b. Yunnan Key Laboratory for Wild Plant Resources, Kunming 650201, Yunnan, China;
c. University of Chinese Academy of Sciences, Beijing 100049, China;
d. Lijiang Forest Biodiversity National Observation and Research Station, Kunming Institute of Botany, Chinese Academy of Sciences, Lijiang 674100, Yunnan, China
Many species in genus Dendrobium (Orchidaceae) are used as medicinal and nutraceutical herbs in China and other Asian countries (Guo et al., 2020; Wang et al., 2020). The main active ingredients in these species are polysaccharides and alkaloids. Previous studies have shown that Dendrobium polysaccharide is complex composed of glucose, mannose, rhamnose, xylose and arabinose, having immunomodulatory and hepatoprotective properties (Xu et al., 2011; Yue et al., 2020), while alkaloids from several Dendrobium species have antioxidant and neuroprotective properties (Mou et al., 2021). In addition, several metabolites such as amino acids, flavonoids, bibenzyls and several trace elements in the Dendrobium stem have anti-mutagenesis and anti-cytotoxicity benefits (Ng et al., 2012; He et al., 2020). However, our understanding of the metabolic pathways that regulate the biosynthesis of these compounds in Dendrobium remains limited.
The biosynthesis of polysaccharides and alkaloids in Dendrobium species have partially characterized. The putative genes involved in polysaccharide biosynthesis in Dendrobium include glycosyltransferase genes (GTs), glycoside hydrolases (GHs), carbohydrate esterases (CEs), carbohydrate-binding modules (CBMs) and polysaccharide lyases (PLs) (He et al., 2015; Shen et al., 2017, Yuan et al., 2019). Phytochemical studies have revealed that Dendrobium alkaloids include sesquiterpene, imidazole, phthalide, pyrrolidine and indolizidine alkaloids, which have complicated chemical structures (Jiao et al., 2018; Ng et al., 2012). Transcriptome analyses have indicated that Dendrobium alkaloids are derived from the terpenoid-forming and indole pathway of the mevalonate (MVA) pathway, the methylerythritol phosphate (MEP) pathway, or the shikimate pathway (Wang et al., 2020), however, details of these biosynthetic pathways to synthesize alkaloids are not thoroughly studied. Furthermore, a series of genes related to these pathways are identified in Dendrobium species, including 1-deoxy-D-xylulose-5-phosphate synthase (DXS), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR), phosphomevalonate kinase (PMK), diphosphomevalonate decarboxylase (MVD). However, the link between polysaccharide and alkaloid biosynthesis in Dendrobium remains largely unknown.
Dendrobium species accumulate both polysaccharides and alkaloids in their stems, but polysaccharide and alkaloid contents differ significantly across species and at different stages of growth. For example, the polysaccharide content in the stem of D. officinale is higher than those in the stem of D. nobile, while the alkaloid content shows an opposite trend (Shen et al., 2017). Furthermore, polysaccharide and alkaloid contents change as the plants of Dendrobium mature. For example, in both D. officinale and D. moniliforme, the ratios of polysaccharide and alkaloid contents reverse as the plants mature (Yuan et al., 2019). In one-year-old stems of D. candidum, the polysaccharide and alkaloid contents are higher in autumn and winter than in spring and summer (Zheng et al., 2012). These age-related differences in polysaccharide and alkaloid contents provide an ideal opportunity to identify metabolites and enzymes involved in carbohydrate and alkaloid biosynthesis.
In this study, our objective was to identify the metabolites and key enzymes that regulate the biosynthesis of carbohydrates to secondary metabolites in Dendrobium nobile. For this purpose, we investigated the metabolomes and transcriptomes in the stems of D. nobile at two growth stages to identify differentially accumulated metabolites and differentially expressed genes. We then predicted a biosynthetic pathway of carbohydrates, alkaloids and other secondary metabolites in the stems of D. nobile.
2. Materials and methods 2.1. Plant materialsDendrobium stems contain higher levels of polysaccharides and alkaloids in autumn and winter than in spring and summer (Zheng et al., 2012). Thus, we harvested stem tissue from one-year-old (immature stem) and more than one-year-old (mature stem) plants of D. nobile from August to October (Fig. 1). Before sampling, the plants were grown in a greenhouse at the Kunming Institute of Botany (102°41'E, 25°01'N), Chinese Academy of Sciences, with an average of ambient temperature of 22 ℃/20 ℃ (day/night), 65% relative humidity, a photoperiod of 16 h/8 h (day/night), and 400 μmol m-2s-1 of photosynthetically active radiation. All stems were immediately frozen in liquid nitrogen and stored at -80 ℃. After separating the leaves and roots from the stems, the content of polysaccharide and total alkaloid were determined, and the metabolic and transcriptomic analysis were used to investigate carbohydrate and secondary metabolites biosynthesis. The samples at each growth stage had three biological repetitions.

|
| Fig. 1 Immature (DnS1) and mature (DnS2) stems of Dendrobium nobile. Scale bar = 1 cm. |
The polysaccharide content of Dendrobium nobile stems was determined by the phenol-sulfuric acid method (Yuan et al., 2019a). Briefly, 1g of stem powder was placed into a 500 ml distillation flask per sample and 200 ml distilled water was added. Then, the sample was heated and refluxed for 2 h. After filtered and diluted to 250 ml by distilled water, 2 ml of the solution was placed into a 15 ml centrifuge tube and 10 ml of ethanol was added. This solution was shaken and refrigerated for 1h, and then centrifuged at 4000 r·min-1 for 20 min. After discarding the supernatant, the precipitate was centrifuged with 8 ml of 80% ethanol again. The supernatant was removed, and the precipitate was dissolved in 5 ml of hot water. Then, 2 ml of sample solution, 1 ml of 5% phenol solution and 5 ml of concentrated sulfuric acid were added to a test tube, and shaken and placed in room temperature for 30 min. The absorbance of sample solution was measured at 485 nm using a UV-visible spectrophotometer with 2 ml of water as a blank. The test was performed in three times. The calibration curve was prepared from the glucose reference, and the regression equation was Y = 0.0054X + 0.066 (R2 = 0.9906). Where X was glucose content and Y was absorbance value. The polysaccharide content was calculated from the linear calibration function of the absorbance versus the corresponding concentration. Total alkaloid content was determined following the method of Yuan et al. (2019) with slight modifications. Briefly, 1g of stem powder was soaked in 8 ml of ammonia solution for 1 h and 50 ml of ethyl acetate was added for each sample. The sample was placed in 85℃ water bath and refluxed for 2 h. After the solution was filtered and vaporized, then residuum was diluted to 10 ml by methyl alcohol. Then, 2 ml of sample solution, 8 ml of dichloromethane, 5 ml of potassium hydrogen phthalate buffer (pH 4.5) and 2 ml of 0.04% bromocresol green solution were mixed, and was shaken for 3min. Then, the mixed solution was allowed to stand for 30 min to obtain the lower filtrate. Then, 1 ml of filtrate, 3 ml of dichloromethane and 1 ml of 0.01M NaOH anhydrous ethanol solution was added to a test tube and shaken. The absorbance of sample solution was measured at 621 nm using a UV-visible spectrophotometer with 2 ml of dichloromethane as a blank. The test was performed in parallel three times. The calibration curve was prepared from the dendrobine reference, and the equation of regression was Y = 37.707X + 0.0147 (R2 = 0.9999). Where X was the total alkaloid content and Y was the absorbance value. Total alkaloid content was calculated from the linear calibration function of the absorbance versus the corresponding concentration. For polysaccharide and total alkaloid contents, independent t-test were performed to detect significant differences between immature and mature stems.
2.3. Metabolite extraction, separation, detection, and identificationExtraction of metabolites from the stem of Dendrobium nobile followed De Vos et al. (2007) and Chen et al. (2013) with slight modifications. Briefly, the freeze-dried samples were crushed to powder (60 Hz, 2 min) with Retsch mixer mill (MM 400, Germany). To extract metabolites, 1 ml of extract solution (3:1 methanol/water, containing 1 ppm 2-L-chlorophenylalanine as internal standard) was added to 50 mg of each sample. Samples were then incubated at 4 ℃ on a shaker overnight. Following centrifugation (10,000×g, 15 min, 4 ℃), the supernatant was filtered through a 0.22-μm filter membrane and transferred to LC-MS vials before liquid chromatography-mass spectrometry (LC-MS) analysis.
Chromatographic separation was performed by ultra-high performance liquid chromatography (UHPLC) using EXIONLC AD System (AB Sciex, USA) with a Waters Acquity UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 μm; Waters Chromatography Division, USA). The mobile phase A and B were 0.1% formic acid (v/v) in water and 100% acetonitrile, respectively. The mobile phase was delivered at 400 μL/min, and the elution gradient (A: B) was 98:2 (v: v) at 0.5 min, 50:50 at 10 min, 5:95 at 13 min, and 98:2 at 15 min. The injection volume was 2 μL, and the temperatures of the column and auto-sampler were set to 40 ℃ and 4 ℃, respectively.
An AB SCIEX QTRAP 6500+ System (AB Sciex, USA) equipped with an IonDrive Turbo V electrospray ionization (ESI) interface and operated in both positive and negative ion mode, controlled by SCIEX Analyst Work Station Software (v.1.6.3, AB Sciex, USA), was applied for assay development with multiple reaction monitoring (MRM) model. The operation parameters were as follows: IonSpray Voltage as +5500 or -4500 V, Curtain Gas as 35 psi, temperature as 400 ℃, Ion Source Gas 1 as 60 psi, Ion Source Gas 2 as 60 psi.
2.4. Metabolome analysisThe Proteo Wizard MSconventer tool was used to convert MS raw data to the TXT format (Holman et al., 2014). Metabolites were identified by searching the mass-to-charge ratio (m/z) of parent (Q1) and daughter ions (Q3) and retention time (RT) in BIOTREE in-house database of Shanghai BIOTREE Biological Technology Co., Ltd (Shanghai, China). Relative concentrations of the metabolites were determined by peak area (mm2). SIMCA software (v.16.0.2, Sartorius Stedim Data Analytics AB, Sweden) was used to perform principal component analysis (PCA) and orthogonal projections to latent structures-discriminate analysis (OPLS-DA). Metabolites were considered to be differentially accumulated between immature and mature stems of D. nobile when the variable importance in projection (VIP) score was ≥ 1 and the P-value was < 0.05. Pathway topology analysis and enrichment analysis was conducted using MetaboAnalyst 5.0 (http://www.metaboanalyst.ca/) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/). Heatmaps were generated in pheatmap R package (https://cran.r-project.org/web/packages/pheatmap/).
2.5. RNA extraction, library construction and RNA-seqTotal RNA was extracted using TRIZOL reagent (TaKaRa, Japan) and its quality was detected by Agilent Bioanalyzer 2100 system (Agilent Technologies, CA). Sequencing libraries were generated with an Agilent Strand Specific RNA Library Preparation Kit (Agilent Technologies, CA) according to the manufacturer's recommendations. Briefly, mRNA was purified from total RNA using Oligo (dT) beads and then randomly fragmented to 200 nt. After first-strand cDNA and second-strand cDNA were synthesized, the ends were repaired, the tails were added, and the adaptors were ligated. The sequencing cDNA libraries were amplified via PCR. The library quality was assessed by Aglient Bioanalyzer 2100 system (Agilent Technologies, CA), and the library was sequenced with Illumina Hiseq 4000 platform (Illumina, USA).
2.6. Transcriptome analysisClean reads were obtained from raw data by removing adapters and low-quality reads. With approximately 75% of clean reads mapped to the reference genome of Dendrobium catenatum (GenBank Assembly ID ASM160598v2) using HISAT2 (https://daehwankimlab.github.io/hisat2/), the un-mapped reads were assembled using StringTie (https://ccb.jhu.edu/software/stringtie/) and annotated using HMMER based on Protein family (Pfam) database (http://pfam.xfam.org/). The reads were normalized to calculated FPKM values by StringTie (Pertea et al., 2015) to obtain gene expression level of each sample. PCA analysis was performed in gmodels R package (http://www.r-project.org/) and visualized by ggplot2 R package (http://had.co.nz/ggplot2/). DESeq2 (Love et al., 2014) was used to identify differentially expressed genes (DEGs) according to false discovery rate (FDR) < 0.05 and absolute value of fold change ≥ 1.5. Clusterprofile R package (Yu et al., 2012) and KEGG databases were employed to perform KEGG enrichment analysis based on DEGs.
2.7. Quantitative real-time PCR (qRT-PCR)To validate the accuracy of RNA-seq data, the genes involved in starch and sucrose metabolism (SPS), shikimate pathway (ADT) and phenylpropanoid biosynthesis (TAT) were selected for qRT-PCR analyses. The samples used in qRT-PCR analysis were the same as RNA-seq and performed on QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific, USA). Total RNA isolation, cDNA synthesis and qRT-PCR analysis followed the manufacturer's instructions for Eastep® Super Total RNA Extraction Kit, Eastep® RT Master Mix Kit and Eastep® qPCR Master Mix Kit (Promega, China), respectively. The β-actin gene was used as the internal control to calculate relative expression with the 2−ΔΔCT method (Livak and Schmittgen 2001). The primers used in this study are given in Table S1. Each sample was measured for three biological replicates. The results were analyzed in Microsoft Excel (2007) (Microsoft Corp., USA) and visualized using ggplot2 R package (http://had.co.nz/ggplot2/).
2.8. Pathway visualizationWe used Adobe Illustrator (v.2021, Adobe Inc., USA) to create the maps of the biosynthetic pathways for carbohydrates, alkaloids and other secondary metabolites of integrated enrichment maps, the content of differentially accumulated metabolites (DAMs) and DEGs. The heatmap of gene expression in the pathway was generated using pheatmap R package (https://cran.r-project.org/web/packages/pheatmap/).
2.9. Correlation analysis of metabolic and transcriptomic dataTo identify the candidate genes that regulate key biosynthetic pathways for carbohydrate, alkaloid, and other secondary metabolites, we searched for the correlations between genes and metabolites that were differentially expressed/accumulated between the stems of Dendrobium nobile at two growth stages. Matrices of differentially accumulated metabolites and differentially expressed genes were generated by reshape2 R package (https://cran.r-project.org/web/packages/reshape2/). Pearson correlation coefficients were calculated by psych R package (https://cran.r-project.org/web/packages/psych/). Metabolites and genes were considered to be significantly correlated when Pearson correlation coefficients were > 0.95 or < -0.95. We calculated degree of network and generated connection network with Cytoscape software (Shannon et al., 2003).
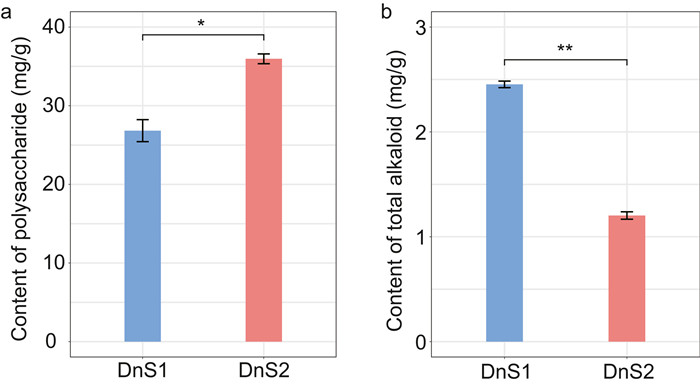
3. Results 3.1. Polysaccharide and alkaloid contentTo verify the changes in the main active components of Dendrobium nobile stems, we measured the contents of polysaccharide and total alkaloid at two growth stages (Figs. 2a and 2b). The polysaccharide and alkaloid contents varied greatly between mature and immature stems, indicating that metabolic activity shifted rapidly when the stems produced bioactive compounds. The polysaccharide content was higher in mature D. nobile stems (35.96 mg g-1) than in immature stems (26.83 mg g-1); in contrast, the total alkaloid content was lower in mature stems (1.20 mg g-1) than in immature stems (2.40 mg g-1). These results indicate that the polysaccharide and alkaloid contents of D. nobile have significances between mature and immature stems (Fig. 2).
|
| Fig. 2 Differences in polysaccharide and total alkaloid contents at two growth stages of Dendrobium nobile stem. a, polysaccharide content at the two growth stages. b, total alkaloid contents at two growth stages. The data are the means ± standard error (n = 6). The significantly changes in polysaccharide and total alkaloid contents between mature and immature stem were indicated by "*" (P < 0.05) and "**" (P < 0.01) according independent t-test, respectively. |
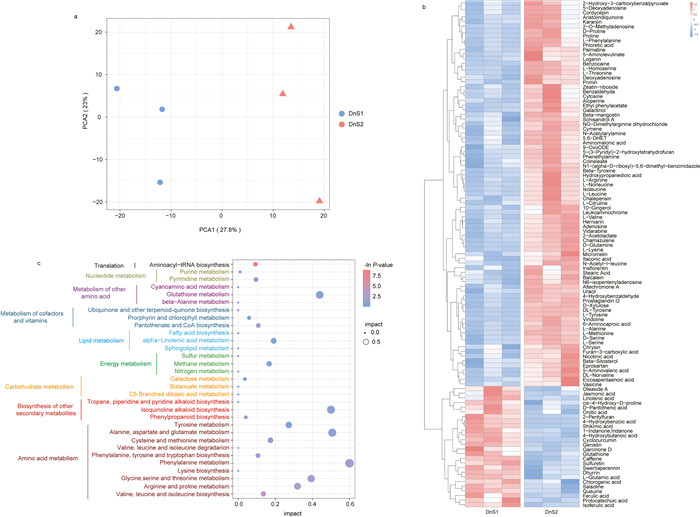
Metabolome analysis was conducted to identify metabolites differentially accumulated between mature and immature stems of Dendrobium nobile. A total of 1005 annotated metabolites were detected in the stems of D. nobile (Table S2). PCA divided the metabolites along PC1 (27.8%) and PC2 (23%) axes (Fig. 3a), suggesting that there was substantial variation in the metabolites of mature and immature stems of D. nobile. OPLS-DA identified 109 metabolites that were differentially accumulated between mature and immature stems (Fig. 3b). More than half of the differentially accumulated metabolites (DAMs) had higher levels in mature stems, indicating that a large number of metabolites were synthesized in D. nobile stems. KEGG enrichment analysis found that 31 pathways associated with differentially accumulated metabolites were enriched (enrichment factor > 1 and P-value < 0.1) (Fig. 3c). According to the secondary categorization of pathway type in KEGG database, these differentially accumulated metabolites were mainly involved in amino acid metabolism (e.g., phenylalanine metabolism, alanine, aspartate and glutamate metabolism), energy metabolism (e.g., methane metabolism), and biosynthesis of other secondary metabolites (e.g., isoquinoline alkaloid biosynthesis) (Fig. 3c).
|
| Fig. 3 Difference in the main metabolites of Dendrobium nobile at two growth stages. a, PCA analysis of total metabolites in D. nobile stem at two growth stages. b, heatmap of the differentially accumulated metabolites at two growth stages (VIP score were ≥ 1 and P-value were < 0.05 according OPLS-DA). Three columns of DnS1 and DnS2 represented three replicates. Color scale on the top right of the heat map represented normalized values of each identified metabolites content. c, bubble plot of KEGG enrichment of the differentially accumulated metabolites. The y-axis indicates the pathway names, and the x-axis indicates the impact score of topology analysis corresponding to the pathway. The P-value is represented by the color of the dot. The score of impact is represented by the size of the dot. Different colors of KEGG pathways represented different classes of pathways according to the secondary classification of KEGG Pathway Maps. DnS1, immature stems; DnS2, mature stems. |
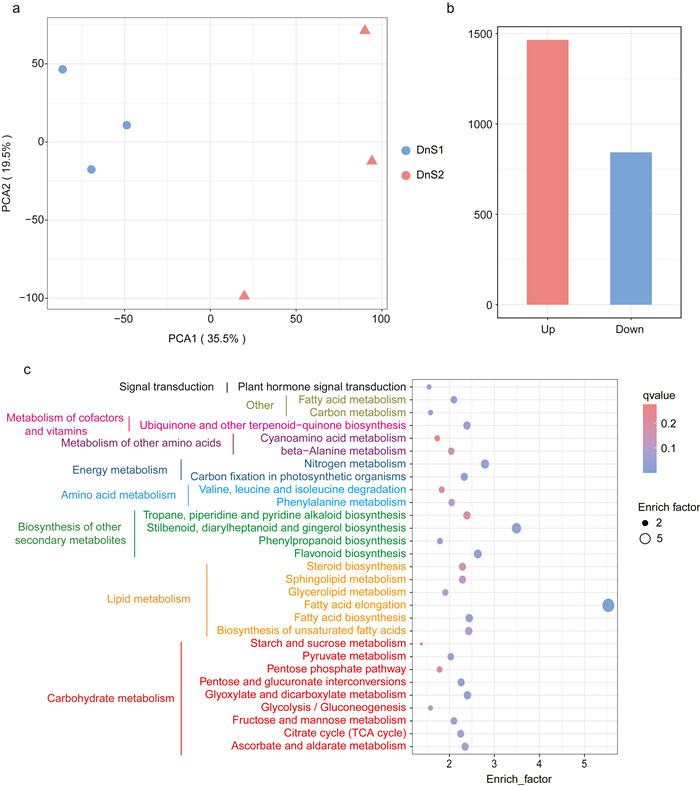
Transcriptome analysis was utilized to detect the global transcriptional alterations involved in carbohydrate and secondary metabolite biosynthesis in the mature and immature stems of Dendrobium nobile. A total of 31,745 genes were detected in the stems of D. nobile. PCA revealed the presence of two distinct transcriptomic clusters along the PC1 (35.5%) and PC2 (19.5%) axes (Fig. 4a). A total of 2308 genes were differentially expressed between mature and immature stems, with 1465 up-regulated and 843 down-regulated (Fig. 4b). To confirm these results, qRT-PCR was used to measure relative gene expression of three differentially expressed genes (SPS, ADT and TAT) involved in starch and sucrose metabolism, shikimate pathway and phenylpropanoid biosynthesis. The consistency of qRT-PCR and RNA-seq results (Fig. S1) suggested that the RNA-seq data in this study were reliable. KEGG enrichment analysis indicated that 29 pathways associated with differentially expressed genes were enriched (enrich factor > 1 and P-value < 0.1) (Fig. 4c). Most of the differentially expressed genes (DEGs) were involved in carbohydrate metabolism (e.g., starch and sucrose metabolism and pyruvate metabolism), lipid metabolism (e.g., steroid biosynthesis) and biosynthesis of other secondary metabolites (e.g., tropane, piperidine and pyridine alkaloid biosynthesis) (Fig. 4c).
|
| Fig. 4 Variation in transcriptome of Dendrobium nobile at two growth stages. a, PCA analysis of total genes expressed in D. nobile stems at two growth stages. b, differentially expressed genes (FDR were < 0.5, fold change were ≥ 1.5 or ≤ -1.5) at two growth stages. c, bubble plot of KEGG enrichment of the differentially expressed genes. The y-axis indicates pathway names, and the x-axis indicates the ratio of the number of genes in the pathway of the DEGs and all genes (enrich factor). The q-value is represented by the color of the dot. The score of enrich factor is represented by the size of the dot. Different colors of KEGG pathways represented different classes of pathways according to the secondary classification of KEGG Pathway Maps. DnS1, immature stems; DnS2, mature stems. |
The main bioactive components of Dendrobium species are polysaccharides. Our metabolome and transcriptome analyses showed that genes and metabolites differentially expressed or accumulated between mature and immature stems of D. nobile were enriched in the pathways related to carbohydrate metabolism. Construction of a carbohydrate biosynthesis pathway using DAMs and DEGs revealed that when D-xylulose levels increased in D. nobile stems, numerous genes involved in carbohydrate metabolism were detected (Fig. 5). We found that nine genes encoding enzymes (SPS, OTSA, GLGC, GLGA, TREP, BGLX, SORD, PFK9 and XYLB) were significantly up-regulated in mature stems, 18 genes (INV, EGLC, CELB, AMYA, GALM, GLPE, ACEE, DLD, PGK, PYK, ALDH, TIGAR, AKR1B, GMUG, TKTB, PMM, GMPP and ALDO) were down-regulated in mature stems.

|
| Fig. 5 Expression changes of genes involved in carbohydrate biosynthesis in the stems of Dendrobium nobile. Red letters represent up-regulated expression (FDR were < 0.5 and fold change were ≥ 1.5), green letters represent down-regulated expression (FDR were < 0.5 and fold change were ≤ -1.5), and black letters represent no significant change in metabolite accumulation and gene expression. The orange box and letters indicate a pathway. Heatmaps show the level of increase and decrease of enzyme gene expression. Color scale on the top left represented normalized values of FPKM values and six boxes of each enzyme gene represented three biological replicates of DnS1 and DnS2, respectively. SPS, sucrose-phosphate synthase; SPP, sucrose-6-phosphatase; SUS, sucrose synthase; UGP2, UTP-glucose-1-phosphate uridylyltransferase; INV, β-fructofuranosidase; BGLX, β-glucosidase; CELB, cellulase; CELS, cellulose synthase; GTP, glucose-1-phosphate guanylyltransferase; GAS, 1, 3-beta-glucan synthase; EGLC, glucan endo-1, 3-beta-D-glucosidase; PGT, phenol beta-glucosyltransferase; GLGC, glucose-1-phosphate adenylyltransferase; GLGA, starch synthase; GLGB, 1, 4-alpha-glucan branching enzyme; AMYA, α-amylase; MGAM, maltase-glucoamylase; ISA, isoamylase; MAPA, maltose phosphorylase; OTSA, trehalose 6-phosphate synthase; OTSB, trehalose 6-phosphate phosphatase; TREP, alpha, alpha-trehalose phosphorylase; ALDO, fructose-bisphosphate aldolase; PFP, diphosphate-dependent phosphofructokinase; PFK9, 6-phosphofructokinase; TIGAR, fructose-2, 6-bisphosphatase; FRUBP, fructose-2, 6-bisphosphate 6-phosphatase; DFPP, D-fructose 2-phosphate phosphatase; SORD, L-iditol 2-dehydrogenase; AKR1B, aldehyde reductase; MI, mannose isomerase; GMUG, mannan endo-1, 4-beta-mannosidase; GM, GDPmannose: mannan 1, 4-beta-D-mannosyltransferase; GMPP, mannose-1-phosphate guanylyltransferase; PMM, phosphomannomutase; MANA, mannose-6-phosphate isomerase; MANX, mannose PTS system EIIAB; KHK, ketohexokinase; TKTB, transketolase; XFP, xylulose-5-phosphate; XYLB, xylulokinase; GALM, aldose 1-epimerase; GLPE, glucose-6-phosphate 1-epimerase; GCK, glucokinase; PGM, phosphoglucomutase; GPI, glucose-6-phosphate isomerase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PGAM, 2, 3-bisphosphoglycerate-dependent phosphoglycerate mutase; ENO, enolase; PYK, pyruvate kinase; ACEE, pyruvate dehydrogenase E1; PDC, pyruvate decarboxylase; ALDH, aldehyde dehydrogenase; ACDA, acetate-CoA ligase subunit α; DLD, dihydrolipoamide dehydrogenase; DLAT, pyruvate dehydrogenase E2; DAK, dihydroxyacetone kinase. |
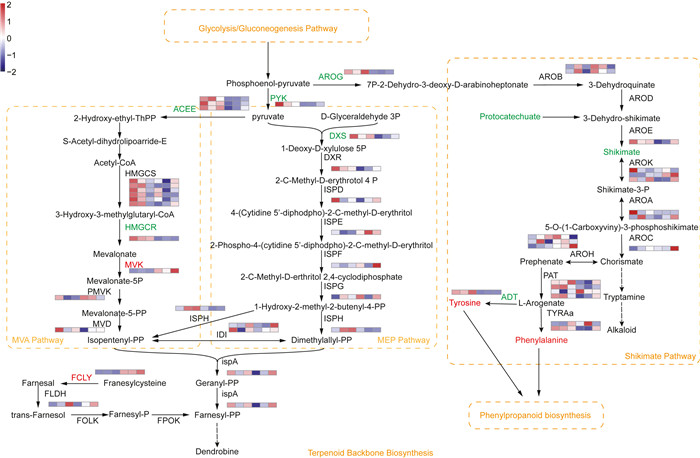
Alkaloids were another kind of important bioactive components in Dendrobium species. The main group of bioactive alkaloids in Dendrobium species are sesquiterpenoid or terpenoid indole alkaloids that initiate from the MVA, MEP, or shikimate pathways (Wang et al., 2020). In our study, metabolites involved in terpenoid backbone biosynthesis (i.e., the MVA and MEP pathways) were not differentially accumulated between mature and immature stems. However, we found that MVK and FCLY were expressed at higher levels in mature than in immature stems, whereas HMGCR and DXS were down-regulated (Fig. 6). Metabolites such as protocatechuate and shikimate were present at lower levels in mature stems; similarly, AROG and ADT were expressed at lower levels in these same stems. In contrast, tyrosine and phenylalanine were present at higher levels in mature stems. Furthermore, it was noticeable that the alkaloid biosynthesis was downstream of carbohydrate metabolism because the metabolites of phosphoenol-pyruvate (PEP) and pyruvate originate from the glycolysis/gluconeogenesis pathway (Fig. 6).
|
| Fig. 6 Alkaloid biosynthesis in the stems of Dendrobium nobile. Red letters represent up-regulated expression (FDR were < 0.5 and fold change were ≥ 1.5), green letters represent down-regulated expression (FDR were < 0.5 and fold change were ≤ -1.5), and black letters represent no significant change in metabolite accumulation or gene expression. The orange box and letters indicate a pathway. Heatmaps show the level of increase and decrease of enzyme gene expression. Color scale on the top left represented normalized values of FPKM values and six boxes of each enzyme gene represented three biological replicates of DnS1 and DnS2, respectively. Dashed arrows indicate unverified steps in the pathway. HMGCS, hydroxymethylglutaryl-CoA synthase; HMGCR, hydroxymethylglutaryl-CoA reductase; MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; MVD, diphosphomevalonate decarboxylase; ISPH, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; ISPD, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; ISPE, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; ISPF, 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate synthase; ISPG, 4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; IDI, isopentenyl-diphosphate Delta-isomerase; ISPA, farnesyl diphosphate synthase; FCLY, farnesylcysteine lyase; FOLK, farnesol kinase; FLDH, NAD+-dependent farnesol dehydrogenase; FPOK, farnesyl phosphate kinase; AROG, 3-deoxy-7-phosphoheptulonate synthase; AROB, 3-dehydroquinate synthase; AROD, 3-dehydroquinate dehydratase I; AROE, shikimate dehydrogenase; AROK, shikimate kinase; AROA, 3-phosphoshikimate-1-carboxyvinyltransferase; AROC, chorismate synthase; PAT, bifunctional aspartate aminotransferase and glutamate; TYRAa, arogenate dehydrogenase; ADT, arogenate/prephenate dehydratase; AROH, chorismate mutase. |
Secondary metabolites and genes that regulate their biosynthesis were significantly enriched in the stems of Dendrobium nobile (Figs. 3c and Fig. 4c). For example, phenylalanine and tyrosine, which play roles in both the shikimate and the phenylpropanoid biosynthesis pathways, were enriched. D-phenylalanine, phenylethylamine, 2-hydroxyphenlpropanoate, β-tyrosine and chrysin were accumulated at higher levels in mature stems than in immature stems; in contrast, 4-hydroxybenzoate, caffeoylquinic acid and ferulic acid were accumulated at lower levels in mature stems than in immature stems (Fig. 7). In addition, the gene expression of CYP73A, PAL, HCT and CYP98A was up-regulated, while the expression of TAT and CHS was down-regulated, and 4CL and TYNA expression was both up- and down-regulated (Fig. 7).

|
| Fig. 7 Regulatory network of other secondary metabolites in the stems of Dendrobium nobile. Red letters represent up-regulated expression (FDR were < 0.5 and fold change were ≥ 1.5), green letters represent down-regulated expression (FDR were < 0.5 and fold change were ≤ -1.5), blue letters represent both up- and down-regulated expression, and black letters represent no significant change in metabolite accumulation or gene expression. The orange box and letters indicate a pathway. Heatmaps show the level of increase and decrease of enzyme gene expression. Color scale on the top right represented normalized values of FPKM values and six boxes of each enzyme gene represented three biological replicates of DnS1 and DnS2, respectively. TAT, tyrosine aminotransferase; PPDC, phenylpyruvate decarboxylase; TYNA, primary amine oxidase; DDC, l-tryptophan decarboxylase; PAL, phenylalanine ammonialyase; TC2M, trans-cinnamate 2-monooxygenase; CR, 2-coumarate reductase; PAR, phenylalanine racemase; 4CL, 4-coumarate-CoA-ligase; CYP73A, trans-cinnamate 4-monooxygenase; PTAL, phenylalanine/tyrosine ammonia-lyase; HCT, shikimate hydroxycinnamoyltransferase; CYP98A, 5-O-(4-coumaroyl)-D-quinate 3'-monooxygenase; COMT, caffeic acid 3-O-methyltransferase; TAM, tyrosine 2, 3-aminomutase; TAT, tyrosine aminotransferase; HPPR, hydroxyphenylpyruvate reductase; 4HCT, 4-hydroxybenzoyl-CoA thioesterase; PAH, phenylalanine-4-hydroxylase; CHS, chalcone synthase; CHI, chalcone isomerase; FNSI, flavone synthase I. |
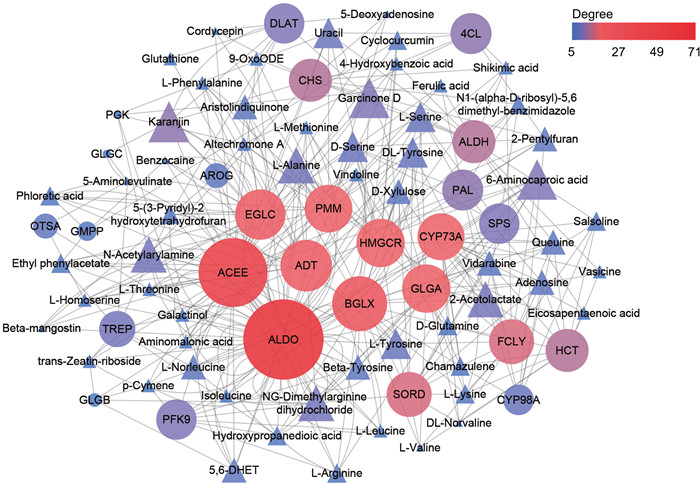
To identify the major enzymes involved the biosynthesis of bioactive metabolites in the stems of Dendrobium nobile, we examined correlations between metabolite accumulation and gene expression data. Enzymes with a degree of correlation greater than 20 with a metabolite was considered to play a key role in the relevant biosynthetic pathways. We found that six genes (e.g., ACEE, ALDO, EGLC, BGLX, PMM, GLGA) were correlated with carbohydrate biosynthesis, whereas two genes (ADT and HMGCR) were correlated with alkaloid biosynthesis. In addition, one gene (CYP73A) was correlated with biosynthesis of other secondary metabolites (Fig. 8). These putative genes involved in biosynthetic pathways in the stems of D. nobile were summarized in Fig. S2.
|
| Fig. 8 Correlations between carbohydrate, alkaloids and other secondary metabolites related differentially accumulated metabolites (triangles) and differentially expressed genes (circles) (Pearson correlation coefficients were > 0.95 or < -0.95, and P-value were < 0.01). The gray lines represent edges describing the interaction between metabolites and genes. Color scale on the top right and node size represented the weight of edges between them. Red circles represent key genes which degree value are top nine in the network. |
For over 2,000 years, Dendrobium species have been used as medical herbs in some Asian countries (Mou et al., 2021). The most well-known active ingredients in Dendrobium are polysaccharides and alkaloids (Yuan et al., 2019). Here, we predicted a biosynthetic pathway for carbohydrates and alkaloids in the stems of D. nobile. To construct these pathways, we first verified that polysaccharide and alkaloid levels differed between mature and immature stems of D. nobile. The total alkaloid content was lower in mature stems than in immature stems, while the polysaccharide content showed an opposite pattern. These differences allowed us to use metabolomic and transcriptomic approaches to identify metabolites and genes involved in biosynthetic pathways. Our biosynthetic pathway indicated that carbohydrate biosynthesis occurred the upstream of alkaloid biosynthesis, suggesting that carbohydrate and alkaloid metabolism were tightly linked.
One compound that appears to connect carbohydrate and alkaloid biosynthesis in Dendrobium is PEP, which is the last product of glycolysis/gluconeogenesis in carbohydrate biosynthesis and a substrate of alkaloid biosynthesis. Glycolysis transforms glucose to PEP, which is subsequently transferred by the rate-limiting enzymes AROG to 7P-2-Dehydro-3-deoxy-D-arabinoheptonate in the shikimate pathway and to pyruvate and ATP in the MEP pathway via the pyruvate kinase (PYK) (Ambasht and Kayastha 2002; Sato et al., 2006; Cai et al., 2018). The metabolite accumulation and gene expression in D. nobile indicated that total alkaloid content and the expression of PYK and AROG was lower in the mature stems. These findings suggested that several polysaccharides were transported from carbohydrate biosynthesis to alkaloids biosynthesis, part of them were controlled by the consumption of PYK and AROG to synthetize alkaloids. We also found that DXS, ACEE and HMGCR expression was lower in the mature stems. Previous studies have reported that these genes are crucial rate-limiting enzymes (e.g., DXS in the MEP pathway, ACEE and HMGCR in the MVA pathway); in addition, a positive correlation has been previously found between DXS expression levels and monoterpenoid and tetraterpenoid alkaloid content (Pan et al., 2019; Tian et al., 2022). Thus, these genes may play important rate-limiting roles in the transition from polysaccharide to alkaloid biosynthesis in D. nobile.
Previous studies have suggested that metabolism of fructose and mannose is active in Dendrobium officinale (Shen et al., 2017; Zhang et al., 2016). Here, the expression of genes involved in fructose, mannose, glucose, xylulose and starch metabolism differed between mature and immature stems of D. nobile (Fig. S2). We found that GLGA expression was higher in the mature stems than in immature stems, where it likely transfers glucose to starch (Pfister and Zeeman 2016). Our findings found that β-gulcanase (BGLX) expression level was higher in the mature stems than in immature stems in D. nobile, while endo-β-1, 3-glucanase (EGLC) expression levels were lower, this suggested that these genes might jointly regulate cell wall expansion in the stem, because of their key roles in cell wall expansion and in D-glucose synthesis in other species (Kang et al., 2019). In addition, xylulokinase (XYLB) expression levels were higher in mature stems. Because XYLB expression is selectively induced by xylulose (Liu et al., 2015), this finding suggested that xylulose content was higher in mature stems of D. nobile. Both phosphomannomutase (PMM) and the enzyme fructose-bisphosphate aldolase (ALDO) were expressed at lower levels in the mature stems of D. nobile. PMM catalyzes the interconversion of mannose-1-phosphate and mannose-6-phosphate in the production of GDP-mannose, which is required for cell metabolites (Yu et al., 2010, 2015). ALDO plays a central role in glycolysis pathway and is required for the conversion of fructose 1-6-diphosphate to glyceraldehyde-3-phosphate (Ziveri et al., 2017). This implies that PMM and ALDO might play important roles in mannose synthesis and glycolysis pathway in D. nobile.
Secondary metabolites not only play a vital role in plant defense against environmental stresses but also serve as therapeutic components for humans. According to our biosynthetic pathway, the shikimate pathway has an important role in alkaloid biosynthesis, and provides critical precursors for a wide range of secondary metabolites (Tohge et al., 2013). The final product of the shikimate pathway is tyrosine, which can be converted into an amino acid (such as β-tyrosine), hydroxycinnamic acid (such as ferulic acid), benzoic acid (such as 4-hydroxybenzoate), or flavone (such as chrysin). As the plants of D. nobile mature, tyrosine content increased, but the arogenate dehydrogenases (ADT) expression decreased. A previous study has showed that ADT expression is strongly inhibited by tyrosine in the growth of Arabidopsis thaliana (de Oliveira et al., 2019). Thus, the tyrosine in D. nobile might inhibited the expression of ADT. Meanwhile, it is likely converted to β-tyrosine, a component of vitamin B5 required for coenzyme A function as well as defense against biotic and abiotic stresses in plants (Jander et al., 2020). Our analyses identified an additional gene that involves in the metabolism of secondary metabolites. Trans-cinnamate 4-monooxygenase (CYP73A) was up-regulated in mature stems of D. nobile stem. It belongs to the cytochrome P450 family (P450s), which are key enzymes in secondary metabolite biosynthesis (Yuan et al., 2020). Studies in closely related orchid species (D. officinale and D. huoshanense) have also characterized the expression of CYP family genes (Guo et al., 2013; Yuan et al., 2019). Thus, we speculate that CYP73A plays a crucial role in secondary metabolites biosynthesis in D. nobile.
5. ConclusionsWe used transcriptome and metabolome analysis to identify metabolites and regulatory genes involved in carbohydrate and secondary metabolite biosynthesis, and predicted a regulatory network linked to these processes in the stems of D. nobile. The significant changes in the expression of genes involved in carbohydrate metabolism (i.e., ALDO, PMM, BGLX, EGLC, XYLB and GLGA) indicated that fructose, mannose, glucose, xylulose and starch were actively metabolized in the mature stems of D. nobile. We also found that polysaccharide and total alkaloid content were inversely related, with high polysaccharide and low alkaloid content in the mature stems of D. nobile. Our results indicated that the transition from carbohydrate to alkaloid metabolism was likely regulated by five genes, AROG, PYK, DXS, ACEE and HMGCR. Secondary metabolites, including amino acids, hydroxycinnamic acid, benzoic acid and flavones, appeared to be principally regulated by ADT and CYP73A, and were synthesized from the shikimate pathway. These findings provide important insights into the regulation of metabolites in Dendrobium species.
AcknowledgementsThis work is supported by the Project for Innovation Team of Yunnan Province (202105AE160012), the Project for Construction of International Flower Technology Innovation Center and Achievement Industrialization (2019ZG006), the Project for the Germplasm Bank of Wild Species, and the KC Wong Education Foundation, CAS, and the project for High-level Talent Training Plan of Yunnan Province.
Author contributions
YZ and SZ conceived the experiments, YZ and YS performed the experiments and analyzed the data. YZ wrote the paper, YZ and SZ revised the paper. All authors have read and approved the manuscript.
Data availability
The transcriptome datasets generated during the current study are available in the National Center for Biotechnology Information under the BioProject number PRJNA810608.
Declaration of competing interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.10.004.
Ambasht, P.K., Kayastha, A.M., 2002. Plant pyruvate kinase. Biol. Plant. (Prague), 45: 1-10. DOI:10.1023/A:1015173724712 |
Cai, Y.C., Li, S.F., Jiao, G.A., et al., 2018. OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnol. J., 16: 1878-1891. DOI:10.1111/pbi.12923 |
Chen, W., Gong, L., Guo, Z.L., et al., 2013. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant, 6: 1769-1780. DOI:10.1093/mp/sst080 |
de Oliveira, M.V.V., Jin, X., Chen, X., et al., 2019. Imbalance of tyrosine by modulating TyrA arogenate dehydrogenases impacts growth and development of Arabidopsis thaliana. Plant J., 97: 901-922. DOI:10.1111/tpj.14169 |
De Vos, R.C., Moco, S., Lommen, A., et al., 2007. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc., 2: 778-791. DOI:10.1038/nprot.2007.95 |
Guo, L.H., Qi, J.X., Du, D., et al., 2020. Current advances of Dendrobium officinale polysaccharides in dermatology: a literature review. Pharm. Biol., 58: 664-673. DOI:10.1080/13880209.2020.1787470 |
Guo, X., Li, Y., Li, C.F., et al., 2013. Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene, 527: 131-138. DOI:10.1016/j.gene.2013.05.073 |
He, C.M., Zhang, J.X., Liu, X.C., et al., 2015. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol., 88: 219-231. DOI:10.1007/s11103-015-0316-z |
He, L., Su, Q., Bai, L., et al., 2020. Recent research progress on natural small molecule bibenzyls and its derivatives in Dendrobium species. Eur. J. Med. Chem., 204: 1-17. DOI:10.1016/j.ejmech.2020.112530 |
Holman, J.D., Tabb, D.L., Mallick, P., 2014. Employing proteowizard to convert raw mass spectrometry data. Curr. Protoc. Bioinformatics, 46: 1-9. DOI:10.1002/0471250953.bi1324s46 |
Jander, G., Kolukisaoglu, U., Stahl, M., et al., 2020. Physiological aspects of non-proteinogenic amino acids in plants. Front. Plant Sci., 11: 1-3. DOI:10.3389/fpls.2020.519464 |
Jiao, C.Y., Song, C., Zheng, S.Y., et al., 2018. Metabolic profiling of Dendrobium officinale in response to precursors and methyl jasmonate. Int. J. Mol. Sci., 19: 728-747. DOI:10.3390/ijms19030728 |
Kang, L.Q., Zhou, J.S., Wang, R., et al., 2019. Glucanase-induced stipe wall extension shows distinct differences from chitinase-induced stipe wall extension of Coprinopsis cinerea. Appl. Environ. Microbiol., 85: e01345-19. DOI:10.1128/aem.01345-19 |
Liu, Y.H., Rainey, P.B., Zhang, X.X., 2015. Molecular mechanisms of xylose utilization by Pseudomonas fluorescens: overlapping genetic responses to xylose, xylulose, ribose and mannitol. Mol. Microbiol., 98: 553-570. DOI:10.1111/mmi.13142 |
Livak, K.J., Schmittgen, T.D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25: 402-408. DOI:10.1006/meth.2001.1262 |
Love, M.I., Huber, W., Anders, S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15: 550-571. DOI:10.1186/s13059-014-0550-8 |
Mou, Z.M., Zhao, Y., Ye, F., et al., 2021. Identification, biological activities and biosynthetic pathway of Dendrobium alkaloids. Front. Pharmacol., 12: 1-14. DOI:10.3389/fphar.2021.605994 |
Ng, T.B., Liu, J., Wong, J.H., et al., 2012. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol., 93: 1795-1803. DOI:10.1007/s00253-011-3829-7 |
Pan, X.H., Li, Y.T., Pan, G.T., et al., 2019. Bioinformatics study of 1-deoxy-d-xylulose-5-phosphate synthase (DXS) genes in Solanaceae. Mol. Biol. Rep., 46: 5175-5184. DOI:10.1007/s11033-019-04975-5 |
Pertea, M., Pertea, G.M., Antonescu, C.M., et al., 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol., 33: 290-295. DOI:10.1038/nbt.3122 |
Pfister, B., Zeeman, S., 2016. Formation of starch in plant cells. Cell. Mol. Life Sci., 73: 2781-2807. DOI:10.1007/s00018-016-2250-x |
Sato, K., Mase, K., Nakano, Y., et al., 2006. 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase is regulated for the accumulation of polysaccharide-linked hydroxycinnamoyl esters in rice (Oryza sativa L. ) internode cell walls. Plant Cell Rep., 25: 676-688. DOI:10.1007/s00299-006-0124-7 |
Shannon, P., Markiel, A., Ozier, O., et al., 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13: 2498-2504. DOI:10.1101/gr.1239303 |
Shen, C.J., Guo, H., Chen, H.L., et al., 2017. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep., 7: 187-198. DOI:10.1038/s41598-017-00292-8 |
Tian, S.K., Wang, D.D., Yang, L., et al., 2022. A systematic review of 1-Deoxy-D-xylulose-5-phosphate synthase in terpenoid biosynthesis in plants. Plant Growth Regul., 96: 221-235. DOI:10.1007/s10725-021-00784-8 |
Tohge, T., Watanabe, M., Hoefgen, R., et al., 2013. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci., 4: 1-13. DOI:10.3389/fpls.2013.00062 |
Wang, Z.C., Zhao, M.L., Cui, H.Q., et al., 2020. Transcriptomic landscape of medicinal Dendrobium reveals genes associated with the biosynthesis of bioactive components. Front. Plant Sci., 11: 391-401. DOI:10.3389/fpls.2020.00391 |
Xu, J., Guan, J., Chen, X.J., et al., 2011. Comparison of polysaccharides from different Dendrobium using saccharide mapping. J. Pharmaceut. Biomed., 55: 977-983. DOI:10.1016/j.jpba.2011.03.041 |
Yu, C.M., Li, Y.W., Li, B., et al., 2010. Molecular analysis of phosphomannomutase (PMM) genes reveals a unique PMM duplication event in diverse Triticeae species and the main PMM isozymes in bread wheat tissues. BMC Plant Biol., 10: 214-230. DOI:10.1186/1471-2229-10-214 |
Yu, C.M., Liu, X.Y., Zhang, Q., et al., 2015. Molecular genetic analysis of phosphomannomutase genes in Triticum monococcum. Crop J., 3: 29-36. DOI:10.1016/j.cj.2014.07.003 |
Yu, G., Wang, L.G., Han, Y., et al., 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16: 284-287. DOI:10.1089/omi.2011.0118 |
Yuan, Y.D., Yu, M.Y., Jia, Z.H., Song, X.E., Liang, Y.Q., Zhang, J.C., 2019a. Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genomics, 19: 978-984. DOI:10.1186/s12864-018-5305-6 |
Yuan, Y.D., Yu, M.Y., Zhang, B., Liu, X., Zhang, J.C., 2019b. Comparative nutritional characteristics of the three major Chinese Dendrobium species with different growth years. PLoS One, 14: e0222666. DOI:10.1371/journal.pone.0222666 |
Yuan, Y.D., Zhang, J.C., Liu, X., et al., 2020. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics, 112: 1781-1794. DOI:10.1016/j.ygeno.2019.10.010 |
Yue, H., Zeng, H., Ding, K., 2020. A review of isolation methods, structure features and bioactivities of polysaccharides from Dendrobium species. Chin. J. Nat. Med., 18: 1-27. DOI:10.1016/S1875-5364(20)30001-7 |
Zhang, J.X., He, C.M., Wu, K.L., et al., 2016. Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci., 7: 5-18. DOI:10.3389/fpls.2016.00005 |
Zheng, Y.P., Jiang, W., Silva, E.N., et al., 2012. Optimization of shade condition and harvest time for Dendrobium candidum plants based on leaf gas excachange, alkaloids and polysaccharides contents. Plant Omics, 5: 253-260. |
Ziveri, J., Tros, F., Guerrera, I.C., et al., 2017. The metabolic enzyme fructose-1, 6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat. Commun., 8: 853-868. DOI:10.1038/s41467-017-00889-7 |



