b. Plant Resources Center, Vietnam Academy of Agricultural Sciences, Hanoi, 100803, Vietnam;
c. Thailand Institute of Nuclear Technology, Bangkok, 10900, Thailand;
d. Pang U Bon Waterlily Garden, Muang Nonthaburi, Nonthaburi, 11000, Thailand;
e. Chinese Society of Landscape Architecture, Beijing, 100835, PR China;
f. Zhejiang Humanity Landscape Co., Ltd., Hangzhou, Zhejiang, 310013, PR China
Lotus (Nelumbo Adans.), a perennial aquatic angiosperm with a very long evolutionary history, has only two surviving species in the world, the Asian lotus (N. nucifera Gaertn.), native throughout Asia and northern Australia (Li et al., 2014), and the American lotus (N. lutea Willd.), distributed mainly in Central and North America (Hall and Penfound 1944). The Asian lotus is widely used as food, an ornamental plant, and medicine in Asia (Guo 2009; Sharma et al., 2016; Chen et al., 2019; Lin et al., 2019), especially in China, where it is one of the most economically important plants. According to its different agricultural uses, Nelumbo cultivars are usually divided into three types: ornamental lotus, rhizome lotus, and seed lotus (Wang and Zhang 2004; Guo 2009).
The wild Asian lotus is grouped into temperate and tropical types (Zhang and Wang 2006), or temperate, subtropical, and tropical types according to climate zone and morphological traits. In general, the temperate lotus, mainly located in East and Northeast Asia, has a short flowering period and obviously expanded rhizomes, whereas the tropical lotus distributed in Southeast and South Asia has a longer flowering period and thinner rhizomes (Zhang and Wang 2004, 2006; Li et al., 2010; Zhang et al., 2011; Liu et al., 2020b). Coincidentally, flowering time and rhizome growth are crucial traits for ornamental and rhizome lotus, respectively. Thus, the ever-flowering tropical lotus is a good material for creating new cultivars with longer flowering lifespan than the temperate lotus. However, little is known about germplasm diversity of either the subtropical or tropical lotus (Chaveerach et al., 2007; Tian et al., 2019; Mekbib et al., 2020). Field surveys and assessment of genetic background are required to understand the diversity of tropical lotus in Southeast Asia (e.g., Thailand and Vietnam) and provide a reference for its conservation and utilization.
Common molecular markers such as random amplification of polymorphic DNA (RAPD), inter-simple sequence repeats (ISSR), sequence-related amplified polymorphism (SRAP), simple sequence repeats (SSR), expressed sequence tag-simple sequence repeats (EST-SSR), and amplified fragment length polymorphism (AFLP) have been successfully employed to assess genetic diversity among lotus cultivars (Chen et al., 2008; Tian et al., 2008) or wild populations (Xue et al., 2006; Han et al., 2007, 2009) from China (Yang et al., 2011; Zheng et al., 2019), Northeast Asia (Kim et al., 1998; Kubo et al., 2009), and America (Li et al., 2015; Islam et al., 2020). Genetic analysis has revealed close relationships between tropical lotus, including 80 accessions of five populations in mainland Thailand (Mekbib et al., 2020), 15 accessions of five populations from northeastern and central Thailand (Chaveerach et al., 2007), 21 accessions of two Thai populations, and seven populations from Heilongjiang Province, China (Yang et al., 2013). Interestingly, Thai and Indian tropical lotus have often been used as outgroups for assessing Chinese lotus diversity (Li et al., 2010; Pan et al., 2011; Hu et al., 2012; Xu et al., 2015). Although our previous work has surveyed wild lotus and cultivars from Vietnam (Tian et al., 2019), the genetic diversity of Vietnamese lotus remains unclear. Therefore, it is imperative to conduct a comprehensive study on the diversity of tropical lotus, both wild and cultivated, from Southeast Asia.
Thailand and Vietnam, two of the central distribution regions of tropical lotus, might represent a center of origin for the Asian wild lotus (Liu et al., 2020b). In this study, we aimed to clarify the genetic relationships and identify the pure pedigrees of tropical lotus from Thailand and Vietnam. For this purpose, we used highly polymorphic EST-SSR markers (Zhang et al., 2014; Xu et al., 2015; Liu et al., 2018, 2020a) and SRAP makers to assess the genetic diversity and genetic structure of representative wild and improved lotus from the two countries. This work provides insights into the conservation and utilization of tropical lotus germplasms and parent selection for breeding novel cultivars.
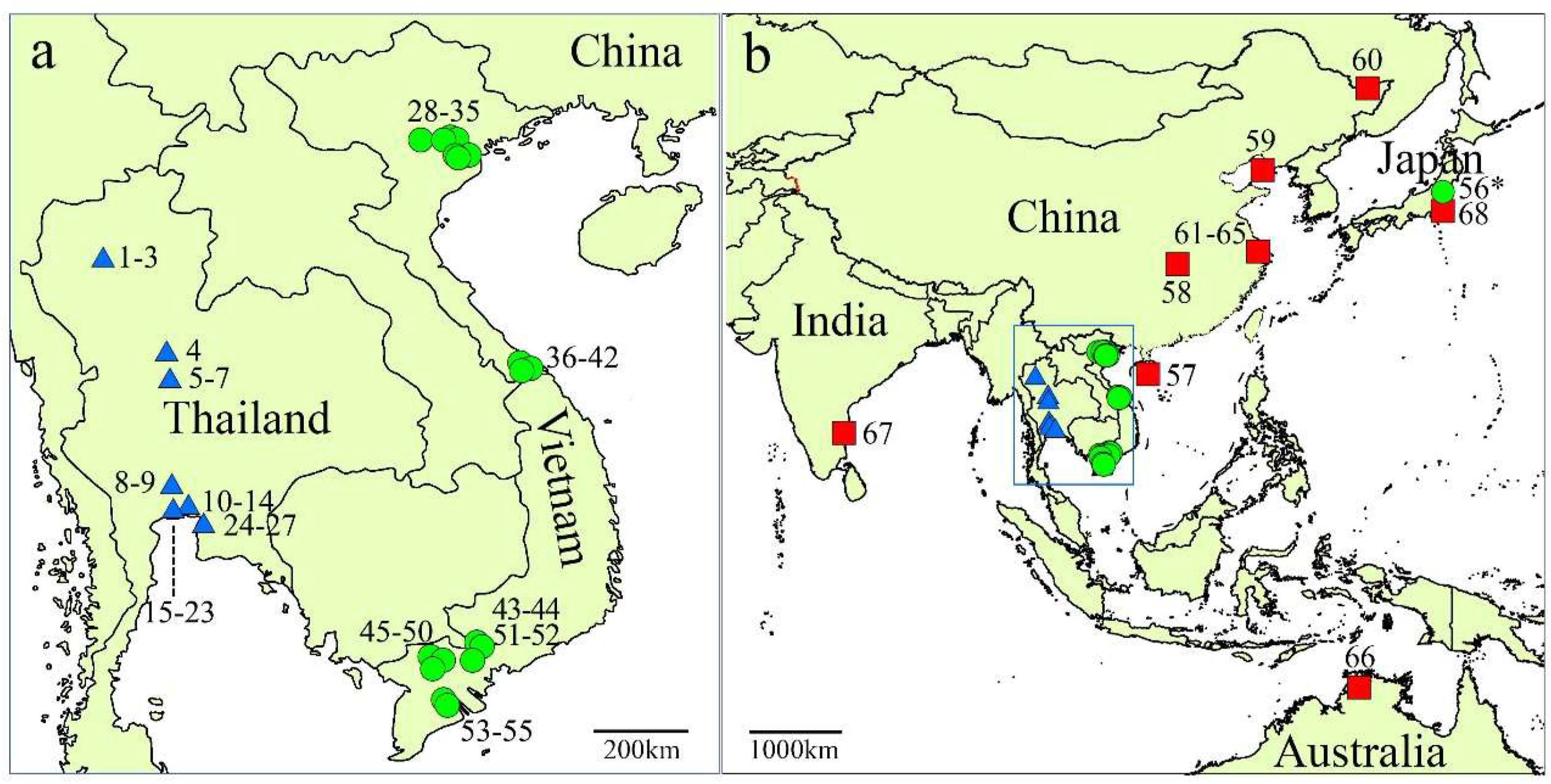
2. Material and methods 2.1. Plant materialsA total of 69 Nelumbo accessions were used in this study (Table 1; Fig. 1), among which, 27 accessions were collected from Thailand in 2015, and 29 accessions were collected from Vietnam in 2016 (Fig. 2a). The remaining 13 accessions (Fig. 2b), including the antique Asian lotus, wild lotus and cultivars, were used as reference materials, which were collected from the International Nelumbo Collection (certified by the International Waterlily & Water Gardening Society in 2016) in Shanghai Chenshan Botanical Garden. The young leaves of each accession were picked and then dried immediately by silica gel desiccant in zipped plastic bags stored at room temperature. The voucher specimens of most sampled accessions were deposited at the Herbarium of Chenshan Botanical Garden (CSH).
| No. | Sampling locality | Flower form | Flower color | Accession type | Longitude and latitude | Voucher specimen |
| 1 | Chiang Mai, THN | Double | White | ACL | 98.990321, 18.706555 | TDK-2781 |
| 2 | Chiang Mai, THN | Single | White | WAL | 98.990321, 18.706555 | TDK-2782 |
| 3 | Chiang Mai, THN | Single | Purple red | WAL | 98.990321, 18.706555 | TDK-2783 |
| 4 | Phitsanulok, THN | Single/semi-double | Purple red | WAL | 100.266139, 16.821678 | TDK-2786 |
| 5 | Phichit, THN | Single | Purple pink | WAL | 100.334412, 16.275754 | TDK-2787 |
| 6 | Phichit, THN | Single | Purple pink | WAL | 100.334412, 16.275754 | TDK-2790 |
| 7 | Phichit, THN | Single | White | WAL | 100.334412, 16.275754 | TDK-2791 |
| 8 | Lat Bua Luang, THN | Single | Purple pink | WAL | 100.372013, 14.159977 | TDK-2771 |
| 9 | Lat Bua Luang, THN | Single | White | WAL/ACL | 100.372013, 14.159977 | TDK-2772 |
| 10 | Suburb of Bangkok, THN | Single | Purple red | WAL | 100.699929, 13.735650 | TDK-2793 |
| 11 | Suburb of Bangkok, THN | Double | Purple pink | ACL | 100.699929, 13.735650 | TDK-2795 |
| 12 | Suburb of Bangkok, THN | Double | Purple pink | ACL | 100.699929, 13.735650 | TDK-2797 |
| 13 | Suburb of Bangkok, THN | Single | Purple red | WAL/ACL | 100.699929, 13.735650 | TDK-2798 |
| 14 | Suburb of Bangkok, THN | Single | White | WAL/ACL | 100.699929, 13.735650 | TDK-2799 |
| 15 | Bang U Bon, Bangkok, THN | Single | Unavailable | Asian-American hybrid | 100.399616, 13.660761 | TDK-2800 |
| 16 | Bang U Bon, Bangkok, THN | Double | Purple pink | Asian-American hybrid | 100.399616, 13.660761 | TDK-2801 |
| 17 | Bang U Bon, Bangkok, THN | Double | Purple pink | ACL | 100.399616, 13.660761 | TDK-2802 |
| 18 | Bang U Bon, Bangkok, THN | Single | Purple pink | WAL/ACL | 100.399616, 13.660761 | TDK-2803 |
| 19 | Bang U Bon, Bangkok, THN | Single | Purple red | ACL | 100.399616, 13.660761 | TDK-2804 |
| 20 | Bang U Bon, Bangkok, THN | Single | Purple red | WAL/ACL | 100.399616, 13.660761 | TDK-2806 |
| 21 | Bang U Bon, Bangkok, THN | Double | Green white | ACL | 100.399616, 13.660761 | TDK-2807 |
| 22 | Bang U Bon, Bangkok, THN | Single | White | WAL | 100.399616, 13.660761 | TDK-2808 |
| 23 | Bang U Bon, Bangkok, THN | Single | Purple red | WAL | 100.399616, 13.660761 | TDK-2809 |
| 24 | Waterlily and Lotus Research Station, Chonburi, THN | Double | Purple pink | ACL | 101.008100, 13.367195 | TDK-2756 |
| 25 | Waterlily and Lotus Research Station, Chonburi, THN | Single | Purple red | WAL | 101.008100, 13.367195 | TDK-2757 |
| 26 | Waterlily and Lotus Research Station, Chonburi, THN | Double | Purple pink | ACL | 101.008100, 13.367195 | TDK-2758 |
| 27 | Waterlily and Lotus Research Station, Chonburi, THN | Single | White | WAL | 101.008100, 13.367195 | TDK-2760 |
| 28 | Tu Son, Bac Ninh, VIE | Double | White | ACL | 105.956301, 21.115495 | TDK-2921 |
| 29 | Thuan Thanh, Bac Ninh, VIE | Single | Purple red | ACL | 106.089394, 21.068686 | TDK-2920 |
| 30 | Ba Vi, Hanoi, VIE | Single | Purple pink | ACL | 105.366647, 21.059721 | TDK-2913 |
| 31 | Ba Vi, Hanoi, VIE | Single | Purple pink | ACL | 105.366647, 21.059721 | TDK-2914 |
| 32 | Tay Ho, Hanoi, VIE | Double | Purple red | ACL | 105.819585, 21.055710 | TDK-2922 |
| 33 | Ninh Giang, Hai Duong, VIE | Single | Purple red | ACL | 106.333802, 20.753897 | TDK-2919 |
| 34 | Kim Dong, Hung Yen, VIE | Single | Purple red | ACL | 106.058175, 20.745049 | TDK-2917 |
| 35 | Tien Lu, Hung Yen, VIE | Single | Purple red | ACL | 106.118613, 20.699408 | TDK-2916 |
| 36 | Phong Dien, Hue, VIE | Single | Purple red | ACL | 107.352916, 16.587165 | TDK-2930 |
| 37 | Phong Dien, Hue, VIE | Single | Purple red | ACL | 107.352916, 16.587165 | TDK-2931 |
| 38 | Phong Dien, Hue, VIE | Single | White | ACL | 107.352916, 16.587165 | TDK-2933 |
| 39 | Tinh Tam, Hue, VIE | Single | Purple red | ACL | 107.578089, 16.469898 | TDK-2924 |
| 40 | Tinh Tam, Hue, VIE | Single | White | ACL | 107.578089, 16.469898 | TDK-2925 |
| 41 | Tinh Tam, Hue, VIE | Single | Purple red | ACL | 107.578089, 16.469898 | TDK-2926 |
| 42 | Huong Tra, Hue, VIE | Single | Purple red | ACL | 107.397759, 16.420760 | TDK-2928 |
| 43 | Cu Chi, Ho Chi Minh, VIE | Double | Purple pink | ACL | 106.484975, 10.969396 | TDK-2935 |
| 44 | Hoc Mon, Ho Chi Minh, VIE | Single | Purple pink | WAL/ACL | 106.590185, 10.880188 | TDK-2934 |
| 45 | Tram Chim, Dong Thap, VIE | Single | White | WAL | 105.556006, 10.686478 | TDK-2947 |
| 46 | Thap Muoi, Dong Thap, VIE | Single | Purple red | ACL | 105.812741, 10.610979 | TDK-2945 |
| 47 | Thap Muoi, Dong Thap, VIE | Single | Purple red | WAL/ACL | 105.812741, 10.610979 | TDK-2946 |
| 48 | Cao Lanh, Dong Thap, VIE | Single/semi-double | Purple pink | ACL | 105.605958, 10.438341 | TDK-2940 |
| 49 | Cao Lanh, Dong Thap, VIE | Double | White | ACL | 105.605958, 10.438341 | TDK-2941 |
| 50 | Cao Lanh, Dong Thap, VIE | Double | Purple pink | ACL | 105.605958, 10.438341 | TDK-2942 |
| 51 | Tan Thanh, Long An, VIE | Single | Purple pink | ACL | 106.402622, 10.602170 | TDK-2943 |
| 52 | Tan Thanh, Long An, VIE | Single | White | WAL/ACL | 106.402622, 10.602170 | TDK-2944 |
| 53 | Nga Bay, Hau Giang, VIE | Single | Purple pink | WAL | 105.818866, 9.812718 | TDK-2937 |
| 54 | Chau Thanh, Soc Trang, VIE | Single | Purple pink | ACL | 105.895327, 9.706268 | TDK-2938 |
| 55 | Chau Thanh, Soc Trang, VIE | Single | White | ACL | 105.895327, 9.706268 | TDK-2939 |
| 56 | Chiba, Japan (Introduced from VIE) | Single | Purple pink | WAL/ACL | 140.108646, 35.610691 | NA |
| 57 | Wanning, Hainan, CHN | Single | Purple pink | WAL | 110.396703, 18.805035 | TDK-3870 |
| 58 | INC (Hong Lake, Hubei Province, CHN)#1 | Single | Purple red | WAL | 113.353481, 29.898918 | TDK-3571 |
| 59 | INC (Pulandian, Liaoning Province, CHN) | Single | Purple red | WAL; antique | 121.954225, 39.407456 | TDK-1977 |
| 60 | INC (Tongjiang, Heilongjiang Province, CHN) | Single | Purple red | WAL | 132.527569, 47.670510 | NA |
| 61 | INC (Nelumbo 'China Antique', CHN)#2 | Single | Purple red | WAL; antique | 121.481001, 31.220305 | NA |
| 62 | INC (Nelumbo 'Dan Sajin', CHN) | Single | Variegated | ACL | 121.481001, 31.220305 | NA |
| 63 | INC (Nelumbo 'Feng Bao', CHN) | Double | Multicolor | ACL | 121.481001, 31.220305 | NA |
| 64 | INC (Nelumbo 'Jing Nü', CHN) | Double | Purple pink | ACL | 121.481001, 31.220305 | NA |
| 65 | INC (Nelumbo 'Jianxuan 35', CHN) | Single | Pink white | ACL | 121.481001, 31.220305 | TDK-2076 |
| 66 | INC (The Mary River, AUS) | Single | Purple red | WAL | 131.651030, -12.800220 | NA |
| 67 | INC (Kanchipuram, Chennai, Tamil Nadu, IND) | Single | Purple pink | WAL | 79.690339, 12.870549 | TDK-3236 |
| 68 | Chiba, Nelumbo 'Ohga', JPN | Single | Purple pink | WAL; antique | 140.108646, 35.610691 | NA |
| 69 | INC (N. lutea, introduced from Florida, USA) | Single | Light yellow | Wild American lotus | −80.819244, 26.921052 | TDK-642 |
| ACL, Asian cultivated lotus (not necessarily cultivar). AUS, Australia. CHN, China. INC, the nursery of International Nelumbo Collection, Shanghai, China. JPN, Japan. NA, Not Available. THN, Thailand. USA, the United States of America. VIE, Vietnam. WAL, wild Asian lotus. Single flower, tepal number usually ≤30 without petaloid tepals. Semi-double flower, tepal number between 30 and 60. Double flower > 60 tepals. #1, #2 INC was the nursery where the leaves of reference lotus were collected, and where the accession lives or its name was placed in the brackets. | ||||||

|
| Fig. 1 Flowers and rhizome from 46 of the 69 accessions. The number on each image corresponds to the accession No. in Table 1. Accession 55 is a rhizome type lotus with a single white flower. |
|
| Fig. 2 Sampling sites in Thailand, Vietnam (a), and the 13 reference accessions except for Nelumbo lutea (b). Accession 69 N. lutea from America is not displayed on the map. Blue triangles represent Thai lotus; green circles represent Vietnamese lotus; red squares represent the reference lotus. The blue box in panel b shows the location of panel a. *, Accession 56 was originally introduced from Vietnam to Japan. |
Genomic DNA was isolated from the dried leaves of each individual following the modified cetyltrimethyl ammonium bromide (CTAB) protocol (Doyle and Doyle, 1987), in which sugar removal was performed three times. DNA quality and concentration were measured by a NanoDrop 2000c and Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA). All DNA samples were stored at 4 ℃ after dilution to 50 ng·μL−1.
2.3. EST-SSR and SRAP analysisBased on our previous studies (Zhang et al., 2014; Xu et al., 2015; Liu et al., 2018, 2020a), 42 pairs of EST-SSR primers producing high-ratio polymorphic bands in Nelumbo were screened to amplify DNA. The PCR reaction was performed in a 10 μL mixture consisting of 5 μL 2 × ES Taq mix (Kangwei Biotech, Beijing, China), 1.0 μL DNA template, 0.5 μL each primer (10 μM), and 3.0 μL ddH2O. PCR conditions were as follows: initial denaturation at 94 ℃ for 5 min, 30 cycles of denaturation at 94 ℃ for 30 s, annealing at 52 ℃ for 30 s, and extension at 72 ℃ for 30 s, with a final elongation step at 72 ℃ for 5 min.
Thirty SRAP primer combinations from five forward and six reverse primers of the SRAP markers (Li and Quiros, 2001) were used (Table 1). Reactions were performed in a 10 μL PCR reaction system: 5 μL 2 × ES Taq mix (Kangwei Biotech, Beijing, China), 2.0 μL DNA template, 0.5 μL each primer (10 μM), and 2.0 μL ddH2O. PCR conditions were as follows: initial denaturation at 94 ℃ for 5 min; first five cycles of 1 min at 94 ℃, 1 min at 35 ℃, and 70 s at 72 ℃; then the annealing temperature was raised to 50–52 ℃ for another 35 cycles; followed by 7 min at 72 ℃.
PCR products were electrophoresed on 6% denaturing polyacrylamide gels, followed by silver staining and sodium hydroxide-formaldehyde staining.
2.4. Data statistics and analysisClear and polymorphic bands were scored with a present (1), absent (0), or missing data (-9) approach for each pair of EST-SST and SRAP primers. Bands of both makers were then combined into a binary matrix. The percentage of polymorphic bands (PPB) and the polymorphism information content (PIC) value were determined to measure the informativeness of a genetic marker. For the co-dominant EST-SSR marker, PIC was computed with Cercus 3.0.7 (Kalinowski et al., 2007), and the dominant SRAP marker with the formula PICi = 2ƒi (1-ƒi), where fi is the frequency of bands presence for locus i (Roldán-Ruiz et al., 2000; Serrote et al., 2020). EST-SSR and SRAP marker data were put into POPGENE 32 software (Yeh et al., 1999) to calculate the following genetic diversity parameters: the observed number of alleles (Na), the effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), the Shannon information index (I), and Nei's gene diversity (H). The genetic distance matrix of the 69 accessions was calculated by POPGENE 32 and then used to construct the dendrogram by the 'Neighbor-Joining tree' clustering procedure of MEGA 7.0.14 software (Kumar et al., 2016).
The films of EST-SSR PCR products were digitally scanned after the staining process, and the molecular mass of DNA bands was calculated using the Quantity One software (Bio-Rad, USA). Based on those molecular masses, the genetic structure of all accessions was assessed using the Bayesian clustering model implanted in the software STRUCTURE v.2.3.4 (Pritchard et al., 2000). To determine fine-scale genetic relationships, we assessed the genetic structure of Asian accessions while excluding the American lotus. The population number K was run from one to seven, with a burn-in time period of 100 000 followed by 1 000 000 Markov chain Monte Carlo (MCMC) iterations. Fifteen independent runs were performed for each simulated value of K. Then, the true K value, representing the most likely number of clusters or subpopulations (Evanno et al., 2005), was identified following the procedure of STRUCTURE Harvester based on the highest value of ΔK (Earl and vonHoldt, 2011). The OriginPro software (https://www.originlab.com/) was used to draw Cluster Images for K-1 and K.
3. Results 3.1. Marker efficiencyOut of the 42 EST-SSR and 30 SRAP primers tested, 36 EST-SSR and seven SRAP primers (Table 2) produced clear and polymorphic bands and were chosen for genetic analysis. In all 69 lotus accessions, 180 and 52 bands were amplified by EST-SSR and SRAP makers, respectively, which contained 164 and 41 polymorphic bands, with average PPBs of 91.1% and 78.8%, and average PICs of 0.487 and 0.215, respectively (Table 2). For the 36 EST-SSR markers, the PPBs ranged from 60% to 100%, and 23 of them reached 100%, and all 36 PICs varied from 0.080 to 0.778. According to PIC classification for co-dominant maker (Botstein et al., 1980), 15 EST-SSR primers were very informative (0.531–0.778), 18 were medium (0.357–0.454), and three was not very informative (0.080–0.207) (Table 2). By contrast, among the seven SRAP makers only the me1-em5 primer had a high PIC value (0.305), and the remaining six produced medium PIC values ranging from 0.117 to 0.268 (Table 2).
| Marker code | Forward primer (5′-3′) | Reverse primer (5′-3′) | TNB | NPB | PPB (%) | PIC |
| EST-SSR | ||||||
| NNFB_83 | CAGCACCATTCTGTAATCGC | GTGTGAGCGTATTTGGATCG | 5 | 5 | 100 | 0.705 |
| NNFB_197 | GGCTTTTTCCCCATTTTAGC | AGCCAGGGTCCTTTTGTTCT | 3 | 3 | 100 | 0.385 |
| NNFB_334 | CAAGGCAACTCCTCTTCTGC | CCACGTTTCATCATCAGGTG | 4 | 3 | 75.0 | 0.405 |
| NNFB_398 | TTCTGCAAGGAGTAGGTGCC | ATCGATTCCGCATCTCAAAG | 5 | 5 | 100 | 0.584 |
| NNFB_510 | CCAGAGCCGTAAGCGTAGTC | GCCCCGAACTCAATTACTCA | 4 | 4 | 100 | 0.417 |
| NNFB_702 | AAGGAAAAGTGACGTGGGTG | AGGGGGTTATCTTCTGTGGG | 8 | 6 | 75.0 | 0.387 |
| NNFB_750 | ACTTTGAGCTTGATCGGCAC | CAGCTGAACGGAACTGAGAA | 5 | 5 | 100 | 0.454 |
| NNFB_797 | CTTCCGTTTTCCTTTCCCTC | GCCTGCTCTGACAGTAACCC | 7 | 7 | 100 | 0.638 |
| NNFB_866 | ATGCGCTGGGAAGATAGAGA | AAGTGAGACGGAGTGATGGG | 6 | 6 | 100 | 0.534 |
| NNFB_984 | AGCACCGACTGATCGAGATT | AGTCCGTTCCGTTGAAAATG | 9 | 9 | 100 | 0.778 |
| NNFB_985 | CCAACCAGAGACAGAGGAGG | GACCGGGTTTGATGGTTAAA | 6 | 6 | 100 | 0.700 |
| NNFB_1115 | TTTTTCTCTTGTCTGTTTCTTGG | TGTCACTAGGCATCATCCCA | 2 | 2 | 100 | 0.080 |
| NNFB_1127 | GGAGGAGTAGGGAAAGGCAG | ATTCGATGCCAGGTGAAAAG | 4 | 3 | 75.0 | 0.406 |
| NNFB_1130 | AGCAAGCTCAGATGATGGGT | GTGGAACCGAAACCAGAAAA | 4 | 4 | 100 | 0.435 |
| NNFB_1140 | CCTGGGGTTTAAGTTGTGGA | TTGGGATTTAGACCCAGCAG | 3 | 3 | 100 | 0.374 |
| NNFB_1142 | TTTGATGTTCCCGTGTTTCA | CTGAGCGACCGATGTATTGA | 7 | 7 | 100 | 0.609 |
| NNFB_1211 | TGGTCTCGACAGCATTGAAG | GGTCAATCATCCCACTTGCT | 3 | 3 | 100 | 0.371 |
| NNFB_1237 | GCTGTCCCACTCCGTACAAT | GCGTCAAAGCTTGATTCTCC | 6 | 6 | 100 | 0.748 |
| NNFB_1296 | TATTTCAACGGCGAAAGAGG | TGAATCCAACGCACACATCT | 4 | 3 | 75.0 | 0.207 |
| NNFB_1333 | TGCTCTTTCTCATGTCGTCG | TGCTGCTACTGCTTCCTTCA | 6 | 6 | 100 | 0.748 |
| NNFB_1390 | ACATGTTGCAAGTGAGCCTG | CCAGCAGCTGTAGTCCCTTC | 3 | 2 | 66.7 | 0.357 |
| NNFB_1454 | TCATCACGCAGGGTATCGTA | GGCAGCTGATTTGTGATTTCT | 5 | 5 | 100 | 0.409 |
| NNFB_1491 | CGAAGTTGAGAACAGGGCTC | GAGAGGAAATGGAAACGCAG | 8 | 7 | 87.5 | 0.618 |
| NNFB_1507 | GAGTAAACCTTTTTCGGCCC | TGACAGCACTGCATCAACTG | 5 | 4 | 80.0 | 0.406 |
| NNFB_1524 | TTCGAATTGTACTCGTGTTTCG | GTGGACAGGAATCCGAGAAA | 5 | 4 | 80.0 | 0.450 |
| NNFB_1557 | GTAGCACGATGACCTGCTGA | TCGTTGACAAAAGGCTTTGC | 5 | 4 | 80.0 | 0.553 |
| NNFB_1727 | CAAAAGGGCCTAAAACACCA | GGCCCACAAGTGAAAGTGAT | 4 | 3 | 75.0 | 0.415 |
| NNFB_1814 | TGAACTGATGTTCATGGGGA | GCAACCCTGCAACTACTGCT | 5 | 5 | 100 | 0.402 |
| NNFB_1971 | TGGTGTGTCATCTTTGCCAT | GAGATGTAGGAGGGAGGGCT | 5 | 3 | 60.0 | 0.384 |
| NNFB_2026 | CCTATCATGCCAATGGGTCT | AACTGCCCCTCTCTCTCTCC | 5 | 3 | 60.0 | 0.587 |
| NNFB_2107 | CAAAACAGAAAAACGGATCACA | CAGGAATATGGACGCAACCT | 4 | 3 | 75.0 | 0.442 |
| NNFB_2332 | CATGGGAATGAAACAAACCA | GGGACTAGGAGGGCGTTTAG | 6 | 6 | 100 | 0.720 |
| NNFB_2646 | CTTTGCTGCTACTCCGAACC | TCCTTCATGCATCTGGTGAG | 4 | 4 | 100 | 0.531 |
| NNFB_2720 | ATGCTCCAATAACTGCACCC | AAAACGCCACTTGGATTACG | 5 | 5 | 100 | 0.386 |
| NNFB_2779 | TTACGAGGCCAAGGTTCATC | AACTCCTTGCAATCCATTCG | 4 | 4 | 100 | 0.194 |
| NNFB_2820 | GGCATACATAACATAGCGTGC | GGGTGCTTTCATTGGTGTTT | 6 | 6 | 100 | 0.719 |
| Total/average | 180 | 164 | 91.1 | 0.487 | ||
| SRAP | ||||||
| me1–em2 | TGAGTCCAAACCGGATA | GACTGCGTACGAATTTGC | 9 | 8 | 88.9 | 0.173 |
| me1–em5 | TGAGTCCAAACCGGATA | GACTGCGTACGAATTAAC | 8 | 7 | 87.5 | 0.305 |
| me1–em6 | TGAGTCCAAACCGGATA | GACTGCGTACGAATTGCA | 7 | 6 | 85.7 | 0.256 |
| me3–em2 | TGAGTCCAAACCGGAAT | GACTGCGTACGAATTTGC | 9 | 6 | 66.7 | 0.117 |
| me3–em3 | TGAGTCCAAACCGGAAT | GACTGCGTACGAATTGAC | 4 | 2 | 50.0 | 0.268 |
| me4–em1 | TGAGTCCAAACCGGACC | GACTGCGTACGAATTAAT | 8 | 7 | 87.5 | 0.242 |
| me4–em2 | TGAGTCCAAACCGGACC | GACTGCGTACGAATTTGC | 7 | 5 | 71.4 | 0.142 |
| Total/average | 52 | 41 | 78.8 | 0.215 | ||
| EST-SSR, expressed sequence tag-simple sequence repeats. SRAP, sequence-related amplified polymorphism. TNB, total number of amplified bands. NPB, number of polymorphic bands. PPB, percentage of polymorphic bands. PIC, polymorphism information content. | ||||||
Thai and Vietnamese lotus was classified into five geographic regions to assess genetic diversity. Based on that, higher values of most indices for genetic diversity were observed in Thai lotus than in Vietnamese lotus, regardless of whether we used EST-SSR or SRAP markers (Table 3), suggesting that Thai lotus contain higher levels of genetic diversity than the latter. In the EST-SSR assessment, the effective number of alleles (Ne) varied from 1.4563 in southern Vietnam to 1.9082 in central and northern Thailand. Similarly, the heterozygosity levels, Ho and He, ranged from 0.1852 and 0.2602 in southern Vietnam to 0.3495 and 0.4462 in Thailand. Although the Shannon information index (I) showed no significant difference between the lotus of southern Thailand (0.6934) and those of central and northern Thailand (0.6824), they increased in lotus resources from southern to northern Vietnam (southern: 0.4427; central: 0.5133; northern: 0.5466) (Table 3). When we used SRAP markers to calculate genetic diversity parameters, the values were lower (e.g., Na, Ne, and I) and showed different patterns (e.g., H and I) in some cases. This may be due to the small SRAP number and poor PCR amplification stability.
| Origin and accession No. | Value | EST-SSR | SRAP | ||||||||
| Na | Ne | Ho | He | I | Na | Ne | H | I | |||
| Central and northern Thailanda | Mean | 2.5278 | 1.9082 | 0.3495 | 0.4462 | 0.6824 | 1.4186 | 1.2744 | 0.1573 | 0.2324 | |
| 1–7 | St. Dev | 1.0278 | 0.6745 | 0.3102 | 0.2169 | 0.3660 | 0.4992 | 0.3762 | 0.2032 | 0.2918 | |
| Southern Thailand | Mean | 3.2500 | 1.8357 | 0.2743 | 0.3865 | 0.6934 | 1.6977 | 1.3217 | 0.1966 | 0.3039 | |
| 8–27 | St. Dev | 1.1802 | 0.7877 | 0.3397 | 0.2061 | 0.3746 | 0.4647 | 0.3429 | 0.1872 | 0.2649 | |
| Northern Vietnam | Mean | 2.3333 | 1.6627 | 0.3051 | 0.3441 | 0.5466 | 1.3488 | 1.1849 | 0.1114 | 0.1699 | |
| 28–35 | St. Dev | 0.9258 | 0.7237 | 0.3250 | 0.2101 | 0.3537 | 0.4822 | 0.3126 | 0.1750 | 0.2556 | |
| Central Vietnam | Mean | 2.2500 | 1.6232 | 0.1852 | 0.3291 | 0.5133 | 1.4419 | 1.2305 | 0.1396 | 0.2141 | |
| 36–42 | St. Dev | 0.8742 | 0.7746 | 0.3360 | 0.2284 | 0.3600 | 0.5025 | 0.3341 | 0.1826 | 0.2660 | |
| Southern Vietnam | Mean | 2.4167 | 1.4563 | 0.2332 | 0.2602 | 0.4427 | 1.4651 | 1.2658 | 0.1551 | 0.2326 | |
| 43–55 | St. Dev | 0.8062 | 0.5117 | 0.3528 | 0.1977 | 0.3095 | 0.5047 | 0.3627 | 0.1980 | 0.2841 | |
| Reference | Mean | 3.5833 | 2.6301 | 0.3274 | 0.6003 | 1.0188 | 1.8837 | 1.4413 | 0.2588 | 0.3960 | |
| 57–69 | St. Dev | 1.3390 | 1.0625 | 0.3087 | 0.1290 | 0.3288 | 0.3244 | 0.3793 | 0.1866 | 0.2465 | |
| H, Nei's gene diversity. Ho and He, observed and expected heterozygosity. I, Shannon's information index. Na, observed number of alleles. Ne, effective number of alleles. a The accessions from central and northern Thailand were combined due to their small numbers. The Vietnamese accession 56 introduced to Japan was not analyzed due to its unknown location. |
|||||||||||
A Neighbor-Joining (NJ) tree, building on the genetic distance matrix from the combined DNA bands of EST-SSR and SRAP makers (Table S1), was constructed and grouped the 69 accessions into five main clusters (Fig. 3). Cluster Ⅰ included 17 accessions, all from Thailand, which accounted for 63% of accessions in the country. Cluster Ⅱ contained 15 accessions: one reference lotus (accession 66 from northern Australia), three Thai lotus (5, 10, 11), and 11 lotus from southern Vietnam (Fig. 3; Table 1). On the opposite side of those two clusters was cluster Ⅲ, which was composed of two reference lotus from China (57, 65), one Thai lotus (19), and 11 Vietnamese lotus. All 14 of these accessions were the type of seed lotus (Table 1), including accession 57, a possibly wild seed lotus from Hainan Province, China, which is geographically close to the sampling sites of the 11 Vietnamese lotus (central and northern Vietnam) (Fig. 3).

|
| Fig. 3 Neighbor-joining dendrogram of 69 lotus accessions based on the combined EST-SSR and SRAP markers. Most Thai and Vietnamese accessions tested in this study were classified into cluster Ⅰ, Ⅱ, and Ⅲ. |
Between cluster Ⅰ/Ⅱ and cluster Ⅲ were positioned several scattered branches, Cluster Ⅳ, and cluster Ⅴ (Fig. 3). Cluster Ⅳ consisted of five reference accessions (58–61, 68) which were wild Asian lotus or the antique lotus, while cluster V consisted of reference accession 69 (American lotus) and two other accessions (15, 16) collected from Thailand but introduced from Europe (Table 1). For the scattered branches, they were three reference accessions (62–64, cultivars from China), three tested accessions with double flowers (17 from Thailand, and 28, 32 from Vietnam), and eight accessions (from Thailand and Vietnam) with single flowers.
Furthermore, two NJ dendrograms were also separately constructed from 36 EST-SSR (Fig. S1) and seven SRAP markers (Fig. S2). Comparing the three NJ dendrograms, a pattern was shown that higher marker numbers increased the resolution of the dendrograms, that is, the branching and clustering was more distinct. The seven SRAP markers generated a 'jellyfish-shaped' dendrogram with short branches among the clusters, and only two distinguishable clusters: cluster 1 and cluster 2 (Fig. S2). When 36 EST-SSR markers were used to construct the dendrogram, two clusters (cluster A and cluster B) were separated clearly, branching at both sides of the tree (Fig. S1). However, in cluster A, which was composed of most Thai lotus and a small number of Vietnamese accessions, and Thai accessions 22 and 23 were still undifferentiated. In fact, the two accessions were only identified in the dendrogram generated by combined EST-SSR and SRAP markers (Fig. 3). Furthermore, seven accessions (30, 35, 38, 40, 41, 42, 57) clustered together in all three dendrograms: cluster 1 in SRAP-tree (Fig. S2), cluster B in EST-SSR-tree (Fig. S1), and cluster Ⅲ in the combined tree (Fig. 3), which indicated a close genetic relationship among these seven accessions.
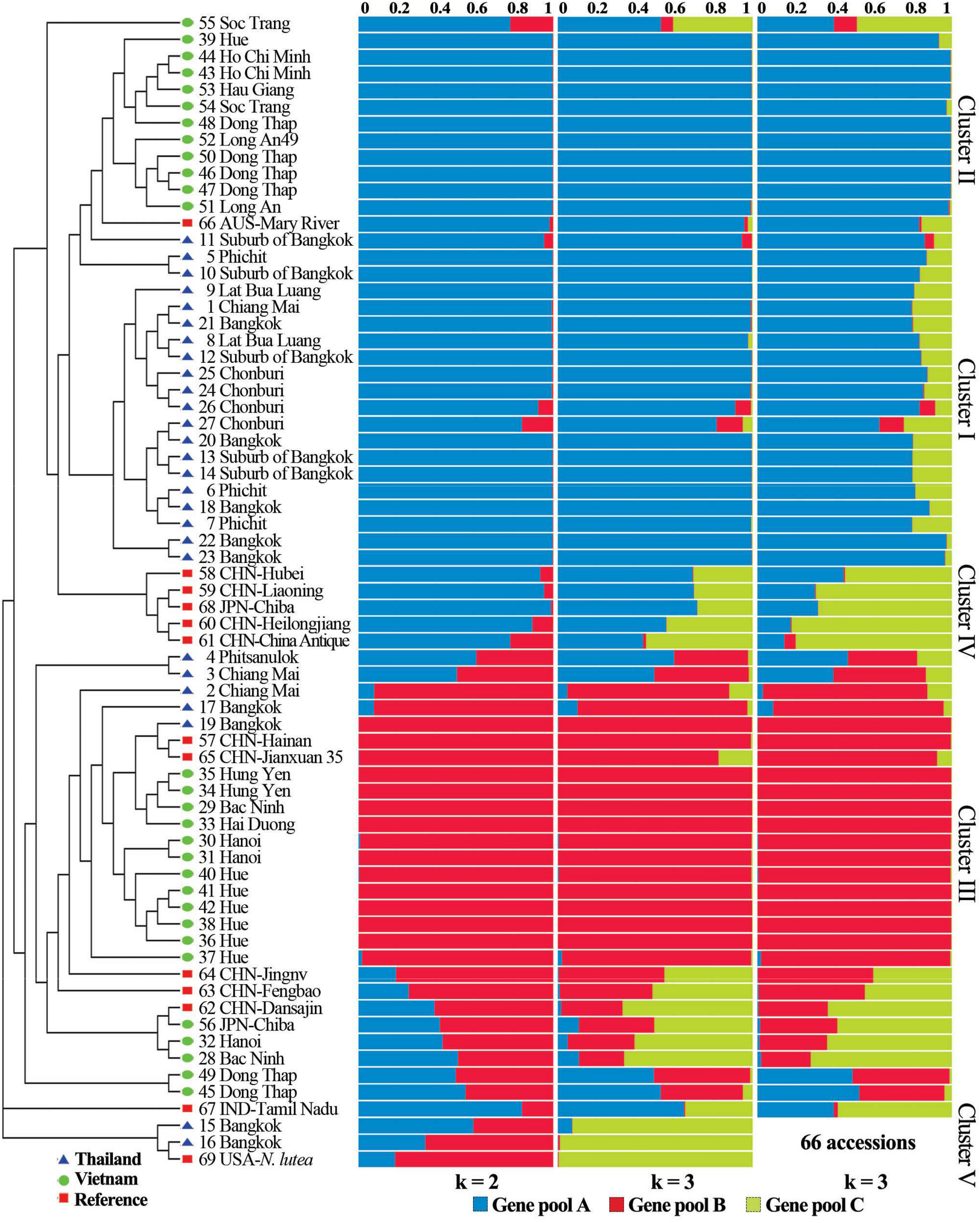
3.4. Inferred ancestry of individualsSTRUCTURE analysis based on ΔK of both all 69 lotus accessions and the 66 Asian accessions without Nelumbo lutea inferred that the lotus clustered optimally into three gene pools (Fig. 4), which was highly consistent with the NJ dendrogram (Fig. 3). In general, the genetic background of most lotus germplasms in Thailand and Vietnam was pure and belonged to two different gene pools or populations. The 11 accessions from southern Vietnam (cluster Ⅱ) and most Thai lotus (cluster Ⅰ) were assigned to gene pool A; cluster Ⅲ, with 12 Vietnamese lotus as the majority, fell into gene pool B; Nelumbo lutea, one of the only two species of the genus, was assigned to gene pool C (Fig. 4). For the five Vietnamese lotus (accessions 28, 32, 45, 49, 56) and all the reference lotus except accessions 57, 66, and 69, at least two gene pools were obviously observed (Fig. 4), which suggest they are heterozygous. In other words, they are the offspring of gene flow among the three gene pools. This fits perfectly with their distribution on the NJ tree (Fig. 3), where they were allocated discretely in the branches between groups A and B.
|
| Fig. 4 Neighbor-joining dendrogram and inferred ancestry of 69 lotus accessions. Each individual was represented by a single cylinder and composed of one to three inferred ancestries in different proportions. Three gene pools or populations were the most reasonable assignation for 69 accessions and 66 accessions when Nelumbo lutea accessions 15, 16, 69, were removed based on the most likely value of K identified by STRUCTURE, which was observed at K = 3 (). |
In this study, we investigated more than ten locations from Bangkok to Chiang Mai, covering the land of Central and Western Thailand, and finally 27 representative lotus accessions from seven sites were selected for genetic diversity analysis. NJ tree classified 20 of them into cluster Ⅰ/Ⅱ, belonging to the same gene pool (Fig. 4). Previous research has shown that genetic similarity values of five lotus populations (three accessions per population) from northeastern and central (near Bangkok) Thailand ranged from 84.1% to 95.0% (Chaveerach et al., 2007). Another study classified 11 populations from America, northeastern China, and Thailand (Bangkok and its suburbs), into a monophyletic cluster (Yang et al., 2013). Although the sampling sites of our study partially overlapped with those of previous studies (e.g., Bangkok), it is not clear whether the samples were duplicated. Regardless, the data mentioned above all showed that Thai lotus resources were closely related each other, which was supported by our finding that the genetic background of the major lotus germplasms in Thailand was highly consistent. It is likely due to the widespread cultivation for both flower and seed production in Thailand before crossbreeding was conducted. The two well-known Asian lotus cultivars commonly used as cut-flowers production in Thailand, 'Roseum Plenum' (double pink) and 'Album Plenum' (double green–white), very possibly originated via natural selection long time ago. If so, it is easily understood that most cultivated lotuses in Thailand were pure wild type plants, and were only translocated by the farmers from wild habitat to the field for agricultural purposes.
In previous work, we systematically surveyed Vietnamese lotus resources (Tian et al., 2019). Its lotus industry, particularly seed and flower production, was highly developed, and lotus was planted nearly everywhere from the north to the deep south of Vietnam. However, naturally growing wild lotus was rare (Tian et al., 2019). Furthermore, the diversity of cultivated lotus in Vietnam is rich. Lotus is primarily grown in Vietnam for seeds, fresh cut flowers, and vegetable runners (stolons). Therefore, in our study, we examined these three types of accessions, focusing especially on seed-lotus type cultivated on a large scale. However, only a small number of seed-lotus cultivars were recognized in Vietnam, and the seed yield was lower than that of those in China (Tian et al., 2019).
In this study, seed-lotus accessions from central and northern Vietnam were grouped together (Fig. 3) and exhibited a genetic background different from that of the lotus in southern Vietnam and Thailand (Figs. 2 and 4). These findings imply that these seed-lotus germplasms have undergone genetic differentiation, which makes them excellent parents for future breeding programs. Similar to Thailand, the majority of Vietnamese lotus germplasms are pure wild type, suggesting that agricultural activities, which have been conducted with scant artificial breeding, have had little influence on gene flow between lotus accessions.
4.2. Relationship of seed lotus, wild lotus and some unknown identitiesPrevious studies on 292 cultivated and wild lotus accessions in Asia led to the speculation that seed lotus and rhizome lotus had not originated from a single source but instead had a more complex multi-source origin (Liu et al., 2020b). We found that seed lotus from central and northern Vietnam clustered together and possessed a gene pool different from other wild or cultivated lotus from Thailand and southern Vietnam. Thus, our findings support the independent origin of seed lotus and rhizome lotus.
In addition, we also found that 'Jianxuan 35' (65) and a seed lotus (57) from Hainan, China, were closely related, and both clustered with the lotus accessions from northern Vietnam. It is likely due to a short geographical distance between northern Vietnam and Hainan Island of China. However, further study is needed to determine whether the two Chinese accessions are native to China or were introduced from northern Vietnam. A similar situation was seen in the close relationship between the lotus of southern Vietnam and Australian lotus. To clarify the nature of these relationships, future studies should expand lotus sampling to South Asian countries, particularly in regions between Vietnam and Australia.
This study also showed that accessions 58 to 61, 68, which have previously been considered wild type in China and Japan, were not pure genotypes but had signs of obvious genomic introgression. Specifically, they formed a branch with close relationships which matched the morphology similarity and adjacent geographic distribution (Figs. 3 and 4). Accession 28 (white double flower-lotus from Bac Ninh, Vietnam) and 32 (pink double seed-lotus from Hanoi of Vietnam) were closely related and formed a small branch with accession 56 (collected from Chiba of Japan but originally introduced from Vietnam possibly in 2013 based on communication with Dr. T.N. Hoang) and 62 ('Dan Sajin', a traditional cultivar commonly seen in both China and Japan). All these four accessions were cultivars with heavy genomic introgression. However, it remains unknown on their origin from either China or Vietnam.
In both Thailand and Vietnam, several morphologically similar cultivars of double-flower lotus is used for flower production, which have pink (accessions 11, 12, 17, 24–26 from Thailand, and 43, 50 from Vietnam) and green–white tepals (1, 21 from Thailand, and 49 from Vietnam). However, these germplasms were separated by country. Both double flower lotus from Thailand, 'Roseum Plenum' (12) and 'Album Plenum' (1, 21), formed a small clade with accession 8 (pink single), indicating they share an origin (Figs. 3 and 4). While accession 11, a cut-flowers lotus collected from the suburb of Bangkok, was separated from 'Roseum Plenum' although both share a high morphological similarity in flower. Vietnamese accession 43 (pink double) and 44 (pink single), collected from the same site of Ho Chi Minh, had the closest relationship, indicating that one possibly originated from the other. A similar situation was also seen on Thailand accession 24 (double), 25 (single), and 26 (double), which were collected at the same site in Thailand (Waterlily and Lotus Research Station, Chonburi), implying that one may originate from the other. According to the records of Dr. Nopchai Chansilpa, accession 25 originated from the seedling of either accession 24 or 26.
Two unidentified accessions (15, 16) were collected from Pang U Bon Waterlily Garden, Nonthaburi, Thailand, but were previously introduced from Europe. Based on leaf morphology only, we tentatively identified them as an American lotus or hybrid, which was supported by molecular data in this study (Figs. 3 and 4).
4.3. Conservation implicationsPopulation structure analysis of 58 Asian wild lotus based on 29, 797 4D SNPs have indicated that when K was set from two to six two pure pedigrees were consistently represented by the nine Thai wild lotus from Chiang Mai, Chonburi, and Bangkok (Liu et al., 2020b). Consistent with that, Thai lotus accessions 19 and 22–23 in our study also exhibited a pure genetic background, respectively, regardless of whether American lotus was used as an outgroup. These findings suggest that these three accessions and their populations might have originated from wild lotus, and, therefore, their conservation should be prioritized. However, our findings contrast with a previous report that examined genetic diversity of 15 lotus populations containing India and America lotus and found that the five populations (Uttaradit, Chon Buri, Phetchaburi, Kalasin, and Ratchaburi) formed a genetic cluster (Mekbib et al., 2020). One explanation for this discrepancy may be the sample selection of this earlier study. Compared with Thai lotus, there were more lotus accessions with pure pedigrees from Vietnam (22/29, 75.9%), including cluster Ⅱ and cluster Ⅲ, which represented two separate gene pools. Conservation priority should be given to these germplasms.
In this study, 13 wild Asian and American lotus, and the antique Asian lotus were selected as a reference group, which guaranteed the geographic span of three continents (Asia, Oceania, America) and a time scale of at least one thousand years (accessions 59 and 68) (Godwin and Willis 1964; Shen-Miller, 2002). However, 11/13 of these accessions showed evidences of multiple gene pools, implying that they might be the offspring of gene exchanges between different populations. By contrast, three Thai accessions, 22 Vietnamese accessions, and one Hainan accession from southern China possessed a single gene pool, suggesting that Southeast Asia may be one of the origin centers of wild lotus, which is in agreement with the inference that wild lotus likely originated in tropical regions (Liu et al., 2020b).
5. ConclusionThe genetic backgrounds of most lotus resources collected from Thailand and Vietnam are uniform, containing one and two gene pools, respectively. This finding implies that the genetic backgrounds of these lotus was less affected by human activities such as artificial breeding and introduction of the cultivars from other countries. Thai accessions 19, 22, 23 and the populations they were introduced from should be prioritized for conservation. In addition, the origin of seed lotus possibly differs from that of rhizome and flower lotus; furthermore, one of the centers of origin of the surviving Asian lotus germplasms may be in Southeast Asia. Thai and Vietnamese lotus germplasms are genetically related to geographical distribution patterns, and the accessions from the same or close location usually show the closest genetic relationships. The origin or genetic relationship of some unidentified lotus sources could be evaluated by comparing morphological characteristics and molecular marker analysis.
AcknowledgementsThe study was supported by Shanghai Landscaping Administration Bureau (Grant number G182412) and the grant from Zhejiang Humanity Landscape Co., Ltd., Hangzhou, China. The authors thank Mr. Dong-Bei Yu from Zhejiang Humanity Landscape Co., Ltd. and Mr. Xian-Bao Zeng from China Lotus Research Center for their kind help on lotus survey in Thailand.
Author contributions
DKT and YCC conducted funding acquisition and conceived the plot design; DKT, TNH, VP, YCC and PWC collected the field data and reviewed the draft; FLL and YLD performed the genotyping, data analysis, and visualization; FLL, MQ and YRF completed the original draft. All authors contributed to the manuscript and gave final approval for publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.05.004.
Chaveerach, A., Sudmoon, R., Tanee, T., et al., 2007. Genetic relationships in a population of Nelumbo nucifera Gaertn. (Nelumbonaceae). J. Biol. Sci., 7: 5-13. DOI:10.3923/jbs.2007.1388.1393 |
Chen, G.L., Zhu, M.Z., Guo, M.Q., 2019. Research advances in traditional and modern use of Nelumbo nucifera: phytochemicals, health promoting activities and beyond. Crit. Rev. Food Sci. Nutr., 59: S189-S209. DOI:10.1080/10408398.2018.1553846 |
Chen, Y.Y., Zhou, R.C., Lin, X.D., et al., 2008. ISSR analysis of genetic diversity in sacred lotus cultivars. Aquat. Bot., 89: 311-316. DOI:10.1016/j.aquabot.2008.03.006 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Earl, D.A., vonHoldt, B.M., 2011. Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour., 4: 359-361. DOI:10.1007/s12686-011-9548-7 |
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Godwin, H., Willis, E.H., 1964. The viability of lotus seeds (Nelumbium nucifera Gaertn.). New Phytol., 63: 410-412. DOI:10.1111/j.1469-8137.1964.tb07391.x |
Guo, H.B., 2009. Cultivation of lotus (Nelumbo nucifera Gaertn. ssp. nucifera) and its utilization in China. Genet. Resour. Crop Evol., 56: 323-330. DOI:10.1007/s10722-008-9366-2 |
Hall, T.F., Penfound, W.T., 1944. The biology of the American lotus, Nelumbo lutea (Willd.) Pers. Am. Midl. Nat., 31: 744-758. DOI:10.2307/2421417 |
Han, Y.C., Teng, C.Z., Wahiti, G.R., et al., 2007. Genetic variation and clonal diversity in populations of Nelumbo nucifera (Nelumbonaceae) in central China detected by ISSR markers. Aquat. Bot., 86: 69-75. DOI:10.1016/j.aquabot.2006.09.007 |
Han, Y.C., Teng, C.Z., Wahiti, G.R., et al., 2009. Mating system and genetic diversity in natural populations of Nelumbo nucifera (Nelumbonaceae) detected by ISSR markers. Plant Syst. Evol., 277: 13-20. DOI:10.1007/s00606-008-0096-x |
Hu, J.H., Pan, L., Liu, H.G., et al., 2012. Comparative analysis of genetic diversity in sacred lotus (Nelumbo nucifera Gaertn.) using AFLP and SSR markers. Mol. Biol. Rep., 39: 3637-3647. DOI:10.1007/s11033-011-1138-y |
Islam, M.R., Zhang, Y., Li, Z.Z., et al., 2020. Genetic diversity, population structure, and historical gene flow of Nelumbo lutea in USA using microsatellite markers. Aquat. Bot., 160: 103162. DOI:10.1016/j.aquabot.2019.103162 |
Kalinowski, S.T., Taper, M.L., Marshall, T.C., 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol., 16: 1099-1106. DOI:10.1111/j.1365-294x.2007.03089.x |
Kim, H., Song, M.J., Kim, K.J., et al., 1998. Genetic variation analysis of Korean lotus (Nelumbo nucifera) using randomly amplified polymorphic DNA (RAPD) markers. Korean J. Plant Taxon., 28: 343-355. |
Kubo, N., Hirai, M., Kaneko, A., et al., 2009. Classification and diversity of sacred and American Nelumbo species: the genetic relationships of flowering lotus cultivars in Japan using SSR markers. Plant Genet. Resour., 7: 260-270. DOI:10.1017/S1479262109356580 |
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Li, C., Mo, H.B., Tian, D.K., et al., 2015. Genetic diversity and structure of American lotus (Nelumbo lutea Willd.) in North America revealed from microsatellite markers. Sci. Hortic., 189: 17-21. DOI:10.1016/j.scienta.2015.03.026 |
Li, G., Quiros, C.F., 2001. Sequence-related amplified polymorphism(SRAP), a new marker system based on a simple PCR reaction its application to mapping and gene tagging in Brassica. Theor. Appl. Genet., 103: 455-461. DOI:10.1007/s001220100570 |
Lin, Z.Y., Zhang, C., Cao, D.D., et al., 2019. The latest studies on lotus (Nelumbo nucifera) – an emerging horticultural model plant. Int. J. Mol. Sci., 20: 3680. DOI:10.3390/ijms20153680 |
Liu, F.L., Qin, M., Liu, Q.Q., et al., 2020a. Detection of genetic variation between broad and narrow-tepalled American lotus (Nelumbo lutea Willd.) by EST-SSR markers. Acta Agric. Boreali-Occident. Sin., 29: 306-314. DOI:10.7606/j.issn.1004-1389.2020.02.017 |
Liu, F.L., Qin, M., Zhang, D.S., et al., 2018. Comparison and analysis of the difference of variegated-petalled cultivars of sacred lotus (Nelumbo nucifera) based on morphological traits and EST-SSR marker. Mol. Plant Breed., 16: 1607-1618. DOI:10.13271/j.mpb.016.001607 |
Liu, Z.W., Zhu, H.L., Zhou, J.H., et al., 2020b. Resequencing of 296 cultivated and wild lotus accessions unravels its evolution and breeding history. Plant J., 104: 1673-1684. DOI:10.1111/tpj.15029 |
Li, Y., Svetlana, P., Yao, J.X., et al., 2014. A review on the taxonomic, evolutionary and phytogeographic studies of the lotus plant (Nelumbonaceae: Nelumbo). Acta Geol. Sin. (Engl Ed), 88: 1252-1261. DOI:10.1111/1755-6724.12287 |
Li, Z., Liu, X.Q., Gituru, R.W., et al., 2010. Genetic diversity and classification of Nelumbo germplasm of different origins by RAPD and ISSR analysis. Sci. Hortic., 125: 724-732. DOI:10.1016/j.scienta.2010.05.005 |
Mekbib, Y., Huang, S.X., Ngarega, B.K., et al., 2020. The level of genetic diversity and differentiation of tropical lotus, Nelumbo nucifera Gaertn. (Nelumbonaceae) from Australia, India, and Thailand. Bot. Stud., 61: 15. DOI:10.1186/s40529-020-00293-3 |
Pan, L., Quan, Z.W., Hu, J.H., et al., 2011. Genetic diversity and differentiation of lotus (Nelumbo nucifera) accessions assessed by simple sequence repeats. Ann. Appl. Biol., 159: 428-441. DOI:10.1111/j.1744-7348.2011.00509.x |
Pritchard, J.K., Stephens, M., Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1093/genetics/155.2.945 |
Roldán-Ruiz, I., Dendauw, J., Van-Bockstaele, E., et al., 2000. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed., 6: 125-134. DOI:10.1023/A:1009680614564 |
Serrote, C.M.L., Reiniger, L.R.S., Silva, K.B., et al., 2020. Determining the polymorphism information content of a molecular marker. Gene, 726: 144175. DOI:10.1016/j.gene.2019.144175 |
Sharma, B.R., Gautam, L.N.S., Adhikari, D., et al., 2016. A comprehensive review on chemical profiling of Nelumbo nucifera: potential for drug development. Phytother. Res., 31: 3-26. DOI:10.1002/ptr.5732 |
Shen-Miller, J., 2002. Sacred lotus, the long-living fruits of China Antique. Seed Sci. Res., 12: 131-143. DOI:10.1079/SSR2002112 |
Tian, D.K., Chen, Y.C., Hoang, T.N., 2019. Understanding current status of germplasm, research and industry of lotus (Nelumbo) in Vietnam through a survey. J. Changjiang Veg., 6: 40-45. DOI:10.3865/j.issn.1001-3547.2019.06.014 |
Tian, H.L., Xue, J.H., Wen, J., et al., 2008. Genetic diversity and relationships of lotus (Nelumbo) cultivars based on allozyme and ISSR markers. Sci. Hortic., 116: 421-429. DOI:10.1016/j.scienta.2008.02.011 |
Wang, Q.C., Zhang, X.Y., 2004. Lotus Flower Cultivars in China. Bao MZ trans. Beijing: China Forestry Publishing House.
|
Xue, J.H., Zhuo, L.H., Zhou, S.L., 2006. Genetic diversity and geographic pattern of wild lotus (Nelumbo nucifera) in Heilongjiang Province. Chin. Sci. Bull., 51: 421-432. DOI:10.1007/s11434-006-0421-0 |
Xu, Y.X., Zhang, W.W., Mo, H.B., et al., 2015. Genetic diversity analysis of Nelumbo accessions based on EST-SSR markers. Plant Diver. Resour., 37: 595-604. DOI:10.7677/ynzwyj201515004 |
Yang, M., Fu, J., Xiang, Q.Y., et al., 2011. The core-collection construction of flower lotus based on AFLP molecular markers. Sci. Agric. Sin., 44: 3193-3205. DOI:10.3864/j.issn.0578-1752.2011.15.015 |
Yang, M., Liu, F., Han, Y.N., et al., 2013. Genetic diversity and structure in populations of Nelumbo from America, Thailand and China: implications for conservation and breeding. Aquat. Bot., 107: 1-7. DOI:10.1016/j.aquabot.2013.01.001 |
Yeh, F.C., Yang, R.C., Boyle, T., 1999. Microsoft window-based freeware for population genetic analysis (Popgene Version 1.31). University of Alberta, Edmonton, Canada.
|
Zhang, W.W., Tian, D.K., Huang, X., et al., 2014. Characterization of flower-bud transcriptome and development of genic SSR markers of in Asian louts. PLoS One, 9: e112223. DOI:10.1371/journal.pone.0112223 |
Zhang, X.Y., Chen, L.Q., Wang, Q.C., 2011. New Lotus Flower Cultivars in China. Beijing: China Forestry Publishing House: pp 36-41.
|
Zhang, X.Y., Wang, Q.C., 2004. Research of winter lotus varieties' selection, cultivation, and planting – Sanshui Lotus World as a base. Chinese Landsc. Archit., 10: 62-65. DOI:10.3969/j.issn.1000-6664.2004.10.015 |
Zhang, X.Y., Wang, Q.C., 2006. Preliminary study of the eco-types of genetic resources of tropical lotus. Chinese Landsc. Archit., 7: 82-85. DOI:10.3969/j.issn.1000-6664.2006.07.018 |
Zheng, X.W., Cheng, T., Yang, L.B., et al., 2019. Genetic diversity and DNA fingerprints of three important aquatic vegetables by EST-SSR markers. Sci. Rep., 9: 14074. DOI:10.1038/s41598-019-50569-3 |



