b. Institute of Marine Drugs, Guangxi University of Chinese Medicine, Nanning 530004, Guangxi, PR China
Fruit is one of the most valuable and diverse angiosperm structures. It substantially impacts seed predation and dispersal and is crucial in driving species diversification (Herrera, 1989; Lu et al., 2019). The evolution of fleshy fruits is considered an important driver of angiosperm diversity in the late Cretaceous and early Cenozoic (Herrera, 1989; Eriksson and Bremer, 1991; Tiffney and Mazer, 1995; Dodd et al., 1999; Smith, 2001; Beaulieu and Donoghue, 2013), and it has long been suggested that the evolution of various fruit colours might be related to diversification patterns.
Previous research has shown that fruit colour is related to habitat, geographical distribution, latitude, and elevation. For example, it has been estimated that more than 70% of woody species in the tropics have fleshy fruits. In addition, a comprehensive survey of fleshy fruit traits showed that fruit colour diversity is highest in the tropics; specifically, the major tropical plant groups with colourful fleshy fruits include Rubiaceae, Myrtaceae, Solanaceae, and Arecaceae (Sinnott-Armstrong et al., 2020). Studies have also found that the proportion of fleshy-fruit species declines along increasing latitudinal gradients (Willson et al., 1989, 1990b; Vittoz et al., 2009) and that fruit colour and fruit size vary with latitudinal gradients (Sinnott-Armstrong et al., 2018). On a large scale, the spatial distribution of different fruit colours exhibits regularity. For example, at higher latitudes red-fruited communities occupy larger areas than do communities of plants with other fruit colours (Nakanishi, 1996; Sinnott-Armstrong et al., 2018). Blue fruits occur in New Zealand, Papua New Guinea, eastern Australia, Chiloé Island in Chile, and temperate and boreal North America (Nakanishi, 1996; Sinnott-Armstrong et al., 2018). In contrast, black fruits are primarily prevalent in low-latitude regions (Sinnott-Armstrong et al., 2018). These findings show that geographical and topographical gradients are greatly influenced by climate and thus potential factors that drive fruit colour diversification. Thus, understanding the evolution of fruit colour in plant groups is aided greatly by studying plant taxa of different fruit colours that have a near-global spatial distributions.
The evolution of fruit colour has been driven by both biotic and abiotic factors. The dispersal syndrome hypothesis posits that animal dispersers drive diversification of fruit colours due to differing perceptual abilities. Birds tend to spread smaller and brightly-coloured fruits, whereas mammals prefer larger fruits with mysterious colours and odours (Janson, 1983; Sinnott-Armstrong et al., 2018; Valenta and Nevo, 2020). Accordingly, birds play an indispensable role in seed dispersal and establishment, influencing species diversity across various regions. Among the 24, 455 mutualistic interactions between flowering plants and seed dispersers, (1631 animal species and 3208 plant species), three-quarters of the interactions involve birds, with most of the remaining interactions involving mammals (Fricke and Svenning, 2020). Furthermore, phylogenetic analysis has shown that long-distance seed dispersal is the cardinal reason for the diversification and broad distribution of fruit colour on the evolutionary branches (Herrera, 1989; Bremer, 1992; Tiffney and Mazer, 1995; Shanahan et al., 2001). Specifically, red fruits, which are favoured by birds, are strongly correlated with large-scale dispersal events (Landis et al., 2013, 2018; Sotomayor, 2014; Van Dam and Matzke, 2016; Cantley et al., 2016; Chen et al., 2017; Lu et al., 2019; Klaus and Matzke, 2020). Although specific fruit colours may promote plant evolution and distribution, most ecological studies have not tested statistical relationships between fruit colour and dispersal at the genus level in a phylogenetic context. In addition to dispersal by animals, fruit-colour evolution might also be influenced by other factors, such as anthocyanin synthesis, pathogens (Cazetta et al., 2008), solar radiation (Willson et al., 1990a; Burns, 2015), wet season temperatures, and growth periods (Willson et al., 1989; Sinnott-Armstrong et al., 2021). Among these, anthocyanin synthesis is especially critical in influencing fruit colour variance. Anthocyanins also help plants adapt to variable habitats by reducing the propagation of fungi and preventing photoinhibition (Chalker-Scott, 1999; Steyn et al., 2002; Gould, 2004; Schaefer et al., 2008). Moreover, anthocyanins are affected by temperature (Bogert, 1949; Steyn et al., 2002, 2009; Latti et al., 2008). In North and South America, anthocyanin biosynthesis and its content within the fruit have been observed to vary with elevation and latitude (Latti et al., 2008; Zoratti et al., 2014), although this has yet to be linked to variation in fruit colour, especially at the genus level.
Understanding how fruit colour drives species diversification is essential for macroevolutionary studies and to understand patterns of species richness among clades (Kozak and Wiens, 2016). Evaluation of diversification rates and models provides insights into the evolutionary mechanisms of large lineages that have undergone dynamic changes in diversity over space and time (Jetz et al., 2012; McGuire et al., 2014). Moreover, diversification rates have been shown to differ for plants with distinct fruit colours (Tiffney and Mazer, 1995; Beaulieu and Donoghue, 2013). For example, in palms the highest diversification rates have been associated with yellow fruits, followed by black, purple, red, orange, and white fruits (Arecaceae; Hill et al., 2021). Nonetheless, the mechanisms by which particular fruit colours might influence speciation and extinction remain complicated (Herrera, 1989). Several studies have proposed that an accommodative environment (such as that in the montane tropical regions) may promote diversification of noticeable fruit colours (Hughes and Eastwood, 2006; Spriggs et al., 2015; Hutter et al., 2017). Specifically, violet fruits might accelerate diversification and high species richness at low latitudes in tropical habitats (Lu et al., 2019). However, the diversification pattern by which particular fruit colours might influence biodiversity at the species level has seldom been demonstrated.
Here, we used Callicarpa as an angiosperm model to analyse whether fruit colours promote seed dispersal, distribution, and species diversification. Callicarpa is a typical pantropical flowering plant group with more than 140 species worldwide (Bramley, 2011), most of which have important medicinal and ornamental value (Cantrell et al., 2005; Chung et al., 2005; Tu et al., 2013). Callicarpa species span many continents (Asia, Australia, North and South America etc.), latitudes (50.3°N–37.5°S) and elevations (0–3500 m; Table S1), and the colour of mature fruits (berry) vary, including violet, red, white and black (Chen and Michael, 1994; Fig. S1). Although we speculate that certain fruit colours of Callicarpa might be favoured in environments that promote rapid diversification (Spriggs et al., 2015; Hutter et al., 2017; Lu et al., 2019), it is unclear whether different coloured fruits of this genus show similar patterns of geographic distribution around the world. In addition, it is unknown whether fruit colour in Callicarpa is correlated with latitude or elevation.
In this study, we used a phylogenetic framework of Callicarpa and data on fruit colour and geographic distribution to address four questions. (1) What is the current geographical distribution of the four fruit colours of Callicarpa? (2) Are the frequencies and distances of these four fruit colours involved in dispersal events equal? (3) What animals disperse Callicarpa berries? (4) Is fruit colour correlated with elevation, latitude, and diversification rate? Note that our focus here is on the potential implications of these fruit phenotypes for the dispersal, distribution and diversification of plant species, regardless of whether the various animal species that might feed on these fruits see these colours the same or differently compared to humans. Therefore, we follow standard practice in the published taxonomic literature and flora in characterizing fruit colours (Lu et al., 2019). However, perceptual biases might be relevant (e.g. Kemp et al., 2015), especially if we fail to find significant patterns using human-based characterization of fruit colours.
2. Materials and methods 2.1. Fruit traits and geographic distributionData on fruit colour and geographic distributions for 95 species were compiled from published literature, herbarium records, and field observations (Table S1). Considering that berry is a unique fruit type, we categorised the ripened fruit of Callicarpa into four types (violet, red, white, and black) based on the colour of the epicarp. Furthermore, we used "violet" to encompass purple and purplish red.
We collated geo-referenced specimen records of Callicarpa from 16 herbarium (ALA, L, BM, KRB, NMNH, TNS, GH, NY, K, E, P, FTG, CNS, PE, KUN, IBSC, http://sweetgum.nybg.org/ih/). For species that lacked specific geolocation information (latitude, elevation), we obtained data by using Google Earth Pro to locate the collection site of specimens (Sullivan, 2009). Subsequently, we mapped the current distribution of Callicarpa with different fruit colours using the ArcGIS software (ArcMap v.10.5.1) based on 7711 geographical coordinates (longitude and latitude). Additionally, relevant references were used to calculate the species range and midpoints of latitude and elevation (Table S1).
2.2. Taxa sampling and DNA sequencesA total of 91 samples were collected based on the subgeneric system and its geographical distribution as delineated by Chen and Michael (1994) and Bramley (2009), and the widespread species were sampled repeatedly (e.g., Callicarpa arborea, C. macrophylla, C. longifolia, C. rubella, C. japonica). Taxa sampled with collection sites and vouchers are listed in Table S2. Furthermore, based on Li et al. (2016) and Shaw et al. (2005, 2007), two nuclear genes (ITS, ETS) and eight chloroplast gene fragments (matK, rpl32-trnl, trnD-trnT, trnH-psbA, psbJ-petA, trnQ-5′rps16, 3′trnV-ndhC, trnS-trnG) were used for this study. Topologies derived from nrDNA (two loci) and cpDNA (eight regions) were broadly congruent, but better resolution and stronger branch support was achieved by combining the data sets with 91 samples of Callicarpa and 17 outgroups. Of these 17 outgroups, two were sampled from subfamily Prostantheroideae and 15 were sampled from three tribes within subfamily Nepetoideae of the Lamiaceae (Li et al., 2016). Detailed methods, sequence information, and results are provided in Supporting Information (Appendix S1; Table S3; Figs. S2–S4). The time-calibrated phylogeny estimation of Callicarpa used a combination of nrDNA and cpDNA.
2.3. DNA extraction, PCR amplification, and sequencingTotal genomic DNA was extracted from silica-dried leaf tissue or herbarium specimens utilizing the CTAB method (Doyle and Doyle, 1987). Primers used for polymerase chain reaction (PCR) amplification of 10 gene fragments are listed in Table S4. PCR amplification of the target fragments was performed using a 30 μL reaction system: 1.2 μL template DNA, 6 μL 5 × buffer (Mg2+plus), 2.4 μL 2.5 mM dNTP, 0.3 μL 10 μM primer, 0.3 μL HS Taq polymerase (TaKaRa), 0.6 μL BSA (Bull Serum Albumin), and finally ddH2O was added to a total volume of 30 μL. PCR reaction procedures included pre-denaturation at 94.0 ℃ for 5 min, followed by 35 cycles of denaturation at 94 ℃ for 30 s, annealing at 52 ℃ for 45 s, extension at 72 ℃ for 1 min, which were then followed by a final extension at 72 ℃ for 10 min, where the annealing temperature was adjusted appropriately for each fragment amplification. PCR amplification products were sequenced by Invitrogen (Invitrogen, commercial sequencing facility, Guangzhou, China).
2.4. Estimation of divergence timeWe assembled a 10-gene (ITS, ETS, matK, rpl32-trnl, trnD-trnT, trnH-psbA, psbJ-petA, trnQ-5'rps16, 3'trnV-ndhC, trnS-trnG) concatenated data matrix for 52 species (excluding repeated species) and 17 outgroups to estimate the time-calibrated phylogeny for Callicarpa. Due to the paucity of published fossils ascribed to Callicarpa, molecular dating was performed based on fossils from associated clades in Lamiaceae. Two-point calibrations were used in this study (Drew and Sytsma, 2012; Li et al., 2016). The first calibration point was based on the hexacolpate and three-nucleate pollen fossil of Ocimum Linn. (Nepetoideae) from the early Eocene sediments in India, which has been previously been placed at the crown of Nepetoideae (Kar, 1996). For this calibration point, we used a lognormal distribution, with an offset at 49 Ma, a mean of 2.6 Ma, and a standard deviation (SD) of 0.5 Ma (Drew and Sytsma, 2012). The second calibration point relied on a fruit fossil of Melissa from the Early-Middle Oligocene (Martinez-Millan, 2010), which was assigned to constrain the most recent common ancestor (MRCA) of Melissa L. and Neoeplingia L. with a lognormal distribution (offset: 28.4 Ma, mean: 1.5 Ma, SD: 0.5 Ma). Divergence time analyses were performed using the BEAST2 program (Bouckaert et al., 2014) implemented in the CIPRES portal (Miller et al., 2010). The required XML files were obtained from BEAUti (Drummond et al., 2012). A Yule tree prior and the relaxed exponential clock model were selected, and GTR + G substitution model was set for each partition, except for trnD-trnT and trnH-psbA (HKY + G). Substitution models for each partition were selected using the Akaike information criterion (AIC; Cavanaugh, 1997), and implemented using ModelPie (Jaa, 2004). Markov chain Monte Carlo (MCMC) analysis was run for 400 million generations of Callicarpa, with sampling at every 40, 000th generation. We applied the software package Tracer v.1.7 (Rambaut et al., 2018) (https://beast.community/tracer) to confirm that all parameters had an estimated sample size (ESS) > 200. We combined the tree files of the three runs (burn-in 20% each) in LOGCOMBINER v.2.4.4, and used TREEANNOTATOR v.2.4.4 to produce a maximum clade credibility tree showing mean divergence time estimates with 95% HPD intervals (Bouckaert et al., 2014). The final result was visualised using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
2.5. Diversification through timeWe performed a Bayesian analysis of macroevolutionary mixtures (BAMM v.2.5.0) to analyse the speciation and extinction dynamics with outgroups removed (Rabosky et al., 2014a, b). The appropriate priors on the consensus tree were estimated using the 'setBAMMpriors' function in the BAMMtools R package (Rabosky et al., 2014b). However, it is controversial whether the rate estimated by the above method is sensitive to the prior value, and thus, whether it is reliable (Moore et al., 2016). Thus, we analysed a range of prior settings (poissonRatePrior = 1.0, 0.5, 2.0, 2.5 and 10.0) to evaluate the prior sensitivity. Ultimately, poissonRatePrior = 1.0 was considered appropriate for this study, because other prior values had little impact on the posterior distribution of rate shift events. Missing taxon sampling was considered with a global sampling fraction set to 0.37 (52/140). The analysis was run for 10 million generations, and the MCMC chain was sampled every 1000 generations. Finally, using BAMMtools, we traced the specific clade diversification and plotted the diversification rate (speciation, extinction, and net diversification rate) through time, after discarding 25% of the generations (MCMC) as burn-in. Further, we constructed semi-logarithmic lineage through time (LTT) plots by using the R package APE 3.3 (http://ape-package.ird.fr/) to visualize temporal variations in diversification rates (Mahler et al., 2010). One thousand trees were sampled randomly from the converged BEAST trees with outgroups removed and used to calculate a 95% credibility interval. The specific clade diversification rates by BAMM analysis are listed in Table S5.
2.6. Testing the most common fruit colourWe tested whether the four fruit colours occurred with equal frequency in Callicarpa. First, we implemented the chi-square test in the R software (https://www.r-project.org/) to assess significant differences between the number of all known species (95 species), and the number of known fruit colour species on the consensus tree (46 species; Table S6). Second, we performed the 'prop.test' in R to test the statistical differences in the fruit colour proportion between all known species and the species on the tree (Table S6).
2.7. Testing correlations between fruit colour and dispersal eventsWe divided Callicarpa distribution into eight representative areas (Table S7) that largely referenced Bremer (1992): (A) East Asia; (B) South Asia; (C) Southeast Asia; (D) Oceania; (E) temperate North America; (F) Neotropics; (G) the Pacific Ocean; (H) Other Zones. Ancestral state reconstruction was conducted using the time-calibrated tree. After comparing the AIC values of six models, we implemented the Statistical Dispersal Extinction Cladogenesis (S-DEC) model to reconstruct the ancestral state using the RASP software (Nylander et al., 2008; Harris and Xiang, 2009; Yu et al., 2015). Divergence-time estimation was set across 10, 000 trees generated from the random combination of the four Markov chains. The mean frequency of ancestral variation was calculated using 1000 trees that were stochastically selected from the 10, 000 usable trees. The maximum number of ancestral regions was set to two to obtain the closest realistic value. We conducted an independent run for 10, 000, 000 generations and sampled every 1000 generations. We inferred that dispersal events were the areas with the largest proportional likelihood distinction between adjacent nodes on the clade.
Fruit-colour reconstructions were based on stochastic character mapping (SIMMAP) of discrete traits on phylogenies and were implemented through the 'phytools' package of the R program (Bollback, 2006; Revell, 2012), which was used to estimate the evolution of fruit colour and infer the most likely fruit colour related with every dispersal event depicted by the tree. It was entirely based on the Bayesian analysis in MCMC sampling for computing the posterior probability distribution of trees to adapt uncertain ancestral states, speciation rates, and evolution. Then, "ER" (a single rate for all possible transitions between states) was selected as the best-fitting model for ancestral reconstructions of discrete characters (Paradis et al., 2004). Subsequently, we applied the result of the posterior distribution and the function of 'make.simmap' with 100 generations in the R package 'phytools' to plot stochastic mappings of fruit-colour variation. The 'describe.simmap' function in the R package 'phytools' was used to integrate the posterior distribution of character evolutional history on each branch. In the analysis process, fruit colour was classified into four categories in all species (violet-0, red-1, white-2, and black-3). In addition, we also conducted Bayesian Binary MCMC (BBM) models to evaluate and visualise the fruit-colour evolution in RASP (Fig. S5). We then tested whether the sampling balance among lineages with various fruit colours had a major influence on the evolution of fruit colours (SIMMAP) by randomly selecting 95%, 90%, 85%, 80%, 75%, and 70% of the samples (52 species) for fruit colour ancestral reconstruction, respectively (Fig. S6).
We also tested whether the most common fruit colour participated most frequently in the dispersal events in Callicarpa. First, dispersal events related to each fruit colour were inferred on the reconstructed branch of the trait. We assigned 1.00 (weight) if only one colour was involved in a dispersal event on the reconstructed branch of fruit colour, 0.50, if two colours were involved in a dispersal event on the branch, 0.33, if three colours were involved in a dispersal event on the branch (Lu et al., 2019). No more than three colours were inferred by all branches. Overall weight values were aggregated for each colour on the consensus tree (Table S8). Subsequently, based on the respective weight value of each fruit colour, we carried out the Friedman nonparametric tests to determine whether the four fruit colours had an equal possibility of involvement in dispersal events. Furthermore, boxplots showed the dispersal probabilities of four fruit colours based on the results of each fruit colour associated with dispersal events on the consensus tree.
We tested whether the dispersal distance of each fruit colour in Callicarpa differed significantly. We performed the Kruskal–Wallis H test based on the mean dispersal distance of each fruit colour on the consensus tree (Table S8). Then, we also drew a boxplot based on the dispersal distance of the four fruit colours. We acknowledge that the dispersal events analysed in this study are independent of phylogeny. Furthermore, there is no direct phylogenetic relationship because only the internal branches were included for most analyses.
2.8. Examining relationships among fruit colours, geographic attributes, and diversification ratesWe used binomial logistic regression analyses in R to examine relationships between fruit colour, elevation, and latitude in 95 species (Table S1). Then, we used the species-based binomial logistic regression to analyse fruit colours and their geographic distribution (elevation and latitude) within eight biogeographic regions (Table S1). However, not all fruit colours occurred across all distribution regions. We also conducted region-based regression analyses and selected the best-fitting regression models based on AIC scores to evaluate the positive or negative correlation depending on coefficients. We did not apply standard Bonferroni method to correct p-values, as this approach is controversial (Nakagawa, 2004). Recent studies have suggested that the most preferable approach is controlling FDR as it not only reduces false positives, but also minimises false negatives (Van Iterson et al., 2010; Jafari and Ansari-Pour, 2019). Therefore, we implemented a sequential FDR correction for p-values (Jafari, and Ansari-Pour, 2019) to select the best-fitting models. In addition, we used linear regression analyses in R to test the association between fruit colours and geographic variables of distribution regions based on the proportion of species in each region that had those fruit colours (Table S1). The best-fitting regression models depended on the r2 values.
To test the correlation between fruit colours and diversification rates, the diversification rate was first calculated using the 'tiprates' function in the R package 'BAMMtools' (http://bamm-project.org/index.html). Subsequently, the boxplot was mapped to exhibit the diversification rate of each fruit colour based on the 46 species on the consensus tree. Finally, we used the logistic regression analyses in R to determine which fruit colour may strongly influence the diversification rate of Callicarpa.
3. Results 3.1. Distribution patterns of Callicarpa fruit coloursAccording to our specimen data analysis, Callicarpa are chiefly located in the tropics, subtropics, and a few temperate zones: East Asia (47 species), Southeast Asia (63 species), and Oceania (22 species), with a few species distributed in South Asia (11 species), the Pacific (10 species), the Neotropics (11 species), the temperate zone of North America (6 species), and other regions (7 species; Fig. 1A; Table S9). Moreover, our analysis showed that violet fruits of Callicarpa are distributed most widely, specifically, within eight distribution regions spanning various latitudinal gradients (50.3°N–37.5°S) in China and Southeast Asia (Fig. 1B). Red fruits are generally more common in the tropics, although a moderate numbers of Callicarpa species with red fruit are also present at higher latitudes (Fig. 1C). The distribution of white fruits is similar to that of violet fruits; specifically, Callicarpa species with white fruit are distributed extensively in all eight distribution areas, especially in temperate and tropical regions of East Asia, Southeast Asia, and Oceania (Fig. 1D). In contrast, black fruits are concentrated in the tropics (Fig. 1E).
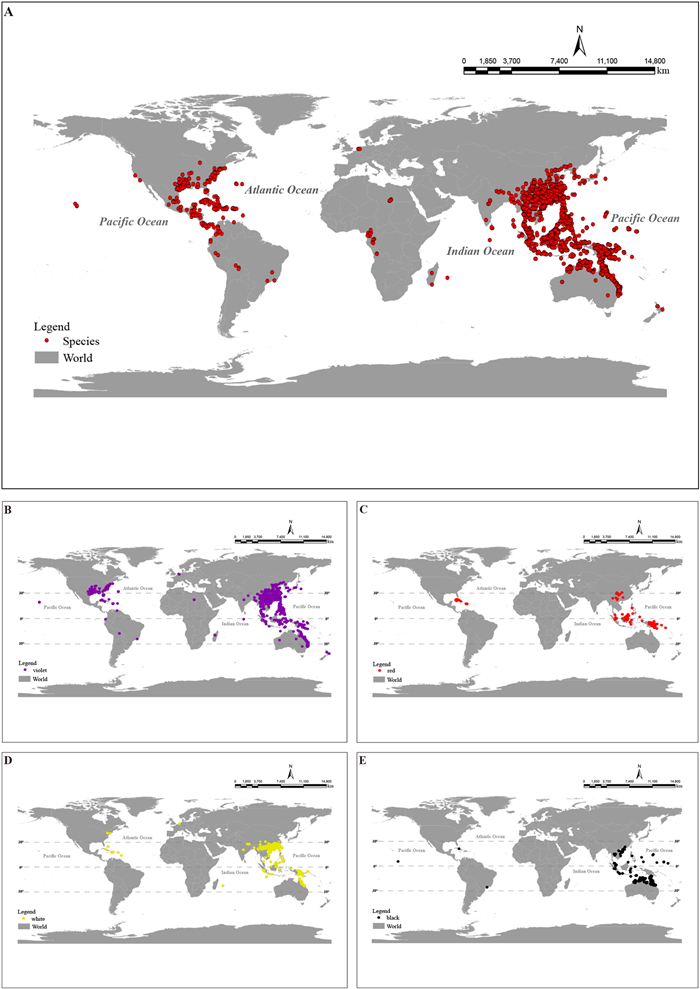
|
| Fig. 1 Global distribution map and the distribution of the four fruit colours of Callicarpa. A, distribution of all known species; B, violet-fruited species; C, red-fruited species; D, white-fruited species; E, black-fruited species. |
The BEAST chronogram indicated that Callicarpa originated around 35.53 Ma in the Eocene (Figs. 2 and S5, node 1, 95% HPD: 18.33–57.46 Ma). Moreover, the divergence of most branches of this genus began in the Miocene and continued into the Pleistocene. Violet-fruited Callicarpa americana (26.87 Ma; Figs. 2 and S5, node 2, 95% HPD: 12.17–43.57 Ma) was the first to diverge from the lineage, in the late Oligocene. During the late Oligocene and early-mid Miocene, some endemic species with red or black fruits evolved in Southeast Asia, with crown ages around 16.92 Ma (Figs. 2 and S5, node 3, 95% HPD: 7.13–27.65 Ma) and 12.50 Ma (Figs. 2 and S5, node 4, 95% HPD: 3.63–32.84 Ma), respectively. Several species with red or violet fruits also diversified in East Asia close to this period, with crown ages started from ca. 23.95 Ma (Figs. 2 and S5, node 5, 95% HPD: 12.56–35.85 Ma), 21.04 Ma (Figs. 2 and S5, node 6, 95% HPD: 10.96–31.26 Ma), and 15.87 Ma (Figs. 2 and S5, node 7, 95% HPD: 7.04–25.90 Ma), respectively. Our findings also indicate that white-fruited Callicarpa started to diversify around 14.59 Ma (Figs. 2 and S5, node 8, 95% HPD: 6.73–23.43 Ma), whereas violet-fruited Callicarpa diversified a little later (Figs. 2 and S5, node 9, 13.23 Ma, 95% HPD: 5.79–21.52 Ma).
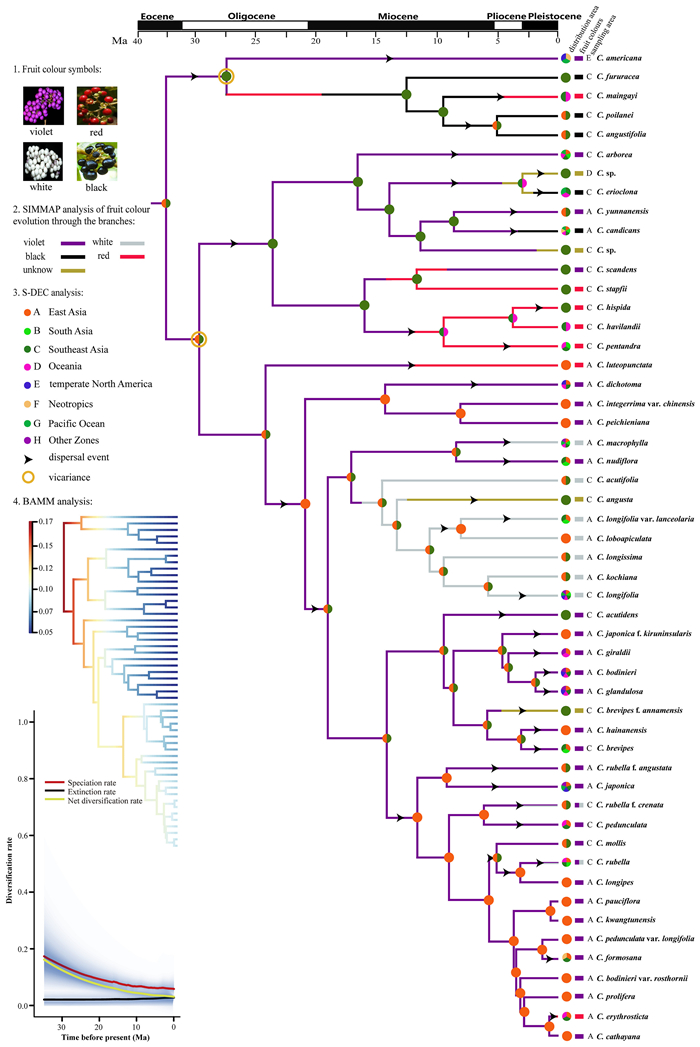
|
| Fig. 2 Fruit-colour evolution, biogeographic dispersal, and BAMM analysis of Callicarpa. Graphs on the left show the Maximum a posteriori probability shift configuration represented as a phylorate plot showing variations in speciation rates along each branch of the Callicarpa phylogeny and the Bayesian reconstruction of rate variations in speciation, extinction and net diversification through time. On the right, the consensus tree was obtained by BEAST analysis. Ancestral-area reconstructions (chromatic cycles at nodes), dispersal events (arrows) and vicariance (brown circles) were inferred by the best-fitting model (S-DEC) in RASP (Detailed information on the dispersal of events is provided in). Fruit colour was reconstructed (based on the colour of branches) using SIMMAP. The pie chart shows the distribution regions of each species. The rectangle indicates the fruit colour of species. The detailed colour-indications are listed in the figure. |
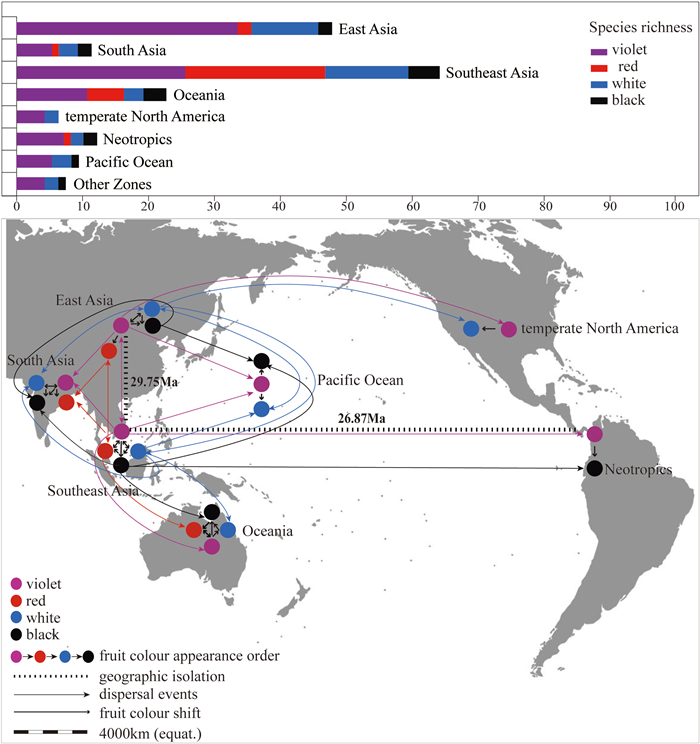
S-DEC analyses indicated that the earliest Callicarpa originated East Asia (A) and Southeast Asia (C) during the Eocene, around 35.53 Ma (Figs. 2 and S5). Two important vicariance events occurred in these regions. The first, which occurred 29.75 Ma (Figs. 2 and S5, 95% HPD: 16.31–45.75 Ma), led to the colonization of East Asia and Southeast Asia by the Callicarpa; the second, at 26.87 Ma (Figs. 2 and S5, node 2, 95% HPD: 12.17–43.57 Ma), initiated a trans-Pacific dispersal event that prompted the formation of two separate Old World and New World lineages in this genus. Subsequently, several dispersal events during the middle Miocene to late Pleistocene contributed to the distribution of Callicarpa in Oceania (Figs. 2 and S5, node 7, 15.87 Ma, 95% HPD: 7.04–25.90 Ma; 9.40 Ma, 95% HPD: 1.55–21.02 Ma; 2.67 Ma, 95% HPD: 0.04–6.39 Ma, etc.). Since 23.95 Ma, multiple dispersal events have occurred between East Asia and Southeast Asia (Figs. 2 and S5, node 5, 95% HPD: 12.56–35.85 Ma; node 6, 21.04 Ma, 95% HPD: 10.96–31.26 Ma; node 8, 14.59 Ma, 95% HPD: 6.73–23.43 Ma), contributing to both regions gradually becoming centers of species diversity for the Callicarpa. In addition, dispersal events have also driven various species to appear in South Asia (Figs. 2 and S5, 8.1 Ma, 95% HPD: 0.56–17.05 Ma; 7.82 Ma, 95% HPD: 2.14–14.92 Ma; 3.42 Ma, 95% HPD: 0.05–7.13 Ma).
3.4. Diversification through timeBAMM analysis indicated that no apparent rate shift occurred in the phylogeny of Callicarpa (Fig. 2). The radiation of Callicarpa species started from the Eocene (~ 35 Ma) at a rate of 0.17 lineages per million years. This analysis indicates that Callicarpa diversification can be characterized by an early burst of speciation with high initial speciation rates followed by a steady decline of speciation (Fig. 2). The rate through-time plot shows that the global speciation and net diversification rates in Callicarpa are gradually decreasing. However, the extinction rate is constant and at low levels (Fig. 2). The semi-logarithmic lineage through time (LTT) plot analysis corroborated results of BAMM analysis (Fig. 3).
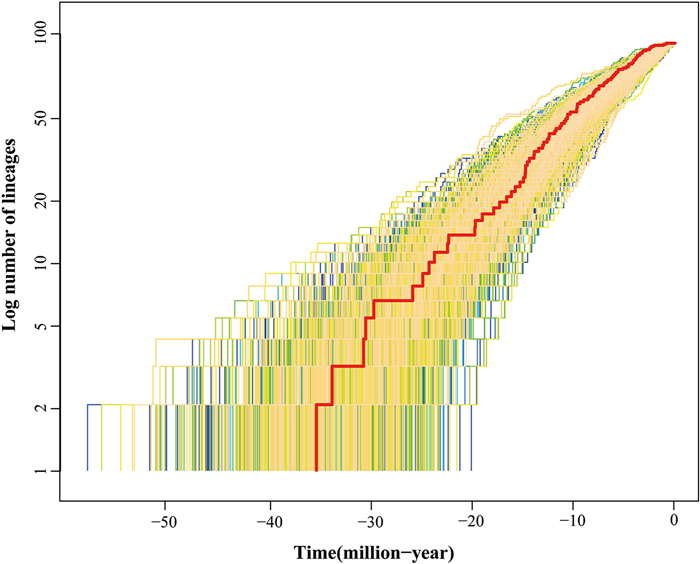
|
| Fig. 3 Semi-logarithmic lineage-through-time (LTT) plots show the cumulative number of lineages over time of Callicarpa. The red line shows the maximum clade credibility tree. |
Chi-square tests indicated that there were significant differences among the species number of different fruit colours, both among all known species and among species on the consensus tree (both p < 0.0001; Table S6). There were similar proportions of species for each fruit colour in both data sets obtained by the 'prop.test' function in R (Table S6). The most common fruit colour of Callicarpa species was violet (53.7% of all Callicarpa species; 58.7% on consensus tree). The second most common was red (25.3% overall, 15.2% on the tree), then white (13.7% overall, 17.4% on the tree), and lastly black (7.3% overall, 8.7% on the tree).
3.6. Relationship between fruit colour and dispersal eventsCombining the divergence time of species and the SIMMAP results, we discovered that the order of occurrence of different fruit colours in Callicarpa was violet, red, white, and then black. Reconstruction of the ancestral fruit colour using SIMMAP placed violet fruit on most of the branches (77 branches, Fig. 2), while red was on 12 branches, white on 16 branches, and black on 9 branches. Most dispersal events were associated with violet-coloured fruit lineages. Biogeographic analyses (S-DEC) indicated that 39 dispersal events occurred among the eight distribution areas (Table S8). Of these, 27 dispersal events were associated with violet fruit, 5 with white fruit, 4 with red fruit, and 3 with black fruit. Dividing the evolutionary history of Callicarpa (35.53–0.79 Ma) into 4-Myr time intervals using the midpoint age of each branch to integrate each fruit colour involved in the dispersal events revealed that violet was the most involved in dispersal events and occupied time intervals, especially from 10 to 0 Ma (35–30 Ma: violet = 1.0; 30–20 Ma: violet = 4.5, red = 0.5; 20–10 Ma: violet = 5.0, red = 0.5, white = 1.0, black = 0.5; 10–0 Ma: violet = 18.5, red = 3.0, white = 4.0, black = 2.5; Table S8). In addition, the results of random sampling remained basically consistent with the results of SIMMAP based on 52 species, mainly in that there was no major change in branch structure, different fruit colour branches still clustered together, and violet fruits still had the highest number of reconstructions. Therefore, sampling of different fruit colour lineages is highly probably balanced (Fig. S6).
Chi-square tests indicated that dispersal frequency of the four fruit colours differed significantly (p < 0.001; Table S10). The fruit colour with the highest dispersal frequency was violet (3.47) followed by white (2.24). Red fruits (2.19) were less frequently associated with dispersal events, and black fruits (2.09) were the least associated with dispersal events (Fig. 4A; Table S10). In addition, the Kruskal–Wallis H test indicated that dispersal distances of the four fruit colours differed significantly (p < 0.001; Table S10). The fruit colour with the greatest dispersal distance was violet, followed by that for white, red, and black (Fig. 4B; Table S10).
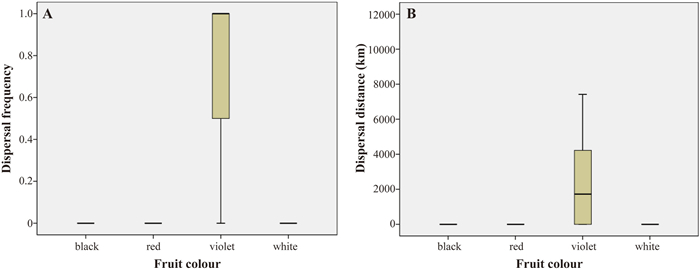
|
| Fig. 4 Boxplots for dispersal probabilities and dispersal distance for each fruit colour based on the DEC and SIMMAP analysis. A, Dispersal probabilities of each fruit colour among the 39 dispersal events linked to fruit colour. B, Geographic distance associated with dispersal events assigned to different fruit colours on each branch with inferred dispersal events. |
We used binomial logistic regression to test the correlation between fruit colours and geographic attributes (elevation and latitude) of all Callicarpa species with known fruit colour (Tables 1 and S11). The results indicated that violet fruits were positively correlated with the combination of mean elevational range and mean latitudinal range (AIC = 108.610). Red fruits were negatively correlated with mean latitudinal range (AIC = 88.840). Black fruits were negatively correlated with the mean latitudinal range (AIC = 48.665), and white fruits were positively correlated with the mean elevational range (r2 = 0.6218; AIC = −18.588; Table S13). In most other analyses fruit colours were correlated with one or more geographic variable. These analyses included both species-based analyses within each biogeographic region and region-based analyses (Tables S12 and S13). Moreover, these results support our findings on the relationships between fruit colour of species and geographical distribution, especially in Southeast Asia.
| Variables | Estimate coefficients | p-values | AIC |
| violet ~ mean elevational range + mean latitudinal range | 0.0016 | 0.0320 | 108.610 |
| 0.0860 | 0.0060 | ||
| red ~ mean elevational range + mean latitudinal range | −0.0015 | 0.1581 | 88.840 |
| −0.0949 | 0.0079 | ||
| black ~ mean latitudinal range | −0.0780 | 0.0443 | 48.665 |
| Note: All results are listed in Table S11. | |||
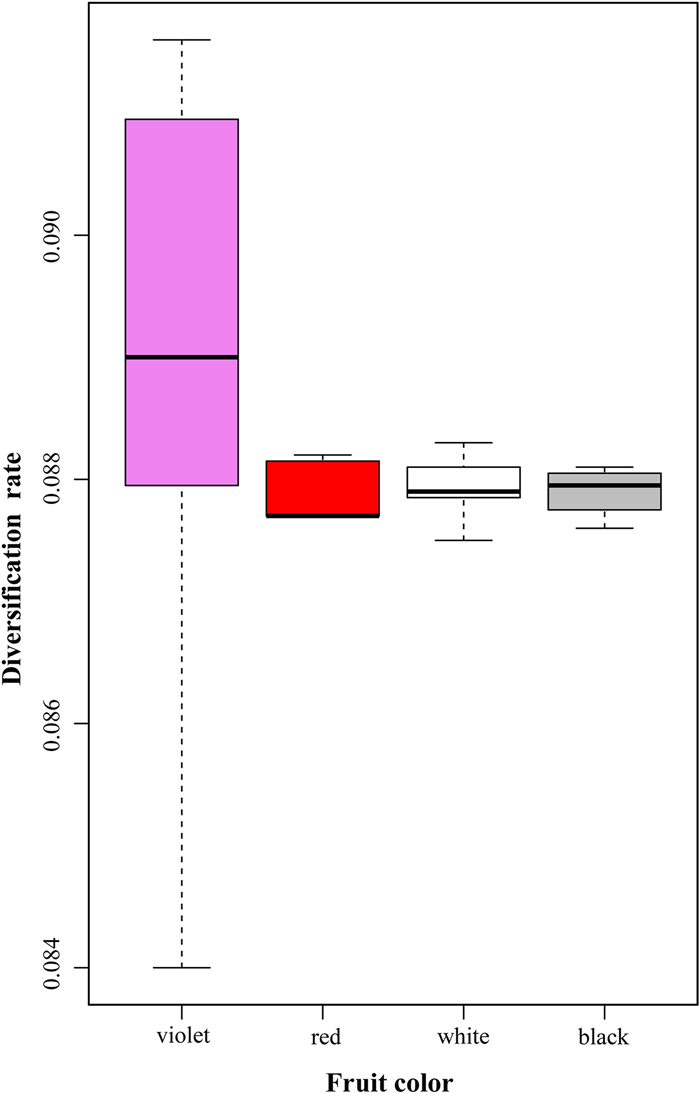
In addition, violet fruits had the highest diversification rate and were positively correlated with the diversification rate of Callicarpa (Fig. 5; Table S14). This finding indicates why violet fruit, which is distributed broadly in the eight regions, is the most common fruit in Callicarpa.
|
| Fig. 5 Diversification rates of different fruit colours based on the consensus tree. |
In this study, we first plotted a global distribution map of Callicarpa species based on four colours using specimen data from 16 significant herbariums (Fig. 1A). We also revealed significant relationships between the four fruit colours and geographic attributes of different regions. Specifically, violet fruit is associated with higher elevations and latitudes and is distributed throughout the eight distribution regions examined; red fruits are largely found at lower latitudes and concentrated mainly in tropical regions, but also occur in moderate numbers at higher latitudes; white fruit is positively correlated with higher elevations and is more widespread; black fruit is correlated with lower latitudes and concentrated in the tropics (Fig. 1; Table 1). These results help us understand the variety of fruit colours on the evolutionary branches of Callicarpa.
How did bioenvironmental factors promote such a distribution pattern? It has been documented that frugivory occurs more frequently at higher latitudes and lower elevations (Willson et al., 1996; Nishi and Tsuyuzaki, 2004; Young et al., 2012). Moreover, avian perceivers prefer darker fruits, including red and violet fruits (Willson et al., 1990a; Schaefer et al., 2014). Another bioenvironmental factor that may explain this pattern is anthocyanin content. Anthocyanins, which absorb high levels of radiation, are synthesised in fruits to prevent frostbite in colder temperate areas (Steyn et al., 2002, 2009; Latti et al., 2008). The correlation between deep-coloured fruits (e.g., violet, red, black) and lower latitudes may also be explained by Gloger's rule, which suggests changes in selection pressure (e.g., UV irradiance) favour darker animals at latitudes closer to the equator (Cuthill, 2015). Taken together, these studies help interpret the spatial distribution of violet fruit investigated in our study. Previous studies on the global distribution of coloured fruit species has shown that red fruit is more common at higher latitudes (Sinnott-Armstrong et al., 2018). However, for Callicarpa, red fruit is mainly distributed in the low latitudes of the tropics, with 23 species in Southeast Asia (mainly in Borneo; Table S9; Bramley, 2009). Furthermore, the red fruit species of Callicarpa tended to spread to higher latitudes; for instance, Callicarpa luteopunctata and C. erythrosticta are distributed in the southwestern temperate regions of China (Chen and Michael, 1994). Additionally, studies have suggested that black fruits are more prevalent in the Neotropics than in Europe (Sinnott-Armstrong et al., 2018). Coincidentally, the black fruit of Callicarpa is also principally centred between the tropics of Cancer and Capricorn (Fig. 1E).
Why has violet fruit adapted to habitats at higher elevations? Anthocyanins not only provide visual cues for animals to induce pollination or seed dispersal, but also act as a light-energy absorption and reflection system that effectively protects plants against the damage caused by UV radiations (Campanella et al., 2014). Plants that grow in habitats at relatively high elevations and low latitudes are generally thought to be exposed to high levels of UV radiation. For instance, studies have indicated that delphinidin, which is involved in the synthesis of purple anthocyanins, was first produced in gymnosperms at high elevation habitats during the Carboniferous period to protect against intense UV radiation damage (Campanella et al., 2014). In addition, studies on skin colour in humans and lizards have shown that populations at higher elevations consist of darker-skinned individuals (Zhang et al., 2012; Reguera et al., 2014).
Intriguingly, white fruit were associated with higher elevation. During our field observations, white fruit species of Callicarpa, such as C. kochiana, C. longifolia, and C. macrophylla, were usually found in shady places of the understory, and appeared more conspicuous in dark environments. These fruits had higher water content than fruits of other colours and were probably favoured by the birds (Lu et al., 2019). Future studies will investigate frugivorous animals based on their preference for Callicarpa fruits of each colour.
Globally, the diversity of fruit colours increases towards the tropics (Jansson and Davies, 2008; Pyron and Wiens, 2013; Rolland et al., 2014); a similar trend was observed in the fruit-colour diversity in Callicarpa (Fig. 1; Table S9).
4.2. Diversification historyThe crown age of Callicarpa was estimated to be 35.53 Ma (Figs. 2 and S5, node 1, 95% HPD: 18.33–57.46 Ma) in the Eocene, which correlates closely with the Ecoene–Oligocene boundary (Fig. 2), followed by subsequent and rapid diversification into the major clades during the late Miocene (Figs. 2 and S5, 15.87–0.79 Ma). This period during the Eocene (~50 Ma) also corresponds to the uplift and formation of the Himalayas and the Tibetan Plateau (Harrison et al., 1992). Further, paleobotanical evidence indicates that significant cooling occurred during the early to middle Eocene (50–48 Ma), followed by two warm intervals (46–43 Ma and 37–34 Ma) during the middle to late Eocene (Wolfe, 1978; Wolfe et al.1997), and this last warm period (37–34 Ma) facilitated the origin and early differentiation of Callicarpa. Our semi-logarithmic lineage-through-time (LTT) plot indicates an accelerated lineage accumulation after ca. 20 Ma (Fig. 3). This acceleration may be attributed to global temperature warming, the shrinkage of Antarctic glaciers during the late Oligocene to Miocene, and a notable temperature peak in the Miocene (Gao et al., 2020). Donoghue and Smith (2004) also proposed that the Miocene was an active period of species differentiation.
Additionally, our results from BAMM analysis revealed that Callicarpa experienced an early burst of diversification followed by a steady decline of diversification, with a relatively constant extinction rate. We estimated diversification rates begining from the Eocene (~ 35 Ma), which had a rate of 0.17 species per million years (species/Myr). This rate is lower than many fast plant radiations in Southwest China, including of alpine bamboos (0.75 species/Myr, Poaceae; Ye et al., 2019), Hirculoideae (Engl. et Irmsch.) Gornall (0.455 species/Myr; Saxifragaceae; Ebersbach et al., 2017), or in other biodiversity groups (Andean Lupinus L., 2.49–3.72 species/Myr; Fabaceae; Hughes and Eastwood, 2006; Andean bellflowers, 1.83 species/Myr; Campanulaceae; Lagomarsino et al., 2016). However, it is higher than rates detected in 17 angiosperm clades (ca. 0.12 species/Myr; Xing and Ree, 2017), and is equivalent to the diversification rate of the tribe Anastaticeae (0.17 species/Myr; Brassicaceae; Qian et al., 2018). In addition, early bursts and subsequent slowdowns in diversification are considered to be the main features of adaptive radiation (Glor, 2010; Yoder et al., 2010; Moen and Morlon, 2014). The early burst of diversification may have been powered by the exploitation of new ecological niches, the result of lineages entering new adaptive zones before the newly available ecological niches reached saturation (Yoder et al., 2010). Biological explanations might also underlie the slowdown of diversification, such as biogeographic events (e.g., vicariance) and variation in how strongly environments promote bursts of speciation (Moen and Morlon, 2014). Importantly, diversification rate would be limited by ecological niche space regardless of how Callicarpa radiated (Kong et al., 2022).
4.3. Fruit colour and biogeographical dispersalWe found that all four fruit colours were involved in dispersal events. Specifically, violet fruit were most significantly correlated with dispersal distance and dispersal frequency (Fig. 4; Table S10). How did violet fruit achieve a wide distribution among regions? Dispersal has been shown to underlie variance in speciation spanning biogeographical areas (Holt et al., 2013). Essentially, animal-mediated dispersal of seeds influences how ecosystems respond to global change, and nearly half of all plant species are dependent on animal activity to migrate seeds in order to adapt to climate change (Holt et al., 2013). Nevertheless, no specific predators of Callicarpa fruit have been reported to date. Our field observations lead us to speculate that birds may be an important medium for spreading the fruits of Callicarpa. There are three birds from North America that most likely prey on the violet fruit of Callicarpa (Fig. 6). Additionally, Zosterops japonicus and Spizixos semitorques have been recorded feeding on violet fruit of Callicarpa in Taiwan and Beijing (China), respectively (http://www.photosharp.com.tw/DGPhoto/PublishView.aspx?id=607370&qid=15&qstr=27178191; https://www.birdnet.cn/forum.php?mod=viewthread&tid=706387&mobile=1). Z. japonicus are found in many areas between Japan and the Philippines (MacKinnon et al., 2000). Therefore, we speculate that the involvement of birds in spreading fruits may be an important factor that explains why violet fruit had the most striking dispersal frequency and distance, further explaining why they occur on the largest number of Callicarpa species (53.7%; Table S6). However, white fruit may also attract birds to achieve significant dispersal frequency and distance (Table S10), as reported in the tribe Gaultherieae (Lu et al., 2019). Obviously, more fieldwork is required to verify this hypothesis.

|
| Fig. 6 Birds that prefer the fruits of Callicarpa in Northern America. A, B, Northern Cardinal (male), C, Northern Cardinal (female), D, White-Crowned Sparrow. |
Long-distance dispersal and overwater dispersal events may be associated with red fruit (Janson, 1983; Wheelwright, 1988; Willson et al., 1989; Lu et al., 2019). Our results also indicate that red fruit may be subject to long-distance dispersal. Although we found no evidence that red fruit is significantly involved in dispersal events in Callicarpa, the dispersal distance was the third highest for red fruit (Table S10). The lack of a correlation between red fruit and dispersal events may be ascribed to insufficient sampling, a lower proportion of species (25.3%), and restricted habitats. Black fruit was also not strongly correlated with dispersal events, which may be a result of the small proportion of species in our study (7.3%).
Does the striking difference in the frequency and distance of four fruit colours relate to differences in the nutrient content within variably coloured fruits? It has been reported that the blue fruits of Viburnum Linn. contain high lipid content, enabling migratory birds to absorb more nutrients to meet their daily energy requirements, while the red and black fruits are quite juicy and contain little sugar, offering a low-quality reward (Sinnott-Armstrong et al., 2020). Future studies are warranted to ascertain what type of nutrients are present in each colour of Callicarpa fruit and whether there are other predators besides birds.
4.4. Fruit colours and species diversificationLu et al. (2019) and Spriggs et al. (2015) observed an important pattern —purple (violet) fruits accelerated diversification rate in contrast to other fruit colours (e.g. red fruits) in tribe Gaultherieae (Ericaceae) and Viburnum (Adoxaceae). Coincidentally, our results support this pattern, as violet fruit accelerated diversification rates in Callicarpa (Fig. 5; Table S14). In addition, previous studies on palms have reported that diversification rates are higher for plant groups with violet-coloured fruit than for those with red or white fruit (Arecaceae; Areces-Berazain and James, 2017). Thus, particular fruit colours might influence the diversification of different angiosperm lineages in a similar way.
Why do violet fruits promote species diversity in Callicarpa? Previous studies have suggested that low-latitude habitats accelerate diversification (e.g., Jansson and Davies, 2008; Pyron and Wiens, 2013; Rolland et al., 2014), especially higher elevation tropical environments (e.g. Hughes and Eastwood, 2006; Hutter et al., 2017). Although the violet fruits of Callicarpa are positively correlated with latitude and elevation (Table 1), a large number of species with violet fruits, including duplicate species, are distributed at low tropical latitudes (Southeast Asia: 23; 10 species in Oceania). Thus, we speculate that violet fruits are more adapted to variable environments, as previously documented for violet fruit in Delphinidin, which can resist intense UV radiation (Campanella et al., 2014), severe cold, and survive in multiple temperature zones and altitude gradients (Steyn et al., 2002, 2009; Latti et al., 2008). In a study on the diversification and evolution of fruit in eumalvoids (Malvaceae), Areces-Berazain and James (2017) proposed that once septic separation of the carpels arises, it is maintained over time, with higher adaptive value. Such morphological characteristics may be one explanation for the strong anti-adversity of the Callicarpa (e.g. two carpels; berries; pericarp has metallic luster; hair on the leaves; erect shrub, woody climber, or trees). In addition, our finding that violet fruits were most frequently involved in dispersal events and dispersed the longest distances (Fig. 4) relative to other fruit colours suggests that violet fruits may have a stronger dispersal ability to promote the diversification rate of Callicarpa. These results may be influenced by important vicariance events (e.g. 26.87 Ma, Fig. S5, node 2, 95% HPD: 12.17–43.57 Ma) that prompted the occurrence of violet fruits (C. americana) in the Pacific, North America and the Neotropics. Simultaneously, our findings may also be attributed to violet fruits being favoured by birds (Fig. 6), which may facilitate seed dispersal, increase the chances of establishing isolated populations, and enhance the rate of species evolution (Herrera, 1989; Tiffney and Mazer, 1995; Smith et al., 2013; Lu et al., 2019). In contrast, red-fruited and black-fruited species may have remained in the tropics due to limitations in habitat and survivability (Fig. 1C and E; Table 1). Furthermore, more large predators have been reported to live at lower latitudes in the tropics than at higher latitudes (Sinnott-Armstrong et al., 2018). Thus, the distribution of red and black fruits of Callicarpa may be affected by the feeding and shorter life routes of these large predators. Nevertheless, such a diverse pattern remains to be studied. The habitat of white fruits is usually restricted to shady places, which does reduce plant–animal interactions to some extent (Lu et al., 2019).
The results of S-DEC and SIMMAP suggest that violet fruits were the primary driving force in the cross-regional diffusion of Callicarpa seeds that increased species richness in each region (Fig. 7). Other fruit colours evolved from violet-fruited species within each region, and subsequently spread to distinct areas. Violet fruits then showed accelerated diversification and high species richness in various habitats. In summary, these patterns are consistent with not only the distribution of Callicarpa but also the biogeographical reconstruction, fruit-colour evolution (Figs. 1 and 2), and relationships between fruit colour and geographic factors (Table 1). Therefore, our findings indicate that at the genus level violet fruits may play a crucial role in promoting species diversity in angiosperms (Spriggs et al., 2015; Lu et al., 2019). However, red fruits are more frequently involved in interregional dispersal events, and violet fruits accelerate the diversification rate to drive variation in fruit colour over macroevolutionary time scales in the tribe Gaultherieae (Ericaceae). Alternatively, the difference between these two diversification patterns may depend on the specific frugivorous dispersers, dispersal patterns, and spatial distribution and dispersal ability of different fruit colours (Jordano et al., 2007; Hill et al., 2021). Therefore, more investigations on other plant groups are needed to conclude whether these patterns apply to other angiosperms. It will be exciting if this pattern of diversification gains support from future ecological studies of more plant groups, as it may help explain the diversification of fruit colour among different regions around the world.
|
| Fig. 7 Species richness and association between fruit colour and dispersal events among eight distribution areas. In the upper section of the figure, species richness is indicated by the species numbers of different fruit colours in each distribution region (Table S9). In the lower part of the figure, BBM (Fig. S5) and DEC analysis summarising associations among fruit colour, dispersal and evolutionary changes between biogeographic regions. Thin lines indicate the dispersal route of each fruit colour, and thick lines indicate the transformation of fruit colours. |
Our study combined the analysis of phylogeny, biogeography, and statistics to reveal the correlations among four fruit colours, geographical distributions, and the dispersal and diversification rates in Callicarpa. Our results indicate that fruit colour is strongly associated with geographical distribution (e.g., violet fruit is correlated with higher latitudes and elevations, red with lower latitudes, white with higher elevations, and black with lower latitudes). Furthermore, all fruit colours were involved in dispersal events; however, the involvement of violet fruit was markedly higher. Most importantly, violet fruit promoted diversification in Callicarpa and drove the evolution and diversity of different fruit colours between regions. Overall, this study sheds new light on why a specific group of angiosperms has variable fruit colours in different locations.
AcknowledgementsWe immensely appreciated the in-depth expertise and precious samples from Prof. Dianxiang Zhang and Dr. Gemma L.C. Bramley (South China Botanical Garden, CAS, China; The Royal Botanic Gardens, Kew, UK). We are also incredibly thankful to post-doctor Lu Sun and Longqian Xiao for guiding data analysis and providing valuable advice on the revisions of the manuscript (Xishuangbanna Tropical Botanical Garden, CAS, China; Huaihua University, China). This research was funded by National Natural Science Foundation of China under Grant [31760045 and 31970220] and Natural Science Foundation of Guangxi Province under Grant [2018GXNSFAA281132].
Author contribution
ZHM and XL conceived and designed the experiments. ZHM, XL, HMC, WQW, and WL collected plant material and species with geographical data and performed the experiments. XL analyzed the data and wrote the manuscript. ZHM and ZWS revised the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.10.002.
Areces-Berazain, Fabiola, James, D.Ackerman, 2017. Diversification and fruit evolution in eumalvoids (Malvaceae). Bot. J. Linn. Soc., 184: 401-417. DOI:10.1093/botlinnean/box035 |
Beaulieu, J.M., Donoghue, M.J., 2013. Fruit evolution and diversification in campanulid angiosperms. Evolution, 67: 3132-3144. DOI:10.1111/evo.12180 |
Bogert, C.M., 1949. Thermoregulation in reptiles, a factor in evolution. Evolution, 3: 195-211. DOI:10.2307/2405558 |
Bollback, J.P., 2006. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics, 7: 88. DOI:10.1186/1471-2105-7-88 |
Bouckaert, R., Heled, J., Kuhnert, D., et al., 2014. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput. Biol., 10: e1003537. DOI:10.1371/journal.pcbi.1003537 |
Bramley, G.L.C., 2009. The genus Callicarpa (Lamiaceae) on Borneo. Bot. J. Linn. Soc., 159: 416-455. DOI:10.1111/j.1095-8339.2009.00907.x |
Bramley, G.L.C., 2011. Distribution patterns in malesian Callicarpa (Lamiaceae). Garden Bull. Singap., 63: 287-298. |
Bremer, K., 1992. Ancestral areas: a cladistic reinterpretation of the center of origin concept. Syst. Biol., 41: 436-445. DOI:10.1093/sysbio/41.4.436 |
Burns, K.C., 2015. The color of plant reproduction: macroecological trade-offs between biotic signaling and abiotic tolerance. Front. Ecol. Evol., 3: 118. DOI:10.4324/9781315664163-13 |
Campanella, J.J., Smalley, J.V., Dempsey, M.E., 2014. A phylogenetic examination of the primary anthocyanin production pathway of the Plantae. Bot. Stud., 55: 1-10. DOI:10.22409/c-legenda.v0i30.26305 |
Cantley, J.T., Markey, A.S., Swenson, N.G., et al., 2016. Biogeography and evolutionary diversification in one of the most widely distributed and species rich genera of the Pacific. AoB Plants, 8: 16. |
Cantrell, C.L., Klun, J.A., Bryson, C.T., et al., 2005. Isolation and identification of mosquito bite deterrent terpenoids from leaves of American (Callicarpa americana) and Japanese (Callicarpa japonica) beautyberry. J. Agric. Food Chem., 53: 5948-5953. DOI:10.1021/jf0509308 |
Cavanaugh, J.E., 1997. Unifying the derivations for the Akaike and corrected Akaike information criteria. Stat. Probab. Lett., 33: 201-208. DOI:10.1016/S0167-7152(96)00128-9 |
Cazetta, E., Schaefer, H.M., Galetti, M., 2008. Does attraction to frugivores or defense against pathogens shape fruit pulp composition?. Oecologia, 155: 277-286. DOI:10.1007/s00442-007-0917-6 |
Chalker-Scott, L., 1999. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol., 70: 1-9. DOI:10.1111/j.1751-1097.1999.tb01944.x |
Chen, S.L., Michael, G.G., 1994. Flora of China, in: Wu Z. Y., & P. H. Raven (eds), Verbenaceae. Science Press & St. Louis: Missouri Botanical Garden Press, Beijing, pp. 25-79.
|
Chen, S.C., Cornwell, W.K., Zhang, H.X., et al., 2017. Plants show more flesh in the tropics: variation in fruit type along latitudinal and climatic gradients. Ecography, 40: 531-538. DOI:10.1111/ecog.02010 |
Chung, I.M., Ali, M., Upadhayay, K., et al., 2005. Isolation and cytotoxic activity of acyclic triterpene callicarpenol from Callicarpa macrophylla. Asian J. Org. Chem., 17: 1907-1914. |
Cuthill, I.C., 2015. Flower colour: gloger's rule isn't just for the birds. Nat. Plants, 1: 1-2. |
Dodd, M.E., J, Silvertown, Chase, M.W., 1999. Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. Evolution, 53: 732-744. DOI:10.2307/2640713 |
Donoghue, M.J., Smith, S.A., 2004. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos. T. R. Soc. B., 359: 1633-1644. DOI:10.1098/rstb.2004.1538 |
Doyle, J.J., Doyle, J.D., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Drew, B.T., Sytsma, K.J., 2012. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot., 99: 933-953. DOI:10.3732/ajb.1100549 |
Drummond, A.J., Suchard, M.A., Xie, D., et al., 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol., 29: 1969-1973. DOI:10.1093/molbev/mss075 |
Ebersbach, J., Schnitzler, J., Favre, A., et al., 2017. Evolutionary radiations in the species-rich mountain genus Saxifraga L. BMC Evol. Biol., 17: 119. DOI:10.1186/s12862-017-0967-2 |
Eriksson, O., Bremer, B., 1991. Fruit characteristics, life forms, and species richness in the plant family Rubiaceae. Am. Nat., 138: 751-761. DOI:10.1086/285247 |
Fricke, E.C., Svenning, J.C., 2020. Accelerating homogenization of the global plant-frugivore meta-network. Nature, 585: 74-78. DOI:10.1038/s41586-020-2640-y |
Gao, J., Liao, P.C., Huang, B.H., et al., 2020. Historical biogeography of Acer L. (Sapindaceae): genetic evidence for Out-of-Asia hypothesis with multiple dispersals to North America and Europe. Sci. Rep., 10: 1-10. DOI:10.1038/s41598-019-56847-4 |
Glor, R.E., 2010. Phylogenetic insights on adaptive radiation. Annu. Rev. Ecol. Evol. Syst., 41: 251-270. DOI:10.1146/annurev.ecolsys.39.110707.173447 |
Gould, K.S., 2004. Nature's Swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol., 2004: 314-320. DOI:10.1155/S1110724304406147 |
Harris, A.J., Xiang, Q.Y., 2009. Estimating ancestral distributions of lineages with uncertain sister groups: a statistical approach to Dispersal-Vicariance Analysis and a case using Aesculus L. (Sapindaceae) including fossils. J. Syst. Evol., 47: 349-368. DOI:10.1111/j.1759-6831.2009.00044.x |
Harrison, T.M., Copeland, P., Kidd, W.S.F., et al., 1992. Raising Tibet. Science, 255: 1663-1670. DOI:10.1126/science.255.5052.1663 |
Herrera, C.M., 1989. Seed dispersal by animals: a role in angiosperm diversification?. Am. Nat., 133: 309-322. DOI:10.1086/284921 |
Hill, A.P., Jiménez, M.F.T., Chazot, N., et al., 2021. Fruit colour and range size interact to influence diversification. bioRxiv. DOI:10.1101/2021.10.26.465838 |
Holt, B.G., Lessard, J.P., Borregaard, M.K., et al., 2013. Response to comment on "An update of Wallace's zoogeographic regions of the world". Science, 341: 343. DOI:10.1126/science.1237541 |
Hughes, C., Eastwood, R., 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. U.S.A., 103: 10334-10339. DOI:10.1073/pnas.0601928103 |
Hutter, C.R., Lambert, S.M., Wiens, J.J., 2017. Rapid diversification and time explain amphibian richness at different scales in the Tropical Andes, Earth's most biodiverse hotspot. Am. Nat., 190: 828-843. DOI:10.1086/694319 |
Jaa, A.A., 2004. MrModeltest v. 2 (program distributed by the author).
|
Jafari, M., Ansari-Pour, N., 2019. Why, when and how to adjust your P values?. Cell J., 20: 604. |
Janson, C.H., 1983. Adaptation of fruit morphology to dispersal agents in a neotropical forest. Science, 219: 187-189. DOI:10.1126/science.219.4581.187 |
Jansson, R., Davies, T.J., 2008. Global variation in diversification rates of flowering plants: energy vs. climate change. Ecol. Lett., 11: 173-183. |
Jetz, W., Thomas, G.H., Joy, J.B., et al., 2012. The global diversity of birds in space and time. Nature, 491: 444-448. DOI:10.1038/nature11631 |
Jordano, P., Garcia, C., Godoy, J.A., et al., 2007. Differential contribution of frugivores to complex seed dispersal patterns. Proc. Natl. Acad. Sci. U.S.A., 104: 3278-3282. DOI:10.1073/pnas.0606793104 |
Kar, R., 1996. On the Indian origin of Ocimum (Lamiaceae): a palynological approach. Palaeobotanist, 43: 43-50. |
Kemp, D.J., Herberstein, M.E., Fleishman, L.J., et al., 2015. An integrative framework for the appraisal of coloration in nature. Am. Nat., 185: 705-724. DOI:10.1086/681021 |
Klaus, K.V., Matzke, N.J., 2020. Statistical comparison of trait-dependent biogeographical models indicates that podocarpaceae dispersal is influenced by both seed cone traits and geographical distance. Syst. Biol., 69: 61-75. DOI:10.1093/sysbio/syz034 |
Kong, H., Condamine, F.L., Yang, L., et al., 2022. Phylogenomic and macro-evolutionary evidence for an explosive radiation of a plant genus in the Miocene. Syst. Biol., 71: 589-609. DOI:10.1093/sysbio/syab068 |
Kozak, K.H., Wiens, J.J., 2016. Testing the relationships between diversification, species richness, and trait evolution. Syst. Biol., 65: 975-988. DOI:10.1093/sysbio/syw029 |
Lagomarsino, L.P., Condamine, F.L., Antonelli, A., et al., 2016. The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytol., 210: 1430-1442. DOI:10.1111/nph.13920 |
Landis, M.J., Matzke, N.J., Moore, B.R., et al., 2013. Bayesian analysis of biogeography when the number of areas is large. Syst. Biol., 62: 789-804. DOI:10.1093/sysbio/syt040 |
Landis, M.J., Freyman, W.A., Baldwin, B.G., 2018. Retracing the Hawaiian silversword radiation despite phylogenetic, biogeographic, and paleogeographic uncertainty. Evolution, 72: 2343-2359. DOI:10.1111/evo.13594 |
Latti, A.K., Riihinen, K.R., Kainulainen, P.S., 2008. Analysis of anthocyanin variation in wild populations of bilberry (Vaccinium myrtillus L.) in Finland. J. Agric. Food Chem., 56: 190-196. DOI:10.1021/jf072857m |
Li, B., Cantino, P.D., Olmstead, R.G., et al., 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci. Rep., 6: 1-18. DOI:10.5089/9781484324752.001 |
Lu, L., Fritsch, P.W., Matzke, N.J., et al., 2019. Why is fruit colour so variable? Phylogenetic analyses reveal relationships between fruit-colour evolution, biogeography and diversification. Global Ecol. Biogeogr., 28: 891-903. DOI:10.1111/geb.12900 |
MacKinnon, J.R., MacKinnon, J., Phillipps, K., et al., 2000. A field guide to the birds of China. Oxford University Press.
|
Mahler, D.L., Revell, L.J., Glor, R.E., et al., 2010. Ecological opportunity and the rate of morphological evolution in the diversification of Greater Antillean anoles. Evolution, 64: 2731-2745. DOI:10.1111/j.1558-5646.2010.01026.x |
Martinez-Millan, M., 2010. Fossil record and age of the Asteridae. Bot. Rev., 76: 83-135. DOI:10.1007/s12229-010-9040-1 |
McGuire, J.A., Witt, C.C., Remsen, J.V., et al., 2014. Molecular phylogenetics and the diversification of hummingbirds. Curr. Biol., 24: 1038. DOI:10.1016/j.cub.2014.04.019 |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: extreme digital discovery, 2011: 1-8. |
Moen, D., Morlon, H., 2014. Why does diversification slow down?. Trends Ecol. Evol., 29: 190-197. DOI:10.1016/j.tree.2014.01.010 |
Moore, B.R., Hohna, S., May, M.R., et al., 2016. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natl. Acad. Sci. U.S.A., 113: 9569-9574. DOI:10.1073/pnas.1518659113 |
Nakagawa, S., 2004. A farewell to Bonferroni: the problems of low statistical power and publication bias. Behav. Ecol., 15: 1044-1045. DOI:10.1093/beheco/arh107 |
Nakanishi, H., 1996. Fruit color and fruit size of bird-disseminated plants in Japan. J. Veg. Sci., 123: 207-218. |
Nishi, H., Tsuyuzaki, S., 2004. Seed dispersal and seedling establishment of Rhus trichocarpa promoted by a crow (Corvus macrorhynchos) on a volcano in Japan. Ecography, 27: 311-322. DOI:10.1111/j.0906-7590.2004.03737.x |
Nylander, J.A., Olsson, U., Alstrom, P., et al., 2008. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst. Biol., 57: 257-268. DOI:10.1080/10635150802044003 |
Paradis, E., Claude, J., Strimmer, K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20: 289-290. DOI:10.1093/bioinformatics/btg412 |
Pyron, R.A., Wiens, J.J., 2013. Large-scale phylogenetic analyses reveal the causes of high tropical amphibian diversity. P. Roy. Soc. B-Biol. Sci., 280: 20131622. DOI:10.1098/rspb.2013.1622 |
Qian, C.J., Shi, Y., Liu, Y., et al., 2018. Phylogenetics and dispersal patterns of Brassicaceae around the Qinghai-Tibet Plateau. J. Syst. Evol., 56: 202-217. DOI:10.1111/jse.12312 |
Rabosky, D.L., Donnellan, S.C., Grundler, M., et al., 2014. Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst. Biol., 63: 610-627. DOI:10.1093/sysbio/syu025 |
Rabosky, D.L., Grundler, M., Anderson, C., et al., 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol., 5: 701-707. DOI:10.1111/2041-210X.12199 |
Rambaut, A., Drummond, A.J., Xie, D., 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol., 67: 901. DOI:10.1093/sysbio/syy032 |
Reguera, S., Zamora-Camacho, F.J., Moreno-Rueda, G., 2014. The lizard Psammodromus algirus (Squamata: Lacertidae) is darker at high altitudes. Biol. J. Linn. Soc., 112: 132-141. DOI:10.1111/bij.12250 |
Revell, L.J., 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol., 3: 217-223. DOI:10.1111/j.2041-210X.2011.00169.x |
Rolland, J., Condamine, F.L., Jiguet, F., et al., 2014. Faster speciation and reduced extinction in the tropics contribute to the mammalian latitudinal diversity gradient. PLoS Biology, 12: e1001775. DOI:10.1371/journal.pbio.1001775 |
Schaefer, H.M., McGraw, K., Catoni, C., 2008. Birds use fruit colour as honest signal of dietary antioxidant rewards. Funct. Ecol., 22: 303-310. DOI:10.1111/j.1365-2435.2007.01363.x |
Schaefer, H.M., Valido, A., Jordano, P., 2014. Birds see the true colours of fruits to live off the fat of the land. P. Roy. Soc. B-Biol. Sci., 281: 20132516. DOI:10.1098/rspb.2013.2516 |
Shanahan, M., So, S., Compton, S.G., et al., 2001. Fig-eating by vertebrate frugivores: a global review. Biol. Rev., 76: 529-572. |
Shaw, J., Lickey, E.B., Beck, J.T., et al., 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot., 92: 142-166. DOI:10.3732/ajb.92.1.142 |
Shaw, J., Lickey, E.B., Schilling, E.E., et al., 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot., 94: 275-288. DOI:10.3732/ajb.94.3.275 |
Sinnott-Armstrong, M.A., Downie, A.E., Federman, S., et al., 2018. Global geographic patterns in the colours and sizes of animal-dispersed fruits. Global Ecol. Biogeogr., 27: 1339-1351. DOI:10.1111/geb.12801 |
Sinnott-Armstrong, M.A., Lee, C., Clement, W.L., et al., 2020. Fruit syndromes in Viburnum: correlated evolution of color, nutritional content, and morphology in bird-dispersed fleshy fruits. BMC Evol. Biol., 20: 1-19. DOI:10.1186/s12862-019-1549-2 |
Sinnott-Armstrong, M.A., Donoghue, M.J., Jetz, W.J., 2021. Dispersers and environment drive global variation in fruit colour syndromes. Ecol. Lett., 24: 1387-1399. DOI:10.1111/ele.13753 |
Smith, J.F., 2001. High species diversity in fleshy-fruited tropical understory plants. Am. Nat., 157: 646-653. DOI:10.1086/320625 |
Smith, S.B., DeSando, S.A., Pagano, T., 2013. The value of native and invasive fruit-bearing shrubs for migrating songbirds. Northeast. Nat., 20: 171-184. DOI:10.1656/045.020.0114 |
Sotomayor, D.A., 2014. Biotic evolution and environmental change in Southeast Asia. Can. Geogr., 58: E58. DOI:10.1111/cag.12114 |
Spriggs, E.L., Clement, W.L., Sweeney, P.W., et al., 2015. Temperate radiations and dying embers of a tropical past: the diversification of Viburnum. New Phytol., 207: 340-354. DOI:10.1111/nph.13305 |
Steyn, W.J., Wand, S.J.E., Holcroft, D.M., et al., 2002. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol., 155: 349-361. DOI:10.1046/j.1469-8137.2002.00482.x |
Steyn, W.J., Wand, S.J.E., Jacobs, G., et al., 2009. Evidence for a photoprotective function of low-temperature-induced anthocyanin accumulation in apple and pear peel. Physiol. Plantarum, 136: 461-472. DOI:10.1111/j.1399-3054.2009.01246.x |
Sullivan, D., 2009. Google earth Pro. EContent, 32: 16-18. |
Tiffney, B.H., Mazer, S.J., 1995. Angiosperm growth habit, dispersal and diversification reconsidered. Evol. Ecol., 9: 93-117. DOI:10.1007/BF01237700 |
Tu, Y.H., Sun, L.N., Guo, M.L., et al., 2013. The medicinal uses of Callicarpa L. in traditional Chinese medicine: an ethnopharmacological, phytochemical and pharmacological review. J. Ethnopharmacol., 146: 465-481. DOI:10.1016/j.jep.2012.12.051 |
Valenta, K., Nevo, O., 2020. The dispersal syndrome hypothesis: how animals shaped fruit traits, and how they did not. Funct. Ecol., 34: 1158-1169. DOI:10.1111/1365-2435.13564 |
Van Dam, M.H., Matzke, N.J., 2016. Evaluating the influence of connectivity and distance on biogeographical patterns in the south-western deserts of North America. J. Biogeogr., 43: 1514-1532. DOI:10.1111/jbi.12727 |
Van Iterson, M., Boer, J.M., Menezes, R.X., 2010. Filtering, FDR and power. BMC Bioinformatics, 11: 1-11. |
Vittoz, P., Dussex, N., Wassef, J., et al., 2009. Diaspore traits discriminate good from weak colonisers on high-elevation summits. Basic Appl. Ecol., 10: 508-515. DOI:10.1016/j.baae.2009.02.001 |
Wheelwright, N.T., 1988. Fruit-eating birds and bird-dispersed plants in the tropics and temperate zone. Trends Ecol. Evol., 3: 270-274. DOI:10.1016/0169-5347(88)90061-4 |
Willson, M.F., Irvine, A.K., Walsh, N.G., 1989. Vertebrate dispersal syndromes in some Australian and New Zealand plant-communities, with geographic comparisons. Biotropica, 21: 133-147. DOI:10.2307/2388704 |
Willson, M.F., Graff, D.A., Whelan, C.J., 1990. Color preferences of frugivorous birds in relation to the colors of fleshy fruits. Condor, 92: 545-555. DOI:10.2307/1368671 |
Willson, M.F., Rice, B.L., Westoby, M., 1990. Seed dispersal spectra: a comparison of temperate plant communities. J. Veg. Sci., 1: 547-562. DOI:10.2307/3235789 |
Willson, M.F., Sabag, C., Figueroa, J., et al., 1996. Seed dispersal by lizards in Chilean rainforest. Rev. Chil. Hist. Nat., 69: 339-342. |
Wolfe, J.A., 1978. A paleobotanical interpretation of Tertiary climates in the Northern Hemisphere: data from fossil plants make it possible to reconstruct Tertiary climatic changes, which may be correlated with changes in the inclination of the earth's rotational axis. Am. Sci., 66: 694-703. |
Wolfe, J.A., Schorn, H.E., Forest, C.E., et al., 1997. Paleobotanical evidence for high altitudes in Nevada during the Miocene. Science, 276: 1672-1675. DOI:10.1126/science.276.5319.1672 |
Xing, Y.W., Ree, R.H., 2017. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A., 114: E3444-E3451. |
Ye, X.Y., Ma, P.F., Yang, G.Q., et al., 2019. Rapid diversification of alpine bamboos associated with the uplift of the Hengduan Mountains. J. Biogeogr., 46: 2678-2689. DOI:10.1111/jbi.13723 |
Yoder, J.B., Clancey, E., Des Roches, S., et al., 2010. Ecological opportunity and the origin of adaptive radiations. J. Evol. Biol., 23: 1581-1596. DOI:10.1111/j.1420-9101.2010.02029.x |
Young, L.M., Kelly, D., Nelson, X.J., 2012. Alpine flora may depend on declining frugivorous parrot for seed dispersal. Biol. Conserv., 147: 133-142. DOI:10.1016/j.biocon.2011.12.023 |
Yu, Y., Harris, A.J., Blair, C., et al., 2015. RASP (reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol., 87: 46-49. DOI:10.1016/j.ympev.2015.03.008 |
Zhang, Y.B., Li, X., Zhang, F., et al., 2012. A preliminary study of copy number variation in Tibetans. PLoS One, 7: e41768. DOI:10.1371/journal.pone.0041768 |
Zoratti, L., Karppinen, K., Escobar, A.L., et al., 2014. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci., 5: 534. |



