b. Qinghai Province Key Laboratory of Crop Molecular Breeding, Xining, China;
c. University of Chinese Academy of Sciences, Beijing, China
Elymus nutans Griseb. is a well-known perennial and caespitose grass belonging to the genus Elymus L. in the tribe Triticeae of the family Poaceae, which comprises approximately 150 perennial and exclusively polyploid species (Löve, 1984). E. nutans is widely distributed in central and eastern Asia including the Himalayas (Lu, 1993; Chen and Zhu, 2006) and grows in a variety of ecological habitats, such as grasslands, bush land, riverbanks, mountain slopes, and swales, at altitudes ranging from 1000 to 5000 m (Lu, 1993). E. nutans is considered a potential forage crop for agriculture on the Qinghai-Tibet Plateau, China, due to its high mass, high quality, and high resistance to various environmental stresses associated with high altitude (Lu and Nie, 2002). E. nutans demonstrates great variability in its morphological traits (Zhang et al., 2009), as shown by ISSR analysis (Chen et al., 2009a), SRAP (Chen et al., 2009b), AFLP (Yan et al., 2010), gliadin polymorphism (Miao et al., 2011), and SSR analysis (Chen et al., 2013).
Elymus nutans was cytogenetically identified as an allohexaploid (2n = 6x = 42) with a genomic constitution of StStHHYY (Lu, 1993), where St is derived from Pseudoroegneria (Neveski) Löve, H comes from Hordeum L., and Y originates from an unknown species (Wang et al., 1994). Chromosome rearrangements in E. nutans were first reported from the analysis of chromosome pairing in a hybrid between parents from two different populations (Lu, 1993). A high frequency of karyotype variations in a domesticated population of E. nutans was observed using sequential FISH and GISH (Dou et al., 2009), and karyotype variation in E. nutans has also been detected in natural populations (Dou et al., 2017). High frequencies of intergenomic translocations, amplification and deletion of repeats have been identified in the studies described above (Dou et al., 2009, 2017). However, chromosome structural variations such as inversion and intragenomic translocation have not been identified because of the limitations of the techniques. Thus, it is possible that variations in the karyotype of E. nutans may involve additional chromosome structural variations. Chromosome rearrangement or chromosome structural variations are largely related to ecotype differentiation, or even to speciation, through recombination suppression (Rieseberg, 2001; Todesco et al., 2020). Although E. nutans is regarded as a self-pollinating plant, intrapopulation differentiation has been somewhat supported by a population structure in which the majority of the total variation occurs within the population rather than among populations (Chen et al., 2009a, 2013; Yan et al., 2010; Miao et al., 2011). It has been suggested that E. nutans may combine a certain level of self-pollination and outcrossing (Miao et al., 2011) or that the population structure of E. nutans is strongly affected by excess gene flow caused by seed dispersal from grazing animals (Yan et al., 2010). However, since highly variable karyotypes have been identified within populations, differentiation resulting from chromosome rearrangement is considered very likely in E. nutans.
Low fertility and even sterile plants, which empirically show no distinct relationship to altitude, are frequently observed in the field in various E. nutans populations. Heterozygotes with rearranged chromosomes, such as inversions and translocations, produce chromatin with duplicated and deficient genes; in addition, disjunction patterns during the first meiotic division can cause varying degrees of sterility in the gametophytic and sporophytic phases (Livingstone and Rieseberg, 2004). Given that intercrossing can occur between individuals with varied karyotypes, it is possible that low fertility in E. nutans may be due to heterozygotes derived from parents with different karyotypes, especially those with distinct chromosome rearrangements. In the present study, molecular cytogenetic analysis of varying fertility plants was carried out by fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH). Heterozygosity of varying-fertility plants was identified cytogenetically, and GISH was used to observe chromosome pairing at the meiotic metaphase (MI) of pollen mother cells (PMCs). In addition, the association between chromosome differentiation, chromosome pairing, and fertility was analyzed. The results of this study will be valuable for assessing whether chromosome structural variation plays a vital role in differentiation in E. nutans on the plateau and may even enable the identification of massive haploblocks associated with adaptation.
2. Materials and methods 2.1. Plant materialsElymus nutans plants with low fertility were morphologically identified first by their shedding of spikelets at maturity in the field in the fallen time. Empirically, the spikelets of plants with high seed set were thoroughly shattered at maturity, while in sterile plants, the spikelets without seeds were still preserved in the field. The vegetative bodies of over 20 target samples, including low-fertility and several high-fertility plants, which were growing along the north-eastern areas of Qinghai Lake (Qinghai, China) at an altitude of approximately 3200 m, were transplanted to an experimental plot in Xining, Qinghai, at an altitude of approximately 2200 m. Twenty-three plants were used in seed setting rate statistic and mitotic chromosome preparation, and ten of the 23 plants were used in pollen fertility observation and meiotic preparation. Finally, plants were classified as low fertility (seed set ≤75%) or normal fertility (seed set > 75%) by checking the seed set in bagged spikes in the next growing season.
2.2. Fertility determinationTo assess pollen fertility, pollen grains of mature anthers were examined after staining in I2-IK solution. Over 500 pollen grains from 5 anthers of 2 different inflorescences were counted from each plant. Pollen fertility was described as the percentage of investigated grains that were fertile (%). The inflorescences were individually bagged to evaluate seed setting after wilting. The seed setting rate was measured as the ratio of the number of seeds to the total number of florets per spike (%) from 3 spikes of each plant.
2.3. Mitotic chromosome preparationNascent secondary roots of the transplanted plants were collected with lengths of 1–2 cm. The harvested root tips were pretreated with N2O at 7 atm for 2 h and fixed in 3:1 (v/v) ethanol: glacial acetic acid. Chromosome spreads and slide preparation were performed as described in Xie et al. (2020).
2.4. Meiotic chromosome preparationInflorescences were collected at the early flowering stage, morphologically when the distance between the flag leaf and the next leaf was approximately 1–2 cm. The collected inflorescences were fixed in CarnoyⅡ solution (ethanol: glacial acetic acid: chloroform = 6:1:3) for 24 h and then stored in 70% alcohol at -20 ℃ until use. First, meiotic metaphase Ⅰ (MⅠ) in pollen mother cells (PMCs) was examined using a phase-contrast microscope after squashing in 45% acetic acid. Further processing was the same as the preparation of mitotic chromosome preparation.
2.5. FISH and GISHFour repetitive sequences were used as markers for chromosomal discrimination, including DNA satellites pAs1 (Rayburn and Gill, 1986) and pSc119.2 (Bedbrook et al., 1980), 45S rDNA, and a microsatellite (AAG)10. Assigned oligonucleotides pAs1-1 plus pAs1-2 representing pAs1 and Oligo-pTa71-2 and Oiogo-pSc119.2-1 plus Oligo-pSc119.2-2 representing 45S rDNA and pSc119.2 (Tang et al., 2014), respectively, were end-labelled using either fluorescein amidite (FAM, green) or carboxy tetramethyl rhodamine (TAMRA, red) (Sangon Biotech Co., Ltd., Shanghai, China) to generate FISH probes. Genomic DNA from Hordeum bogdanii Wilensky (2n = 2x = 14, HH) and Pseudoroegneria stipifolia (Czern. ex Nevski) Á. Löve (2n = 2x = 14, StSt) was treated and labelled with fluorescein-12-dUTP and tetramethyl-rhodamine-5-dUTP, respectively, by the random primer labelling method as described by Dou et al. (2009). FISH and GISH were carried out following Xie et al. (2020). Images were obtained using fluorescence microscopy (BX63, Olympus, Tokyo, Japan).
2.6. StatisticsKaryotype heterozygosity was quantified as the ratio of the number of homologous chromosome pairs with different FISH patterns to the total number of chromosome pairs, multiplied by 100%, in each investigated plant and subgenome. Plants with nonzero karyotype heterozygosity were considered to have heterozygosis. The numbers of univalents, bivalents, trivalents and quadrivalents formed within or between the subgenomes in each PMC were counted and averaged to represent the values of each configuration per plant.
One-way analysis of variance (ANOVA) and linear fitting were carried out using Origin Pro 2019 (https://www.originlab.com/2019). Linear fitting was performed between karyotype heterozygosity, pollen fertility, seed setting rate, univalent fraction of total genomes or each subgenome, and bivalent fraction of total genomes or each subgenome. ANOVA was performed among karyotype heterozygosity, univalent fraction and bivalent fraction of each of the subgenomes.
3. Results 3.1. Statistics and correlation analysis of seed setting rate and pollen fertilityExamination of transplanted plants with different seed setting rates (as observed in the field) revealed the diversity of seed setting rates among individuals under experimental plot conditions (Table S1). The results showed that 7 of 23 (30.44%) had seed setting rates exceeding 70%, 2 plants had seed setting rates of 33.2% and 50%, and the remaining plants (60.87%) had seed setting rates below 10%. Ten plants were randomly selected for pollen fertility statistics. Pollen fertility showed high values in some cases, ranging from 84.95% to 92.99% in 3 plants, and low values (from 2.73% to 47.22%) in the others (Table 1, Fig. S1). Correlation analysis showed a distinct relationship between pollen fertility and seed setting (R2 = 0.94, P = 4.45E-6) in 10 plants.
| Plant | No. of PMCs | No. of univalents | No. of bivalents | No. of trivalents | No. of quadrivalents | No. of pentavalents | No. of hexavalents | No. of decavalents | Pollen grain fertility % | |||||
| Total | Homo-logous | Heterologous | ||||||||||||
| Total | Ring | Rod | Ring | Rod | ||||||||||
| HY-21 | 43 | 0.00 | 21.00 | 20.14 | 0.86 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 92.99 |
| HY-2 | 54 | 0.44 | 20.78 | 19.07 | 1.70 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 9.56 |
| HY-3 | 34 | 0.30 | 19.03 | 17.21 | 1.82 | 0.00 | 0.91 | 0.00 | 0.74(Y/H) | 0.18 | 0.00 | 0.00 | 0.00 | 47.22 |
| HY-6 | 54 | 0.00 | 21.00 | 20.39 | 0.61 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 84.95 |
| HY-22 | 78 | 1.59 | 18.58 | 13.22 | 5.36 | 0.03 | 0.80 | 0.01 | 0.39(S/H) | 0.40 | 0.00 | 0.00 | 0.00 | 26.52 |
| HY-13 | 30 | 0.47 | 20.70 | 18.07 | 2.63 | 0 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 10.84 |
| HY-19 | 34 | 0.24 | 18.97 | 17.35 | 1.62 | 0.06 | 0.91 | 0.00 | 0.53(S/H) | 0.38 | 0.00 | 0.00 | 0.00 | 1.86 |
| HY-24 | 52 | 0.15 | 20.89 | 19.89 | 1.00 | 0.00 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 88.98 |
| HY-8 | 32 | 1.50 | 17.38 | 14.56 | 2.81 | 0.06 | 1.00 | 0.03 | 0.50 (0.34S/H, 0.16Y/H) | 0.47 (0.28S/H, 0.19Y/H) | 0.06 (0.03H/S, 0.03H/Y) | 0.16(H/Y) | 0.03(H/Y) | 8.58 |
| HY-20 | 64 | 1.17 | 18.44 | 15.75 | 2.69 | 0.05 | 0.95 | 0.00 | 0.69(S/H) | 0.27 | 0.00 | 0 | 0 | 2.73 |
| Total | 475 | 311 | 9343 | 8253 | 1090 | 9 | 219 | 4 | 133 | 82 | 2 | 5 | 1 | |
| X | 0.66 | 19.67 | 17.38 | 2.30 | 0.02 | 0.46 | 0.01 | 0.28 | 0.17 | 0.004 | 0.011 | 0.002 | ||
| Rate (%) | 1.56 | 93.67 | 82.74 | 10.93 | 0.14 | 4.39 | 0.08 | 2.67 | 1.64 | 0.05 | 0.15 | 0.05 | ||
| S | 1.40 | 1.48 | 2.96 | 2.11 | 0.14 | 0.53 | 0.09 | 0.45 | 0.39 | 0.07 | 0.10 | 0.05 | ||
| CV (%) | 213.46 | 7.52 | 17.02 | 92.13 | 719.57 | 115.21 | 1085.27 | 162.02 | 228.40 | 1538.01 | 969.54 | 2177.15 | ||
Four chromosomal markers (pTa71-2, pSc119.2, pAs1 and (AAG)10) were used to identify chromosomes in E. nutans. Sequential FISH and GISH were conducted with three rounds of hybridization: first with probes of pTa71-2 and pSc119.2, second with probes of pAs1 and (AAG)10, and third with probes of St and H genomic DNA. Following this procedure, molecular karyotyping was carried out by allocating all identified chromosomes into the St, H and Y subgenomes ((Fig. 1). The nomenclature of the labelled chromosomes followed Dou et al. (2017). Finally, molecular karyotypes were obtained for 23 plants with varying seed setting (Fig. S2 and Table S2). The results showed high karyotype variation among plants and high heterozygosity in varying fertility plants; no identical karyotypes were found among these plants.

|
| Fig. 1 FISH patterns in mitotic cells and karyotypes of Elymus nutans representatives. a: HY-20, b: HY-21. a1 and b1 probed by pSc119.2 (green) and pTa71-2 (red), a2 and b2 reprobed by pAs1 (red) and (AAG)10 (green), a3 and b3 reprobed by genomic DNA of Hordeum bogdanii (green) and Pseudoroegneria stipifolia (red), and a4 and b4 are karyotypes of HY-20 and HY-21, respectively. The scale bars are 10 μm. |
A total of 157 chromosome variants, including 9 intergenomic translocations, were uncovered in the 23 plants investigated by FISH and GISH. These variants were unevenly distributed among the genomes and among homologous chromosome pairs in each genome (Fig. S2; Table S2). The Y genome exhibited the highest number of chromosome variants (59), while the St and H genomes had 51 and 47, respectively. Chromosomes 4H, 2St, and 5Y showed the highest numbers of polymorphisms, and chromosomes 2H, 1St and 2Y showed the fewest polymorphisms in each genome (Table S2, Table 2). The polymorphism pattern found in the present study was similar to that for normal fertility samples (seed setting > 75%) (Dou et al., 2017), which showed high to low numbers of polymorphisms from Y to St to H. However, many more variants were detected in the present 23 plants than in the 27 samples with normal fertility (Dou et al., 2017). In the present study, the proportions of variants in the H, St, and Y genomes were 123.81%, 45.71%, and 34.09% greater, respectively, than those in normal fertility samples (Table 2). In addition, chromosomes 1H, 3H, 4H, 5H, 3St, 5Y and 7Y exhibited values over 130% higher than those for the normal fertility samples, but chromosomes 2H, 1St, 1Y, 2Y and 3Y had similar or even decreasing proportions of variants compared with the normal fertility samples (Table 2).
| Chromosome | H1 | H2 | (H1-H2)/H2 (%) | St1 | St2 | (St1-St2)/St2 (%) | Y1 | Y2 | (Y1-Y2)/Y2 (%) |
| 1 | 9 | 3 | 200.00 | 3 | 5 | -40.00 | 7 | 8 | -12.50 |
| 2 | 2 | 2 | 0.00 | 12 | 9 | 33.33 | 5 | 7 | -28.57 |
| 3 | 8 | 3 | 166.67 | 7 | 3 | 133.33 | 8 | 8 | 0.00 |
| 4 | 10 | 4 | 150.00 | 5 | 4 | 25.00 | 7 | 6 | 16.67 |
| 5 | 5 | 2 | 150.00 | 9 | 5 | 80.00 | 14 | 6 | 133.33 |
| 6 | 5 | 3 | 66.67 | 6 | 3 | 100.00 | 10 | 6 | 66.67 |
| 7 | 8 | 4 | 100.00 | 9 | 6 | 50.00 | 8 | 3 | 166.67 |
| Total | 47 | 21 | 123.81 | 51 | 35 | 45.71 | 59 | 44 | 34.09 |
| Note: H1, St1 and Y1: genomes of plants with varying fertility; H2, St2 and Y2: genomes of normal fertility samples from Dou et al. (2017). | |||||||||
Twenty of the 23 investigated plants were heterozygous karyotypes (Table S1). The proportion of heterozygosis (86.96%) in the present sample of plants was approximately 3 times that in normal fertility samples (25.93%) (Dou et al., 2017). Furthermore, the calculated karyotype heterozygosity values varied among plants and genomes (Table S1). Total karyotype heterozygosity among plants ranged from 0 to 76.19%, with an average of 37.47%. H genome karyotype heterozygosity ranged from 0 to 85.71%, with an average of 36.65%. St genome karyotype heterozygosity ranged from 0 to 71.43%, with an average of 37.27%. Y genome karyotype heterozygosity ranged from 0 to 85.71%, with an average of 38.51% (Table S1). In the present study, comparison with data from normal fertility samples (Dou et al., 2017), which had mean values of total heterozygosity and H, St, and Y genome heterozygosity of 2.12%, 2.64%, 1.59%, and 2.12%, respectively, showed that plants with varying fertility exhibited total genome and subgenome heterozygosity over 13 times greater than in normal fertility samples (Table 3).
| Structural heterozygosity | H | St | Y | Total |
| A (Plants with varying fertility, %) | 36.65 | 37.27 | 38.51 | 37.47 |
| B (Plants with normal fertility, %) | 2.64 | 1.59 | 2.12 | 2.12 |
| A/B | 13.88 | 23.44 | 18.17 | 17.67 |
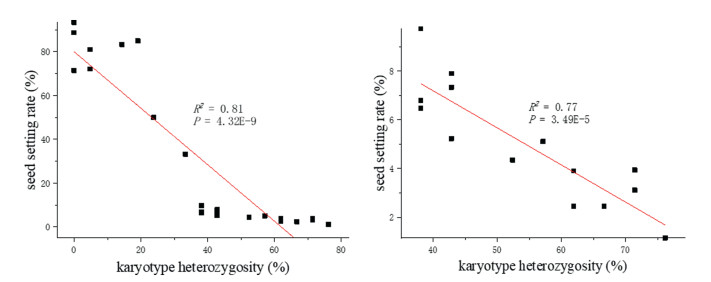
Given that varying seed setting may be related to different karyotype heterozygosities in these plants, correlation analysis and linear fitting were carried out. The fitting results demonstrated that total genome heterozygosity was significantly correlated with the seed setting rate: this was seen not only in the group of 23 plants having seed setting rates of 1.15–93.33% (P = 4.32E-9) but also in the group of 14 plants having seed setting rates of 1.15–9.74% (P = 3.49E-5) (Fig. 2). Nevertheless, when the heterozygosity of each of the genomes was measured, ANOVA revealed no significant differences in heterozygosity among the three genomes (F = 0.03, P = 0.97). However, seed setting rates were significantly correlated with the heterozygosity of the corresponding St (R2 = 0.76, P = 7.04E-8), Y (R2 = 0.75, P = 1.03E-7) and H (R2 = 0.68, P = 1.46E-6) genomes.
|
| Fig. 2 Regression scatterplots of karyotype heterozygosity and seed setting rate. Left: seed setting rate ranged from 1.15% to 93.33%; Right: seed setting rate ranged from 1.15% to 9.74%. |
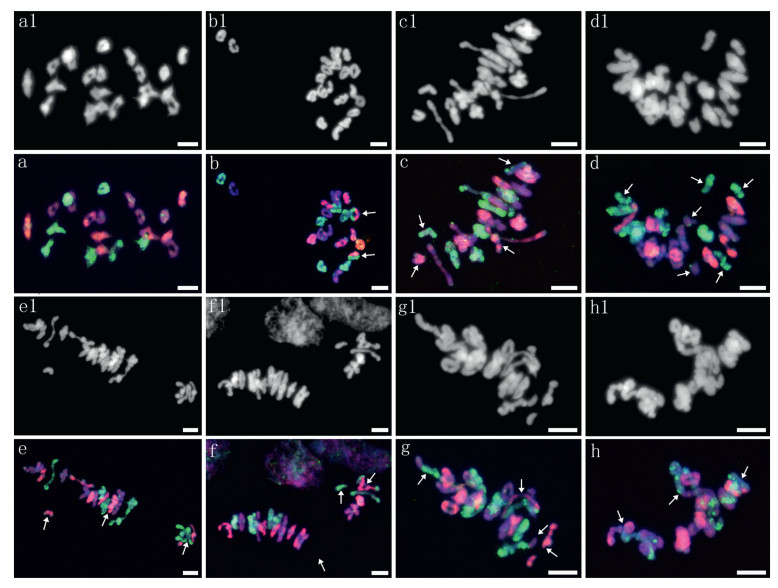
Meiosis in 10 plants selected for pollen fertility statistics was examined by GISH using MI in PMCs (Fig. 3). A total of 475 PMCs (47.5 PMCs per plant) were observed, and the average chromosome configuration was 0.66 Ⅰ + 19.67 Ⅱ (17.38 rings + 2.30 rods) + 0.02 Ⅲ + 0.46 Ⅳ + 0.004 Ⅴ + 0.011 Ⅵ + 0.002 Ⅹ (Table 1). Bivalents were predominant, showing a high frequency of 93.67%, while other forms present at low frequencies included univalents (1.56%), trivalents (0.14%), quadrivalents (4.39%), and other polyvalents (0.25%). The chromosome configuration forms and frequencies varied among the investigated samples. Statistically, bivalents (including both ring and rod bivalents) had the lowest coefficient of variation (CV%) of 7.52, while univalent and multivalent forms showed crucially high CV% ranging from 213.46 to 2177.54. Among the bivalents, rod bivalents with a CV% of 92.13 were remarkably more diverse than ring bivalents with a CV% of 17.02. Nevertheless, heteropolyvalents were observed in 5 low-fertility plants, which were identified as heterozygous intergenomic translocation lines by FISH patterns; no heteropolyvalents were detected in the other two low-fertility plants. Moreover, the chromosome configurations of the univalents and bivalents in each of the St, Y and H genomes could be analyzed exclusively by their distinct GISH patterns in the PMCs at MI (Table 4). This analysis yielded mean formulas of 0.26 Ⅰ + 6.36 Ⅱ (5.54 rings + 0.82 rods), 0.24 Ⅰ + 6.50 Ⅱ (5.64 rings + 0.86 rods) and 0.16 Ⅰ + 6.81 Ⅱ (6.18 rings + 0.63 rods) in the H, St and Y genomes, respectively. Ring bivalents were more varied in the St and H genomes, with CV% values of 22.51 and 24.69, respectively, than in the Y genome, with a CV% of 15.82 between samples (Table 4). ANOVA showed significant variation in the mean number of total bivalents among the three genomes. The average number of ring bivalents in the Y genome was significantly higher than those in both the St and H genomes, while the average number of rod bivalents in the Y genome was significantly lower than those in the St and H genomes. The average number of univalents in the H genome was significantly higher than that of the Y genome (Table 5). This indicates that the Y chromosomes were the most stably paired, whereas the H genome chromosomes were the most variably paired, on average, for plants in meiosis.
|
| Fig. 3 GISH patterns at meiotic metaphase Ⅰ in plants with varying fertility. a1-h1 show the DAPI pattern diagrams; a-h are the corresponding GISH pattern diagrams. The St genome DNA signal is red, the H genome DNA signal is green, and the Y genome DNA signal is purple blue. a: 7 St Ⅱ (ring) + 7 Y Ⅱ (ring) + 7 H Ⅱ (ring), b: 6 St Ⅱ (ring) + 7 Y Ⅱ (6 ring + 1 rod) + 6 H Ⅱ (ring) + 2 St/H Ⅱ (arrows), c: 2 St Ⅰ (arrows) + 6 St Ⅱ (5 ring + 1 rod) + 7 Y Ⅱ (6 ring + 1 rod) + 2 H Ⅰ (arrows) + 6 H Ⅱ (5 ring + 1 rod), d: 7 St Ⅱ (6 ring + 1 rod) + 2 Y Ⅰ (arrows) + 6 Y Ⅱ (ring) + 4 H Ⅰ (arrows) + 5 H Ⅱ (ring), e: 2 St Ⅰ (arrowhead) + 5 St Ⅱ (4 ring + 1 rod) + 7 Y Ⅱ (6 ring + 1 rod) + 6 H Ⅱ (3 ring + 3 rod) + 1 St/St/H/H Ⅳ (chain, arrowhead), f: 6 St Ⅱ (5 ring + 1 rod) + 7 Y Ⅱ (5 ring + 2 rod) + 1 H Ⅰ (arrowhead) + 6 H Ⅱ (5 ring + 1 rod) + 1 H/St/St Ⅲ (chain, arrowhead), g: 6 St Ⅱ (5 ring+ 1 rod) + 7 Y Ⅱ (6 ring + 1 rod, with the rod separating in advance) + 5 H Ⅱ (4 ring + 1 rod) + 1 Y/H Ⅱ (rod, arrowhead) + 1 St/St/H/H Ⅳ (ring, arrowhead), h: 5 St Ⅱ (ring) + 5 Y Ⅱ (ring) + 2 H Ⅱ (1 ring + 1 rod) + 2 St/St/H/H Ⅳ (1 ring + 1 chain, arrowhead) + 1 4Y/6H Ⅹ (ring, arrowhead). The scale bars are 10 μm. |
| Plant | No. of PMCs | H | St | Y | |||||||||||
| No. of univalents | No. of bivalents | No. of univalents | No. of bivalents | No. of univalents | No. of bivalents | ||||||||||
| Total | Ring | Rod | Total | Ring | Rod | Total | Ring | Rod | |||||||
| HY-21 | 43 | 0.00 | 7.00 | 6.77 | 0.23 | 0.00 | 7.00 | 6.65 | 0.35 | 0.00 | 7.00 | 6.72 | 0.28 | ||
| HY-2 | 54 | 0.15 | 6.93 | 6.28 | 0.65 | 0.22 | 6.89 | 6.32 | 0.57 | 0.07 | 6.96 | 6.48 | 0.48 | ||
| HY-3 | 34 | 0.12 | 6.03 | 5.41 | 0.62 | 0.06 | 6.97 | 6.27 | 0.71 | 0.12 | 6.03 | 5.53 | 0.50 | ||
| HY-6 | 54 | 0.00 | 7.00 | 6.87 | 0.13 | 0.00 | 7.00 | 6.87 | 0.13 | 0.00 | 7.00 | 6.65 | 0.35 | ||
| HY-22 | 78 | 0.72 | 5.81 | 3.94 | 1.87 | 0.54 | 5.94 | 4.00 | 1.94 | 0.33 | 6.83 | 5.28 | 1.55 | ||
| HY-13 | 30 | 0.27 | 6.80 | 5.70 | 1.10 | 0.13 | 6.93 | 5.87 | 1.07 | 0.07 | 6.97 | 6.50 | 0.47 | ||
| HY-19 | 34 | 0.12 | 6.00 | 5.29 | 0.71 | 0.12 | 5.97 | 5.24 | 0.74 | 0.00 | 7.00 | 6.82 | 0.18 | ||
| HY-24 | 52 | 0.04 | 6.98 | 6.79 | 0.19 | 0.00 | 7.00 | 6.42 | 0.58 | 0.12 | 6.90 | 6.67 | 0.23 | ||
| HY-8 | 32 | 0.25 | 5.31 | 4.19 | 1.13 | 0.56 | 6.03 | 5.00 | 1.03 | 0.69 | 6.03 | 5.38 | 0.66 | ||
| HY-20 | 64 | 0.52 | 5.77 | 4.78 | 0.99 | 0.50 | 5.75 | 4.84 | 0.91 | 0.16 | 6.92 | 6.13 | 0.80 | ||
| Total | 475 | 124 | 3021 | 2636 | 385 | 114 | 3087 | 2681 | 406 | 74 | 3235 | 2936 | 299 | ||
| X | 0.26 | 6.36 | 5.54 | 0.82 | 0.24 | 6.50 | 5.64 | 0.86 | 0.16 | 6.81 | 6.18 | 0.63 | |||
| Rate (%) | 0.62 | 30.29 | 26.40 | 3.89 | 0.57 | 30.95 | 26.91 | 4.04 | 0.37 | 32.43 | 29.43 | 3.00 | |||
| S | 0.76 | 0.77 | 1.37 | 0.98 | 0.69 | 0.64 | 1.27 | 0.95 | 0.59 | 0.45 | 0.98 | 0.85 | |||
| CV (%) | 291.77 | 12.06 | 24.69 | 121.01 | 286.54 | 9.90 | 22.51 | 111.18 | 380.00 | 6.63 | 15.82 | 134.96 | |||
| ANOVA with multiple comparisons | Univalents | Bivalents | ||
| Total | Ring | Rod | ||
| H | 0.259a | 6.360A | 5.549A | 0.811A |
| St | 0.240ab | 6.499B | 5.644A | 0.855A |
| Y | 0.156b | 6.810C | 6.181B | 0.629B |
| F value | 3.075 | 62.729 | 37.087 | 7.832 |
| P value | 0.046 | 7.811E-27 | 1.993E-16 | 4.144E-4 |
| Note: The same Roman letter in the superscript of each column in the table indicates that the difference between the two averages is not significant (P > 0.05). Otherwise, the difference between the two averages is significant (0.01 ≤ P < 0.05) or very significant (P < 0.01). | ||||
Considering that chromosome pairing deviations can strongly affect pollen fertility, an analysis of correlation between chromosome configuration and pollen fertility was performed in these 10 plants. In the overall genome, pollen fertility was significantly related to the number of ring bivalents (R2 = 0.42, P = 0.04) but not to the numbers of univalents or rod bivalents. However, pollen fertility and the number of ring bivalents were significantly correlated in the St (R2 = 0.45, P = 0.03) and H genomes (R2 = 0.50, P = 0.02) but not in the Y genome; moreover, pollen fertility was significantly related to univalents only in the St genome (R2 = 0.44, P = 0.04) and related to rod bivalents (R2 = 0.47, P = 0.03) only in the H genome. This indicates that pollen fertility was greatly affected by the chromosome pairing behaviour of both the St and H genomes but not the Y genome.
3.4. Associations among chromosome configuration, karyotype heterozygosity and seed settingIn the analysis presented above, significant correlations were found between karyotype heterozygosity and seed setting in each genome. However, significant correlations between chromosome configuration and pollen fertility were detected only in the St and H genomes and not in the Y genome. Correlation analysis showed a distinct relationship between pollen fertility and seed setting (R2 = 0.94, P = 4.45E-6) in 10 plants. Further correlation analysis was carried out among chromosome configuration, karyotype heterozygosity and seed setting, revealing that chromosome configuration (total bivalents, ring and rod bivalents) was significantly correlated with the heterozygosity of each genome and with seed setting in the St and H genomes, respectively, but not in the Y genome (Table 6). This suggests that the effect of heterozygosity on pollen fertility and seed setting in the Y genome was different from its effects in the St and H genomes due to the alteration of chromosome pairing.
| Linear fitting | Heterozygosity and | Seed setting and | ||||||
| Total bivalents | Ring bivalents | Rod bivalents | Total bivalents | Ring bivalents | Rod bivalents | |||
| H | R2 | 0.5676 | 0.7361 | 0.6764 | 0.4542 | 0.6323 | 0.6264 | |
| P value | 0.0119 | 0.0015 | 0.0035 | 0.0326 | 0.0060 | 0.0064 | ||
| St | R2 | 0.7300 | 0.8138 | 0.5802 | 0.4547 | 0.5803 | 0.4835 | |
| P value | 0.0016 | 3.57E-4 | 0.0105 | 0.0325 | 0.0104 | 0.0256 | ||
| Y | R2 | 0.1194 | 0.3193 | 0.2584 | 0.0514 | 0.2198 | 0.2307 | |
| P value | 0.3282 | 0.0888 | 0.1335 | 0.5287 | 0.1717 | 0.1600 | ||
Significant correlation between the karyotype heterozygosity of each genome and fertility (including pollen fertility and seed setting) was found in varying fertility plants. However, significant correlations between heterozygosity and chromosome pairing behaviour at MI in pollen mother cells were detected only in the St and H genomes and not in the Y genome. Chromosome pairing behaviour and pollen fertility are closely related to chromosome structural variation (Stebbins, 1945; Stathos and Fishman, 2014). Chromosome rearrangements such as chromosome inversions, translocations, fusions, and fissions are considered dominant mutations affecting hybrid fitness, whereas heterochromatin addition and deletion are not (Rieseberg, 2001). In the present study, it was difficult to detect chromosome structural variations in mitotic cells due to the use of tandem repeated DNAs as chromosomal markers, except when intergenomic translocation was detected exclusively by GISH. St/H reciprocal translocations in the heterozygous state were detected in several plants with low fertility. In some of these translocation lines, the expected frequencies of heteroquadrivalents as well as frequencies of other chromosomes with irregular pairings were observed. Furthermore, their fertility was significantly lower than that of the expected semisterility. This indicates that additional chromosome structural variations, such as intragenomic translocations or inversions, may still be included in the St and H genomes. The chromosome structural variations in the St and H genomes could be indirectly supported by comparing chromosome variants from plants with normal fertility to plants with varying fertility. The variant accumulation in the St and H genomes was higher than that in the Y genome, although a greater number of chromosome variants were also detected in the Y genome than in the St and H genomes. This suggests that chromosome polymorphisms in the Y genome may be caused by variations in repetitive sequence copies rather than variations in chromosome structure, and chromosome pairing may be less affected by the variation in repeat number.
Chromosome rearrangement is considered more likely to be a major cause of sterility in plants than in animals, perhaps because of differences in male gamete gene expression and in sex determination (Rieseberg, 2001). The fertility of heterozygotes with chromosome arrangements is strongly affected by aberrant meiotic division (Livingstone and Rieseberg, 2004). Furthermore, rearrangements could act synergistically with isolation genes to diminish gene flow over much larger chromosomal regions by reducing recombination (Rieseberg, 2001). In the hybrid between two sympatric sister species of monkey flower, Mimulus cardinalis and M. lewisii, all QTLs for male sterility, including two underdominant loci, mapped to regions of chromosome rearrangements (Fisherman et al., 2013). In this study, a few Elymus nutans plants with low fertility showed much lower fertility than expected from chromosome rearrangements. This indicates the possible contribution of the evolved sterility genes in chromosome structure variation regions. Cytoplasm male sterility (CMS), which results from rearrangements in mitochondrial genomes, is common in plants, but fertility can be restored by nuclear fertility restoration genes (Rieseberg and Blackman 2010; Bohra et al., 2016). The concept of nucleocytoplasmic interaction (NCI) hypothesizes that specific chromosome rearrangement plays an important role in restoring fertility and nucleocytoplasmic compatibility for the genetic stabilization of newly formed hybrids and polyploids (Gill, 1991; Jiang and Gill 1994). In E. nutans, heteroplasmy and variation in mitochondrial genes have been identified among individuals (Liu et al., 2020). Whether the fertility of the heterozygote is affected by NCI still requires further investigation in E. nutans. The heterozygosity of each genome of the plants with varying fertility was distinctly correlated with pollen fertility and seed setting. The St and H genomes exhibited distinctly higher levels of meiotic aberration than the Y genome in these samples. The meiotic chromosome pairing configuration was distinctly associated with pollen fertility and seed setting in the St and H genomes but not in the Y genome in these samples. This suggests that pollen fertility and seed setting rate in E. nutans may be influenced by St and H genome heterozygosity at both the chromosomal and gene levels and influenced by Y genome heterozygosity at the gene level.
Molecular phylogeny has demonstrated that extensive reticular evolution is widespread in Elymus, and interspecific hybridization frequently occurs between Elymus species (Sun, 2014). Large and divergent chromosome regions associated with structural variations represent introgression from other, possibly extinct, congeners (Todesco et al., 2020). E. nutans readily crosses with other Elymus taxa (Lu, 1993), and interspecies hybrids have been frequently observed in the field (Lu et al., 2019). Diverse Elymus species with StY, StH, and StYH genomes as well as StYP are sympatrically distributed with E. nutans on the Qinghai-Tibet Plateau (Lu et al., 1987, 1990, 1999). A few H genome ancestral species, such as Hordeum bogdanii and H. brevisubulatum, are still found on the Qinghai-Tibet Plateau (Bothmer, 1979; Dou et al., 2016). Moreover, several ancestral species of the St genome, such as Pseudoroegneria stipifolia and Ps. strigosa are codistributed in some regions of central and eastern Asia (Dewey, 1984). This suggests that the chromosome structural variations in the E. nutans St and H genomes may be largely the result of introgression hybridization with related or ancestral species. The ancestor of the Y genome remains unknown. Some researchers have suggested that the Y genome is independent of St (Sun and Komatsuda, 2010; Fan et al., 2013); in contrast, the results of other studies suggest that the Y genome originated from the St genome (Liu et al., 2020). The lower structural variation in E. nutans suggests that the ancestral taxa of the Y genome may have had low species diversity, since variation in genome structure may play an important role in speciation (Rieseberg, 2001). Thus, the possibility of Y genome extinction cannot be excluded.
Adaptive genes can be held together in chromosomal regions with structural variations through recombination suppression (Rieseberg, 2001; Livingstone and Rieseberg, 2004). Chromosome structural variations play an essential role in ecotype differentiation (Todesco et al., 2020). Chromosome structural variations in E. nutans have been inferred from observations of mitotic chromosome patterns and meiosis pairing behaviour, as well as from the analysis of associations with fertility. Due to the high variability of the karyotype and wide adaptation of E. nutans, chromosome structural variations may play an important role in the ecological differentiation of this species. Several of the low-fertility plants in this study were in the exclusive intergenomic translocation heterozygous state. They may have been hybrids between differentiated ecotypes of E. nutans. Such differentiated ecotypes might explain why the majority of the overall variation occurred within populations rather than among populations in E. nutans.
In this study, the most important chromosome structural variations, such as inversion and intragenomic translocation, cannot be clearly identified due to limitations of the applied techniques. Advanced molecular cytogenetic methods capable of detecting structural variations using single-copy genes (Danilova et al., 2012; Said et al., 2018) and Oligo-painting are well established in Triticeae (Li et al., 2021). In the future, chromosome structural variations will be widely identified by a combination of molecular cytogenetics and genomics in different habitats, and structural variants associated with ecological traits are expected to be uncovered.
5. ConclusionElymus nutans plants with varying fertility exhibit remarkably higher numbers of chromosome variants and karyotype heterozygosity overall and in each genome than plants with normal fertility. Heterozygosity of the total genome and of each genome is distinctly related to the seed setting rate and pollen fertility. However, chromosome configurations at MI in PMCs are significantly correlated with the heterozygosity of each genome and with seed setting in the St and H genomes but not in the Y genome. This suggests that the effect of heterozygosity on pollen fertility and seed setting in the Y genome is different from its effects in the St and H genomes due to altered chromosome pairing; in addition, the St and H genomes may include many more chromosome structural variations than in the Y genome in E. nutans populations.
Author contributionsBL have done the cytogenetic experiment and analyzed the data. XYT assisted in field work. QWD provided the financial support and designed the experiment.
AcknowledgementsThis research was supported by the Natural Science Foundation of Qinghai Province, China (2020-ZJ-914) and the Second Tibet Plateau Scientific Expedition and Research (STEP) Program (Grant No. 2019QZKK0303).
Declaration of competing interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.12.003.
Bedbrook, J.R., Jones, J., O'Dell, M., et al., 1980. A molecular description of telomeric heterochromatin in secale species. Cell, 19: 545-560. DOI:10.1016/0092-8674(80)90529-2 |
Bothmer, R., 1979. Revision of the Asiatic taxa of Hordeum sect. Stenostachys. Bot. Tidsskr., 74: 117-146. |
Bohra, A., Jha, U.C., Adhimoolam, P., et al., 2016. Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep., 35: 967-993. DOI:10.1007/s00299-016-1949-3 |
Chen, S., Zhang, X., Ma, X., et al., 2013. Assessment of genetic diversity and differentiation of Elymus nutans indigenous to Qinghai-Tibet Plateau using simple sequence repeats markers. Can. J. Plant Sci., 93: 1089-1096. DOI:10.1139/CJPS2013-062 |
Chen, S.L., Zhu, G.H., 2006. Elymus L. In: Flora of China (Poaceae). Science Press and Missouri Botanical Garden, Beijing and St. Louis.
|
Chen, S.Y., Ma, X., Zhang, X.Q., et al., 2009. Genetic variation and geographical divergence in Elymus nutans Griseb. (Poaceae: Triticeae) from west China. Biochem. Systemat. Ecol., 37: 716-722. DOI:10.1016/j.bse.2009.12.005 |
Chen, Z.H., Miao, J.M., Zhong, J.C., et al., 2009. Genetic diversity of wild Elymus nutans germplasm detected by SRAP markers. Acta Pratacult. Siniva, 18: 192-200. |
Danilova, T.V., Friebe, B., Gill, B.S., 2012. Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma, 121: 597-611. DOI:10.1007/s00412-012-0384-7 |
Dewey, D.R., 1984. The genomic system of classification as a guide to intergeneric hybridization with the perennial triticeae. In: Gustafson, J.P. (Ed.), Gene Manipulation in Plant Improvement: 16th Stadler Genetics Symposium. Springer US, Boston, MA, pp. 209-279.
|
Dou, Q., Chen, Z., Liu, Y., Tsujimoto, H., 2009. High frequency of karyotype variation revealed by sequential FISH and GISH in plateau perennial grass forage Elymus nutans. Breed Sci., 59: 651-656. DOI:10.1270/jsbbs.59.651 |
Dou, Q., Liu, R., Yu, F., 2016. Chromosomal organization of repetitive DNAs in Hordeum bogdanii and H. brevisubulatum (Poaceae). Comp. Cytogenet., 10: 465-481. DOI:10.3897/CompCytogen.v10i4.9666 |
Dou, Q., Yu, F., Li, Y., et al., 2017. High molecular karyotype variation revealed in indigenous Elymus nutans in the Qinghai Plateau. Plant Divers., 39: 117-122. DOI:10.1016/j.pld.2017.05.003 |
Fan, X., Sha, L.N., Dong, Z.Z., et al., 2013. Phylogenetic relationships and Y genome origin in Elymus L. sensu lato (Triticeae; Poaceae) based on single-copy nuclear Acc1 and Pgk1 gene sequences. Mol. Phylogenet. Evol., 69: 919-928. DOI:10.1016/j.ympev.2013.06.012 |
Fishman, L., Stathos, A., Beardsley, P.M., et al., 2013. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monky flowers). Evolution, 67: 2547-2560. DOI:10.1111/evo.12154 |
Gill, B.S., 1991. Nucleocytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. In: Sasakuma, T., Kinoshita, T. (Eds.), Nuclear and Organellar Genomes of Wheat Species. Yokohama: Kihara Memorial Foundation for the Advancement of Life Science, pp. 48-53.
|
Jiang, J., Gill, B.S., 1994. Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploid wheats. Chromosome Res., 2: 59-64. DOI:10.1007/BF01539455 |
Li, G., Zhang, T., Yu, Z., et al., 2021. An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J., 105: 978-993. DOI:10.1111/tpj.15081 |
Liu, B., Lu, X.W., Tao, X.Y., et al., 2020. Analysis of cytoplasmic gene variation in Elymus nutants. Chin. J. Grassl., 42: 1-9. |
Liu, Q.L., Liu, L., Ge, S., et al., 2020. Endo-allopolyploidy of autopolyploids and recurrent hybridization—a possible mechanism to explain the unresolved Y-genome donor in polyploid Elymus species (Triticeae: Poaceae). J. Syst. Evol.: 10.1111/jse.12659. DOI:10.1111/jse.12659 |
Livingstone, K., Rieseberg, L., 2004. Chromosomal evolution and speciation: a recombination-based approach. New Phytol., 161: 107-112. DOI:10.1046/j.1469-8137.2003.00942.x |
Löve, Á. , 1984. Conspectus of the triticeae. Feddes Repert., 95: 425-521. DOI:10.1002/fedr.4910950702 |
Lu, B.R., Yan, J., Yang, J.L., 1990. Cytological observations on triticeae materials from Xinjiang, Qinghai and Sichuan. Acta Bot. Yunnanica, 1: 57-66. |
Lu, B.R., 1993. Meiotic studies of Elymus nutans and E. jacquemontii (Poaceae, Triticeae) and their hybrids with Pseudoroegneria spicata and seventeen Elymus species. Plant Systemat. Evol., 186: 193-212. DOI:10.1007/BF00940798 |
Lu, G.P., Nie, B., 2002. Field evaluation of Elymus nutans under alpine grassland conditions. Pratacult. Sci., 9: 13-15. |
Lu, S.L., Sun, Y.H., Liu, S.W., et al., 1987. Flora Republicae Popularis Sinicae. Science Press, Beijing.
|
Lu, S.L., Liu, S.W., Wu, Z.L., et al., 1999. Flora Qinghaiica. People's Publishing Press, Xining.
|
Lu, X.W., Liu, B., Liu, R.J., et al., 2019. Cytogenetic identification on interspecific hybrids in genus Elymus L. of Qinghai plateau. Bull. Bot. Res., 39: 846-852. |
Miao, J.M., Zhang, X.Q., Chen, S.Y., et al., 2011. Gliadin analysis of Elymus nutans Griseb. from the Qinghai–Tibetan plateau and Xinjiang, China. Grassl. Sci., 57: 127-134. DOI:10.1111/j.1744-697X.2011.00219.x |
Rayburn, A.L., Gill, B.S., 1986. Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Rep., 4: 102-109. DOI:10.1007/BF02732107 |
Rieseberg, L.H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol., 16: 351-358. DOI:10.1016/S0169-5347(01)02187-5 |
Rieseberg, L.H., Blackman, B.K., 2010. Speciation genes in plants. Ann. Bot., 106: 439-455. DOI:10.1093/aob/mcq126 |
Said, M., Hřibová, E., Danilova, et al., 2018. The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor. Appl. Genet., 131: 2213-2227. DOI:10.1007/s00122-018-3148-9 |
Stathos, A., Fishman, L., 2014. Chromosomal rearrangements directly cause underdominant F1 pollen sterility in Mimulus lewisii-Mimulus cardinalis hybrids. Evolution, 68: 3109-3119. DOI:10.1111/evo.12503 |
Stebbins, G.L., 1945. The cytological analysis of species hybrids. II. Bot. Rev., 11: 463-486. DOI:10.1007/BF02861140 |
Sun, G., 2014. Molecular phylogeny revealed complex evolutionary process in Elymus species. J. Syst. Evol., 52: 706-711. DOI:10.1111/jse.12080 |
Sun, G., Komatsuda, T., 2010. Origin of the Y genome in Elymus and its relationship to other genomes in Triticeae based on evidence from elongation factor G (EF-G) gene sequences. Mol. Phylogenet. Evol., 56: 727-733. DOI:10.1016/j.ympev.2010.03.037 |
Tang, Z., Yang, Z., Fu, S., 2014. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC. 1 for FISH analysis. J. Appl. Genet., 55: 313-318. DOI:10.1007/s13353-014-0215-z |
Todesco, M., Owens, G.L., Bercovich, et al., 2020. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature, 584: 602-607. DOI:10.1038/s41586-020-2467-6 |
Wang, R.R.-C., von Bothmer, R., Dvorak, J., et al.,1994. Genome symbols in the Triticeae (Poaceae). In: Wang, R.R.-C. (Ed.), Triticeae. Herbarium Publications, Riverside, CA.
|
Xie, J., Zhao, Y., Yu, L., et al., 2020. Molecular karyotyping of Siberian wild rye (Elymus sibiricus L. ) with oligonucleotide fluorescence in situ hybridization (FISH) probes. PLoS One, 15: e0227208. DOI:10.1371/journal.pone.0227208 |
Yan, X.B., Guo, Y.X., Liu, F.Y., et al., 2010. Population structure affected by excess gene flow in self-pollinating Elymus nutans and E. burchan-buddae (Triticeae: Poaceae). Popul. Ecol., 52: 233. DOI:10.1007/s10144-009-0169-x |
Zhang, J.B., Bai, S.Q., Zhang, X.Q., et al., 2009. Study on ear Characters of Elymus nutans Griseb. in the northwestern plateau of Sichuan province. J. Sichuan Univ., 46: 1505-1509. |



