tolerance to V. dahliae by directly targeting JA-associated genes such as AtERF1 and GhERF2.
Recent studies have identified several critical components that control Verticillium wilt resistance in plants. Ve was identified as an important disease resistance gene in tomato, and further studies demonstrated that overexpression of Ve can increase tolerance to V. dahliae in both Arabidopsis and tomato (Fradin et al., 2009). Similarly, GbVe and GbVe1, homologous genes of tomato Ve, also positively regulate plant disease resistance to V. dahliae (Zhang et al., 2011). Interestingly, studies have demonstrated that Ve1-mediated resistance to V. dahliae required JA signaling in both Arabidopsis and tomato (Fradin et al., 2011). Considering the involvement of GhWRKY33 and GhJAZ1 in JA-mediated defense against V. dahliae, GhWRKY33 and GhJAZ1 may also participate in Ve1-mediated resistance to V. dahliae. Overall, we revealed that GhWRKY33 may negatively modulate plant tolerance to V. dahliae through its coordination with JA signaling.
AcknowledgementsThis work was supported by the National key R & D plan (2016YFD0101006), and Yunnan Fundamental Research Projects (2019FA010).
Author contributions
Y.J., M.M., H.Z., R.W., S.W., Y.J., H.Z., L.L., and Z.L. performed the research. L.C. designed the experiments, supervised the study, drafted, and revised the manuscript. All authors read and approved of its content.
Data availability statement
All datasets generated for this study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.04.001.
Bacete, L., Mélida, H., Miedes, E., et al., 2018. Plant cell wall mediated immunity: cell wall changes trigger disease resistance responses. Plant J., 93: 614-636. DOI:10.1111/tpj.13807 |
Chen, L.G., Song, Y., Li, S.J., et al., 2012. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta-Gene Regul. Mech, 1819: 120-128. DOI:10.1016/j.bbagrm.2011.09.002 |
Chen, L.G., Xiang, S.Y., Chen, Y.L., et al., 2017. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant, 10: 1174-1189. DOI:10.1016/j.molp.2017.07.008 |
Chen, L.G., Zhang, L.P., Xiang, S.Y., et al., 2020. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot., 72: 1473-1489. |
Chen, L.G., Zhang, L.P., Yu, D.Q., 2010. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant Microbe Interact., 23: 558-565. DOI:10.1094/MPMI-23-5-0558 |
Dhar, N., Chen, J.Y., Subbarao, K.V., et al., 2020. Hormone signaling and its interplay with development and defense responses in Verticillium-plant interactions. Front. Plant Sci., 11: 584997. DOI:10.3389/fpls.2020.584997 |
Fradin, E.F., Abd-El-Haliem, A., Masini, L., et al., 2011. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol., 156: 2255-2265. DOI:10.1104/pp.111.180067 |
Fradin, E.F., Zhang, Z., Juarez Ayala, J.C., et al., 2009. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol, 150: 320-332. DOI:10.1104/pp.109.136762 |
Glazebrook, J., 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol., 43: 205-227. DOI:10.1146/annurev.phyto.43.040204.135923 |
He, X., Zhu, L.F., Wassan, G.M., et al., 2018. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol., 19: 896-908. DOI:10.1111/mpp.12575 |
Hu, Y., Jiang, Y.J., Han, X., et al., 2017. Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot., 68: 1361-1369. DOI:10.1093/jxb/erx004 |
Huang, H., Liu, B., Liu, L.Y., et al., 2017. Jasmonate action in plant growth and development. J. Exp. Bot., 68: 1349-1359. DOI:10.1093/jxb/erw495 |
Jiang, Y.J., Liang, G., Yang, S.Z., et al., 2014. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell, 26: 230-245. DOI:10.1105/tpc.113.117838 |
Jones, J.D.G., Dangl, J.L., 2006. The plant immune system. Nature, 444: 323-329. DOI:10.1038/nature05286 |
Kazan, K., Manners, J.M., 2012. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci., 17: 22-31. DOI:10.1016/j.tplants.2011.10.006 |
Klosterman, S.J., Atallah, Z.K., Vallad, G.E., et al., 2009. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol., 47: 39-62. DOI:10.1146/annurev-phyto-080508-081748 |
Li, C., He, X., Luo, X.Y., et al., 2014. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium Dahliae by activating JasmonateZim-Domain1 expression. Plant Physiol., 166: 2179-2194. DOI:10.1104/pp.114.246694 |
Li, J., Brader, G., Kariola, T., et al., 2006. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J., 46: 477-491. DOI:10.1111/j.1365-313X.2006.02712.x |
Pandey, S.P., Somssich, I.E., 2009. The role of WRKY transcription factors in plant immunity. Plant Physiol, 150: 1648-1655. DOI:10.1104/pp.109.138990 |
Pauwels, L., Goossens, A., 2011. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell, 23: 3089-3100. DOI:10.1105/tpc.111.089300 |
Pieterse, C.M.J., Van der Does, D., Zamioudis, C., et al., 2012. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol., 28: 489-521. DOI:10.1146/annurev-cellbio-092910-154055 |
Rushton, P.J., Somssich, I.E., Ringler, P., et al., 2010. WRKY transcription factors. Trends Plant Sci., 15: 247-258. DOI:10.1016/j.tplants.2010.02.006 |
Ulker, B., Somssich, I.E., 2004. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol., 7: 491-498. DOI:10.1016/j.pbi.2004.07.012 |
Wang, H.P., Pan, J.J., Li, Y., et al., 2016. The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol., 172: 479-488. DOI:10.1104/pp.16.00891 |
Wang, N.N., Xu, S.W., Sun, Y.L., et al., 2019. The Cotton WRKY Transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci. Rep., 9: 1-13. DOI:10.1038/s41598-018-37186-2 |
Xiong, X.P., Sun, S.C., Li, Y.J., et al., 2019. The cotton WRKY transcription factor GhWRKY70 negatively regulates the defense response against Verticillium dahliae. Crop J., 7: 393-402. DOI:10.1016/j.cj.2018.10.005 |
Xu, X.P., Chen, C.H., Fan, B.F., et al., 2006. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell, 18: 1310-1326. DOI:10.1105/tpc.105.037523 |
Yang, Y.W., Chen, T.Z., Ling, X.T., et al., 2018. Gbvdr6, a gene encoding a receptor-like protein of cotton (Gossypium barbadense), confers resistance to Verticillium wilt in Arabidopsis and Upland Cotton. Front. Plant Sci., 8: 2272. DOI:10.3389/fpls.2017.02272 |
Yu, J., Jung, S., Cheng, C.H., et al., 2021. CottonGen: the community database for cotton genomics, genetics, and breeding research. Plants, 10: 2805. DOI:10.3390/plants10122805 |
Zhang, B.L., Yang, Y.W., Chen, T.Z., et al., 2012. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One, 7: e51091. DOI:10.1371/journal.pone.0051091 |
Zhang, H.Y., Zhang, L.P., Ji, Y.R., et al., 2022. Arabidopsis SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot., 73: 182-196. DOI:10.1093/jxb/erab391 |
Zhang, H.Y., Zhang, L.P., Wu, S.G., et al., 2021. AtWRKY75 positively regulates age-triggered leaf senescence through gibberellin pathway. Plant Divers., 43: 331-340. DOI:10.1016/j.pld.2020.10.002 |
Zhang, L., Zhang, F., Melotto, M., et al., 2017. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot., 68: 1371-1385. |
Zhang, W.W., Zhang, H.C., Qi, F.J., et al., 2016. Generation of transcriptome profiling and gene functional analysis in Gossypium hirsutum upon Verticillium dahliae infection. Biochem. Biophys. Res. Commun., 473: 879-885. DOI:10.1016/j.bbrc.2016.03.143 |
Zhang, Y., Wang, X.F., Yang, S., et al., 2011. Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep., 30: 2085-2096. DOI:10.1007/s00299-011-1115-x |
Zhang, Y., Wang, X.F., Ding, Z.G., et al., 2013. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics, 14: 637. DOI:10.1186/1471-2164-14-637 |
Zhao, J., Chen, Q.H., Zhou, S., et al., 2020. H2Bub1 regulates RbohD-dependent hydrogen peroxide signal pathway in the defense responses to Verticillium dahliae Toxins. Plant Physiol., 182: 640-657. DOI:10.1104/pp.19.00913 |
Zheng, Z.Y., Qamar, S.A., Chen, Z.X., et al., 2006. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J., 48: 592-605. DOI:10.1111/j.1365-313X.2006.02901.x |



