b. Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China
Gaultheria Kalm ex L. (Ericaceae), containing approximately 288 species, are widespread throughout continental areas and islands bordering the Pacific Rim (Lu et al., 2019b; Kron et al., 2020). Some Gaultheria species are commonly called wintergreens because they produce wintergreen oil (methyl salicylate), which is widely used in drugs, pesticides, cosmetics, and foods (Nikolić et al., 2013; Aruna et al., 2014; Luo et al., 2018). In China, nine out of 47 Gaultheria species are widely used as Chinese medicinal wintergreens and traditional medicine by more than nine ethnic minorities (Lu et al., 2019a; Fritsch and Lu, 2020; our own field investigations in 2007). Gaultheria species are polyphyletic and include G. fragrantissima Wall., G. griffithiana Wight, G. hookeri C.B. Clarke, G. leucocarpa var. yunnanensis (Franch.) T.Z. Hsu & R.C. Fang, G. longibracteolata R.C. Fang, G. nummularioides D. Don, G. pyrolifolia J.D. Hooker ex C.B. Clarke, G. semi-infera (C.B. Clarke) Airy-Shaw, and G. sinensis Anth. These species, which all contain methyl salicylate, usually possess a distinct, mint-like aroma (Liu et al., 2013). Some species are also used in the production of Chinese herbal prescriptions, e.g., G. leucocarpa var. yunnanensis is used to produce essential balm, and G. fragrantissima is used in the production of "oil of Indian wintergreen" (Ma et al., 2001; Liu et al., 2013).
Chinese medicinal wintergreen plants differ in the type and content of chemical components, for instance, the content of wintergreen oil (Mukhopadhyay et al., 2016; Luo et al., 2021), but are not easily distinguished by morphological characteristics, particularly when processed into a pharmaceutical preparation. For example, one class of components that varies among Chinese medicinal wintergreens includes lignan glucosides, which have the highest content in G. leucocarpa var. yunnanensis and are known to possess strong anti-arthritic activity (Ma et al., 2001, 2002; Cheng et al., 2009; Hu et al., 2020). In addition, G. longibracteolata is used to treat pulmonary heart disease due to its unique phosphodiesterase-4 (PDE4). However, misidentification of G. longibracteolata for G. leucocarpa var. yunnanensis or G. fragrantissima has resulted in the misuse of G. longibracteolata to treat rheumatism and inflammation (Luo et al., 2021). This misuse of wintergreen plants may decrease medicinal effects or even cause medical malpractice. Therefore, accurate species identification of Chinese medicinal wintergreens is necessary.
Previous pharmaceutical studies on wintergreens have mainly focused on the chemical compositions and their pharmacological actions (Ma et al., 2001, 2002; Liu et al., 2013; Li et al., 2017; Luo et al., 2018; Zhang et al., 2020), whereas precise species identification is often neglected. Phylogenetic analysis has placed these nine Gaultheria species into three clades (Fritsch et al., 2011, Lu et al., 2019a, Zhang et al., 2017): the Gymnobotrys clade (G. leucocarpa var. yunnanensis), the Trichophyllae clade (G. sinensis), and the Leucothoides clade (seven other species). However, this analysis based on multiple-genes (including ITS, matK, rpl16, trnL-trnF and trnS-trnG) has failed to resolve the position of seven species within the Leucothoides clade, which likely due to polyploidy, hybridization/introgression, or recent radiation (Lu et al., 2010, 2019a). Similarly, four universal DNA barcodes (i.e., ITS, matK, trnH-psbA, and rbcL) were unable to provide species-level discrimination in most Gaultheria, particularly among Chinese medicinal wintergreens (Ren et al., 2011). In contrast to a multiple-gene approach, using plastid genomes (plastomes) as DNA barcodes have been extensively adopted to provide increased numbers of DNA markers for taxonomically difficult taxa (Mwanzia et al., 2020; Lv et al., 2022).
The rapid development of high-throughput sequencing technology (Moore et al., 2006) has increased the availability of plastomes and lead to improved phylogenetic resolution at the interspecific level for many genera, e.g., Acacia, Atractylodes, Rhododendron and Salvia (Williams et al., 2016; Wang et al., 2021; Wu et al., 2021a; Fu et al., 2022). Zhang et al. (2017) resolved the inter-specific relationships of 19 species of the series Trichophyllae within Gaultheria, in which one medicinal wintergreen, G. sinensis, is phylogenetically placed. However, this study failed to obtain the complete plastid genome from long PCR sequences due to the presence of long repeats of a certain size, which hinder plastid genome assemblies of most species within Ericaceae (Fajardo et al., 2013; Li et al., 2020; Fu et al., 2022). Structural variations in plastome, including IR expansion or contraction, inversion, and gene duplication or loss, together with DNA sequences can be used as effective tools for plant phylogenetic analyses (Ahmed et al., 2013; Daniell et al., 2016; Liu et al., 2020). Moreover, mutations in plastomes are usually concentrated in certain regions that form mutation 'hotspots' (Dong et al., 2012), suggesting that plastome sequences can be used to exploit short DNA barcodes and markers. To date, complete plastid genomes and plastome structural variation in Gaultheria have yet to be elucidated.
In this study, we sequenced the plastid genomes of 42 individuals from all nine recognized medicinal wintergreen species within Gaultheria. The aims of this study were to (1) resolve phylogenetic relationships among the nine Chinese medicinal wintergreen species; (2) investigate the structural variation of plastid genomes in Gaultheria at both the population and species levels; and (3) propose potential DNA barcodes for future wintergreen species identification.
2. Materials and methods 2.1. Plant materials, DNA extraction, and sequencingA total of 42 samples of Chinese medicinal wintergreens were collected in China. These samples represent nine species within Gaultheria and have distinct distributions. Species identification was primarily based on morphological characteristics and geological distribution. Specifically, we collected six samples of G. fragrantissima (LL20, LL25, LL26, LL27, X4, and X5), five samples of G. griffithiana (LL45, LL47, X16, X17, and X18), four samples of G. hookeri (LL28, LL29, X8, and X9), nine samples of G. leucocarpa var. yunnanensis (LL59, LL61, LL63, LL65, LL66, X26, X27, X28, and X29), two samples of G. longibracteolata (LL67 and X20), six samples of G. nummularioides (LL39, LL41, LL42, LL43, X11, and X14), four samples of G. pyrolifolia (LL33, LL34, LL35, and LL36), three samples of G. semi-infera (LL50, LL51, and X19) and three samples of G. sinensis (XX26, XX27, and XX40). Fresh leaves of each sample were collected in the field and immediately dried with silica gel for subsequent DNA extraction. Voucher specimens were preserved at the herbarium of Kunming Institute of Botany (KUN), and detailed information of the plant samples are presented in Table 1.
| Sample | Size (bp) | LSC (bp) | SSC (bp) | IR (bp) | GC (%) | GenBank accession number | Voucher information |
| G. fragrantissima LL20 | 176, 230 | 107, 783 | 3509 | 32, 469 | 36.6 | OM009272 | LL-060027-KUN |
| G. fragrantissima LL25 | 176, 146 | 107, 710 | 3434 | 32, 501 | 36.6 | OM009273 | LL-0607-KUN |
| G. fragrantissima LL26 | 176, 212 | 107, 738 | 3508 | 32, 483 | 36.6 | OM009274 | LL-0602-KUN |
| G. fragrantissima LL27 | 176, 270 | 107, 834 | 3434 | 32, 501 | 36.6 | OM009275 | LL-S.N.-KUN |
| G. fragrantissima X4 | 176, 230 | 107, 783 | 3509 | 32, 469 | 36.6 | OM009276 | LL-202002-KUN |
| G. fragrantissima X5 | 176, 254 | 107, 783 | 3531 | 32, 470 | 36.6 | OM009277 | LL-202022-KUN |
| G. griffithiana LL45 | 178, 045 | 107, 415 | 3642 | 33, 494 | 36.6 | OM048894 | LL-06100-KUN |
| G. griffithiana LL47 | 178, 128 | 107, 473 | 3597 | 33, 529 | 36.6 | OM048893 | LL-S.N.-KUN |
| G. griffithiana X16 | 178, 089 | 107, 441 | 3572 | 33, 538 | 36.6 | OM048892 | LL-201331-KUN |
| G. griffithiana X17 | 178, 116 | 107, 468 | 3572 | 33, 538 | 36.6 | OM048891 | LL-201357-KUN |
| G. griffithiana X18 | 178, 116 | 107, 491 | 3617 | 33, 504 | 36.6 | OM048890 | LL-201362-KUN |
| G. hookeri LL28 | 175, 265 | 106, 831 | 3432 | 32, 501 | 36.6 | OM048889 | LL-07089-KUN |
| G. hookeri LL29 | 176, 287 | 107, 835 | 3510 | 32, 471 | 36.6 | OM048888 | LL-031500-KUN |
| G. hookeri X8 | 176, 230 | 107, 800 | 3488 | 32, 471 | 36.6 | OM048887 | LL-201619-KUN |
| G. hookeri X9 | 175, 306 | 106, 872 | 3432 | 32, 501 | 36.6 | OM048886 | LL-201813-KUN |
| G. leucocarpa var. yunnanensis LL59 | 179, 173 | 106, 807 | 76 | 36, 145 | 36.7 | OM084803 | LL-201701-KUN |
| G. leucocarpa var. yunnanensis LL61 | 179, 842 | 107, 439 | 137 | 36, 133 | 36.6 | OM084804 | LL-201707-KUN |
| G. leucocarpa var. yunnanensis LL63 | 179, 970 | 107, 567 | 137 | 36, 133 | 36.6 | OM084805 | LL-201205-LUN |
| G. leucocarpa var. yunnanensis LL65 | 179, 841 | 107, 438 | 137 | 36, 133 | 36.6 | OM084806 | LJ-Liu1001-KUN |
| G. leucocarpa var. yunnanensis LL66 | 179, 122 | 106, 756 | 76 | 36, 145 | 36.7 | OM084807 | HJ-He001-KUN |
| G. leucocarpa var. yunnanensis X26 | 179, 121 | 106, 745 | 78 | 36, 149 | 36.6 | OM084800 | LL-201936-KUN |
| G. leucocarpa var. yunnanensis X27 | 179, 914 | 107, 364 | 140 | 36, 205 | 36.6 | OM084801 | LL-202025-KUN |
| G. leucocarpa var. yunnanensis X28 | 179, 802 | 107, 399 | 137 | 36, 133 | 36.6 | OM084802 | LL-202037-KUN |
| G. leucocarpa var. yunnanensis X29 | 182, 430 | 107, 408 | 124 | 37, 449 | 36.6 | OM084808 | LL-202039-KUN |
| G. longibracteolata LL67 | 176, 749 | 107, 764 | 3503 | 32, 741 | 36.5 | OM048885 | LL-202045-KUN |
| G. longibracteolata X20 | 176, 637 | 107, 848 | 3473 | 32, 658 | 36.5 | OM048884 | LL-202024-KUN |
| G. nummularioides LL39 | 176, 229 | 107, 766 | 3435 | 32, 514 | 36.6 | OM048883 | LL-07010-KUN |
| G. nummularioides LL41 | 176, 229 | 107, 766 | 3435 | 32, 514 | 36.6 | OM048882 | LL-201143-KUN |
| G. nummularioides LL42 | 176, 200 | 107, 698 | 3432 | 32, 535 | 36.6 | OM048881 | LL-0618A-KUN |
| G. nummularioides LL43 | 176, 229 | 107, 766 | 3435 | 32, 514 | 36.6 | OM048880 | QD-2006-KUN |
| G. nummularioides X11 | 176, 189 | 107, 729 | 3432 | 32, 514 | 36.6 | OM048879 | LL-201457-KUN |
| G. nummularioides X14 | 174, 337 | 107, 340 | 3089 | 31, 954 | 36.7 | OM048878 | LL-201815-KUN |
| G. pyrolifolia LL33 | 175, 895 | 107, 523 | 3538 | 32, 417 | 36.6 | OM048877 | LL- 07117-KUN |
| G. pyrolifolia LL34 | 175, 895 | 107, 523 | 3538 | 32, 417 | 36.6 | OM048876 | LL-07117-KUN |
| G. pyrolifolia LL35 | 175, 895 | 107, 523 | 3538 | 32, 417 | 36.6 | OM048875 | LL-07161-KUN |
| G. pyrolifolia LL36 | 175, 895 | 107, 523 | 3538 | 32, 417 | 36.6 | OM048874 | LL-07161-KUN |
| G. semi-infera LL50 | 175, 905 | 107, 498 | 3477 | 32, 465 | 36.6 | OM048870 | LL-06103-KUN |
| G. semi-infera LL51 | 175, 982 | 107, 471 | 3491 | 32, 465 | 36.6 | OM048869 | LL-06QQ-KUN |
| G. semi-infera X19 | 175, 916 | 107, 509 | 3491 | 32, 458 | 36.6 | OM048868 | LL-201455-KUN |
| G. sinensis XX26 | 176, 659 | 107, 426 | 2585 | 33, 324 | 36.7 | OM048873 | LL-201428-KUN |
| G. sinensis XX27 | 176, 632 | 107, 395 | 2573 | 33, 332 | 36.7 | OM048872 | LL-201802-KUN |
| G. sinensis XX40 | 176, 749 | 107, 500 | 2585 | 33, 332 | 36.7 | OM048871 | LL-202106-KUN |
Genomic DNA was extracted using the modified CTAB method (Doyle and Doyle, 1987), and the quality and quantity of genomic DNA were checked. The Illumina HiSeq 2500 platform was used to sequence the 500 bp insert-size libraries and to generate 2–3 Gb pair-end reads of 150 bp in length for each sample.
2.2. Assembly, annotation, and validation of complex repeat regionsTo assemble the clean reads (processed reads) into complete plastomes, de novo assembly was performed with the GetOrganelle toolkit (Jin et al., 2020b). Connection and annotation were subsequently conducted using Bandage 0.8.1 (Wick et al., 2015) and Geneious 9.1.4 (Biomatters Ltd., Auckland, New Zealand) with Vaccinium macrocarpon Aiton. (GenBank accession number: NC019616) used as the reference. Notably, some plastid regions could not be disentangled in Bandage 0.8.1 (Wick et al., 2015) due to complex or long-size repeats (Fig. 1). These regions were subsequently verified by PCR and Sanger sequencing, using newly designed primers (Table S1).
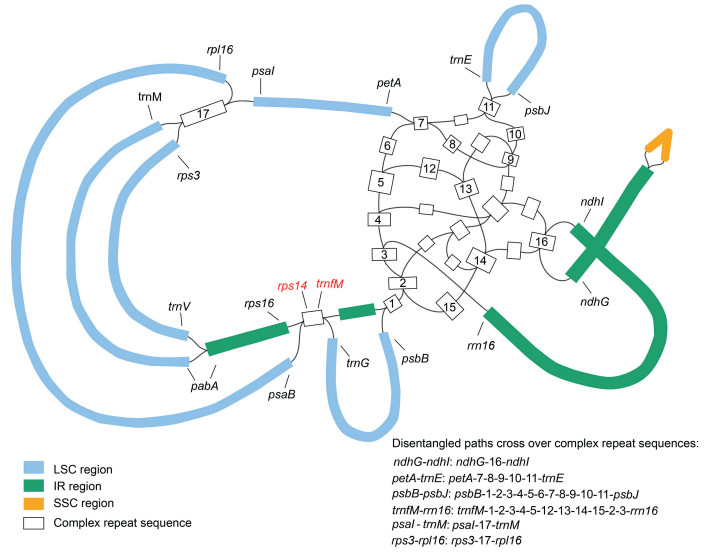
|
| Fig. 1 Schematic diagram showing complex repeat sequences observed in the assembly graph of de novo assembled Gaultheria plastomes. The paths cross over complex repeat sequences are indicated. |
REPuter software (Kurtz et al., 2001) was used to identify forward (F), reverse (R), palindrome (P), and complementary (C) repeats in the resultant plastomes with the setting of a minimum repeat size of 30 bp and 90% or greater sequence identity (Hamming Distance = 3). Tandem Repeats Finder v.4.04 (Benson, 1999) was used to detect tandem repeats in the plastomes, with parameters set to 30 bp for the minimum repeat length, 0% for maximum mismatches and excluding repeats up to 10 bp longer than contained repeats. Simple sequence repeats (SSRs) were predicted using MISA (Beier et al., 2017) with the parameters of mono-, di-, tri-, tetra-, penta- and hexa-nucleotides set as 10, 5, 4, 3, 3, and 3, respectively. The maximum length of the sequence between two SSRs to register as a compound SSR was set to 0 bp.
2.4. Structural variation of Gaultheria plastomesTo investigate the structural characteristics of the Gaultheria plastomes under a broader scale, we downloaded representative plastomes from six related families within Ericales (i.e., Ericaceae, Lecythidaceae, Pentaphylacaceae, Primulaceae, Styracaceae and Theaceae; detailed information is provided in Table S2), and compared the structure of these plastomes with our newly generated plastomes using the MAUVE plug-in in Geneious (Darling et al., 2004).
2.5. Phylogenetic analysesWe downloaded three plastomes of the genus Vaccinium as outgroups (Vaccinium bracteatum GenBank No.: LC521967, V. uliginosum, LC521968 and V. vitis-idaea, LC521969). MAFFT v.7.310 (Katoh and Standley, 2013) was used to align the plastome sequences. Gene-partitioned model estimated using PartitionFinder2 (Lanfear et al., 2017) were used in the maximum-likelihood (ML) and Bayesian inference (BI) analyses. ML analysis was conducted using IQ-tree 1.6.12 (Nguyen et al., 2015) with support values estimated by 10, 000 ultrafast bootstrap replicates. BI analysis was conducted using MrBayes 3.2.6 (Ronquist et al., 2012). Two runs with four chains were performed in parallel for ten million generations, with trees being sampled every 1000 generations.
2.6. DNA barcodes screeningThe nucleotide diversity of 42 plastomes from nine species was calculated based on sliding window analysis using DnaSP v.5.10 (Librado and Rozas, 2009), with the window length set to 600 bp and step size set to 100 bp. Regions with high Pi values (> 0.2) were picked out initially and checked within Geneious v.8.1.7 (Kearse et al., 2012) with PCR success rates considered. The selected regions at this stage were subsequently used in phylogenetic tree reconstruction using RAxML (Stamatakis, 2014). The discriminability of DNA barcodes was evaluated by reconstructing phylogenetic trees. Species in monophyly with a support rate greater than 80% was considered to be identified successfully (Barrett et al., 2016).
3. Results 3.1. Characteristics of plastomes of the Chinese medicinal wintergreensThe plastomes of all sampled Gaultheria share a typical quadripartite structure (Fig. 2), with GC content ranging from 36.5% to 36.7%. The plastomes ranged from 174, 337 bp to 182, 430 bp, with the LSC region ranging from 106, 745 bp to 107, 848 bp, the SSC region from 76 bp to 3642 bp, and the IR regions from 31, 954 bp to 37, 449 bp (Table 1). The total number of annotated genes was 137 in all samples of G. fragrantissima, G. hookeri, G. longibracteolata, G. nummularioides, G. pyrolifolia, G. semi-infera and two samples of G. griffithiana (LL50 and LL51), whereas it was 135 in all samples of G. sinensis, one sample of G. griffithiana (X19), and 138 in all samples of G. leucocarpa var. yunnanensis.
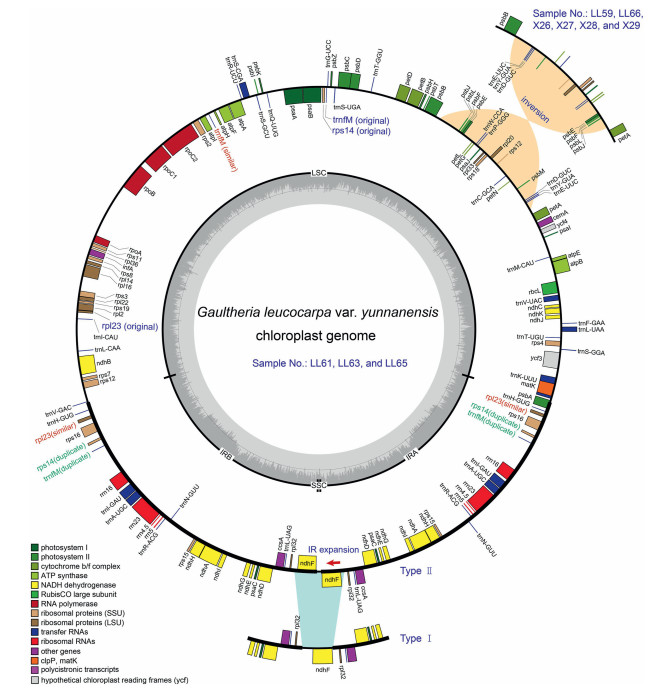
|
| Fig. 2 Schematic plastome maps of Chinese medicinal wintergreens (Gaultheria). There are two basic plastome structure types. Type Ⅰ: the plastomes of eight species, with the ndhF gene located in the SSC region (indicated under the circle). Type Ⅱ: the plastomes of all samples of G. leucocarpa var. yunnanensis; the structure of samples LL61, LL63 and LL65 is denoted by the main circle and the remaining samples (LL59, LL66, X26, X27, X28, and X29), which contain an inversion, are indicated at upper right corner of the circle. Genes located outside the outer rim are transcribed in the counter clockwise direction, whereas genes inside are transcribed in the clockwise direction. The colored bars indicate different functional groups. The dashed gray area in the inner circle denotes the GC content of the corresponding genes. LSC: large single-copy region, SSC: small single-copy region, and IR: inverted repeat region. |
Although the total number of chloroplast genes differed, the number of unique genes in each plastome was identical. Each plastome encoded 110 unique genes, including 76 protein-coding, 30 tRNA and 4 rRNA genes. All the Gaultheria plastomes have substantially enlarged IR regions, and the SSC region contains only one ndhF gene; moreover, the IR region of all samples of G. leucocarpa var. yunnanensis also contain the ndhF genes. We annotated two extra copies of the rpl23 gene (not identical in sequences) in the IR regions of all Gaultheria plastomes except the samples of G. sinensis and one sample of G. griffithiana (X19). We also annotated two extra identical copies of the adjacent trnfM and rps14 genes in the IR regions, and one extra copy of the trnfM gene in the LSC region (not identical in sequence) in all Gaultheria plastomes.
3.2. IR boundary shifts and genome comparisonWe aligned all the resultant plastomes to investigate the expansion and contraction of IR boundaries. The plastomes showed high stability at the IR/LSC boundaries but were more varied at IR/SSC boundaries. The psbA gene spanned the IRb/LSC junction, with 236 bp on the LSC side adjacent to the matK gene and 826 bp on the IRb side adjacent to the trnH gene. For the IRa/LSC junction, the trnV gene was located in the LSC region 51–63 bp away from the junction, whereas the trnH gene was located in the IRa region 1175–1179 bp away from the junction. The boundaries of IR/SSC regions can be classified into two types. Type Ⅰ: the ndhF gene was located in the SSC region, 15–261 bp away from the IRb/LSC boundaries and 261–1296 bp away from IRa/SSC boundaries. This boundary type was observed in all sampled Gaultheria species except G. leucocarpa var. yunnanensis (Fig. 2). Type Ⅱ: the ndhF genes were located in the IR regions due to IR expansion, and located 256–313 bp away from the IRb/SSC junction and 256–313 bp away from the IRa/SSC junction; no genes were present in the SSC region (Fig. 2). This boundary type was only observed in all samples of G. leucocarpa var. yunnanensis. Moreover, six out of nine samples of G. leucocarpa var. yunnanensis (incl., LL59, LL66, X26, X27, X28 and X29) had a unique inversion from the psbJ gene to the trnE-UUC gene (about 14 kb in length) in the LSC region, which resulted in the adjacencies of psbJ with petA and trnE-UUC with psbB (Fig. 2).
In addition, we compared the plastome structure of Gaultheria with plastomes from Ericaceae and five other families of Ericales (Lecythidaceae, Pentaphylacaceae, Primulaceae, Styracaceae, and Theaceae; Table S2). We found that the gene order of Gaultheria plastomes was collinear with Vaccinium (Ericaceae) but not collinear with that of Rhododendron (Ericaceae) (Fig. 3). The gene order of plastomes from five families of Ericales, excluding Ericaceae, are collinear (Fig. 3); however, the gene orders among Gaultheria, Rhododendron, and those five families can be explained by more than ten inversions (Fig. 3).
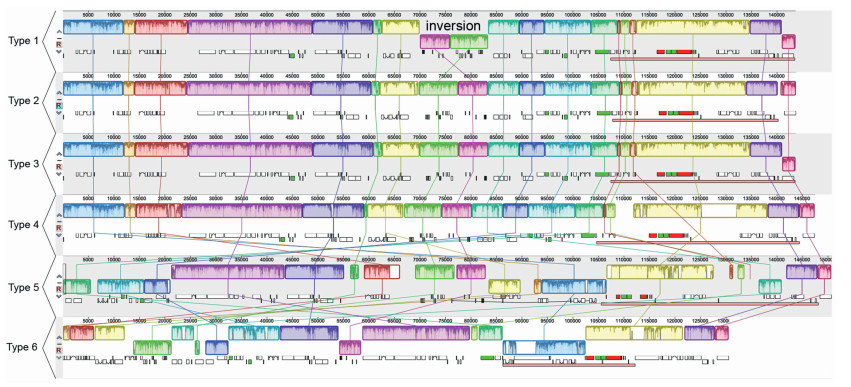
|
| Fig. 3 Syntenies and rearrangements detected in the complete plastomes of nine Chinese medicinal wintergreens (Gaultheria) and the genera used for comparison from six related families within Ericales. Boxes of the same color connected with lines indicate the local collinear blocks and represent syntenic regions. Sequence identity similarity profiles are represented by histograms within each block, and sequences transcribed in reverse directions are indicated by the colored blocks above and below the center line. The taxa studied are divided into six types of plastome arrangements: Type 1, samples LL59, LL66, X26, X27, X28, and X29 of G. leucocarpa var. yunnanensis; Type 2, samples LL61, LL63, and LL65 of G. leucocarpa var. yunnanensis; Type 3, all samples of G. fragrantissima, G. griffithiana, G. hookeri, G. longibracteolata, G. nummularioides, G. pyrolifolia, G. semi-infera, and G. sinensis; Type 4, Vaccinium from Ericaceae; Type 5, Rhododendron from Ericaceae; and Type 6, genera studied from Pentaphylacaceae, Styracaceae, Primulaceae, Lecythidaceae, and Theaceae. |
A total of 2740 SSRs were identified across the 42 wintergreen plastomes (Table S3). The number of SSRs in each plastome of Gaultheria ranged from 57 (G. griffithiana) to 75 (G. longibracteolata) (Table S3). The majority of the SSRs were mononucleotides (52.33%), followed by tetranucleotides (20.80%), trinucleotides (13.40%), dinucleotides (11.50%), and hexanucleotides (1.42%); the smallest number of SSRs were pentanucleotides (0.55%). No hexanucleotide repeats were detected in G. griffithiana, and pentanucleotide repeats were only detected in G. longibracteolata, G. nummularioides, and G. sinensis (Table S3).
The results of long-size repeat (> 30 bp) analysis showed that there were only two types of repeats in the wintergreen samples: forward and reverse. The number of long repeats among the wintergreen plastomes ranged from 182 in Gaultheria nummularioides to 259 in G. leucocarpa var. yunnanensis (Fig. 4A). The maximum length of long repeats in each chloroplast ranged from 706 bp (e.g., G. leucocarpa var. yunnanensis-LL59) to 1722 bp (e.g., G. fragrantissima-LL27). A total of 9475 long repeats were detected in the 42 samples. There were 5321 long repeats with 30–50 bp in length, 2894 repeats of 51–100 bp, 989 repeats of 101–500 bp, 137 repeats of 501–1000 bp, and 134 repeats of 1001–2000 bp (Fig. 4A). The frequency of long repeats was a minimum of 2 repetitions and a maximum of 7 repetitions (Fig. 4B).The long repeats of the wintergreen plastomes were more frequently located in non-coding regions (e.g., atpI-trnfM, ndhF-trnV, ndhI-ndhG, pbf1-psbT, petD-petB, psaI-trnM, psbB-psbJ, rpoA-rpoB, rps3-rpl16, rrn16-trnI, trnE-petA, trnfM-rrn16, trnH-rpl23, trnI-rpl23, trnN-rps15, trnR-trnS, trnT-petD, trnV-rps12, and ycf3-trnK) than in coding regions (e.g., infA, ndhA, ndhF, ndhI, petD, rpl22, rpl23, rpl32, rpoA, rpoC1, rps14, rps3, trnfM, and trnI). Notably, we identified about 600 bp repeats located in both LSC and IR regions that covered the trnfM and rps14 genes, which resulted in two extra copies of these two genes in the IR regions (Fig. 4C).
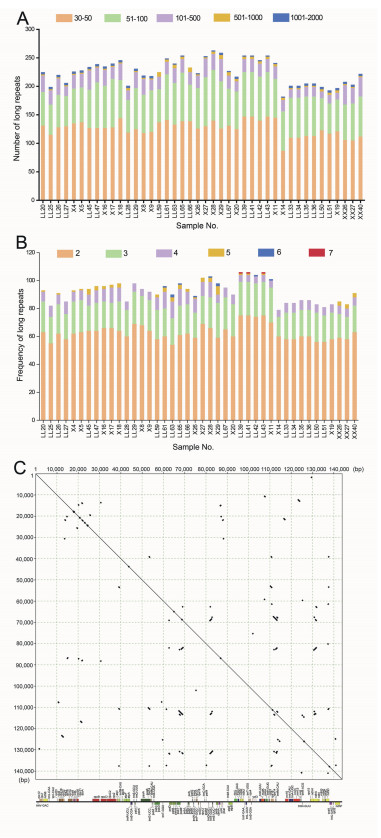
|
| Fig. 4 Repeat sequences in Chinese medicinal wintergreen plastid genomes. (A) Number of long repeats by length; (B) Number of long repeats by frequency; (C) Self comparison of Gaultheria plastome (G. fragrantissima-LL20). |
The resulting ML and BI tree topologies were highly similar to one another. Our phylogenetic analysis using plastomes in which one of the IR copies was removed revealed that all samples of each wintergreen species clustered into a monophyletic group with moderate supports for Gaultheria hookeri (BS = 69, PP = 0.99) and G. semi-infera (BS = 87, PP = 1.00), and strong supports for seven other species (BS = 100, PP = 1.00; Fig. 5). G. leucocarpa var. yunnanensis was sister to the remaining eight wintergreen species. G. sinensis was sister, in turn, to a grade consisting of G. griffithiana, G. pyrolifolia, G. longibracteolata, G. nummularioides, G. fragrantissima, G. semi-infera, and G. hookeri.
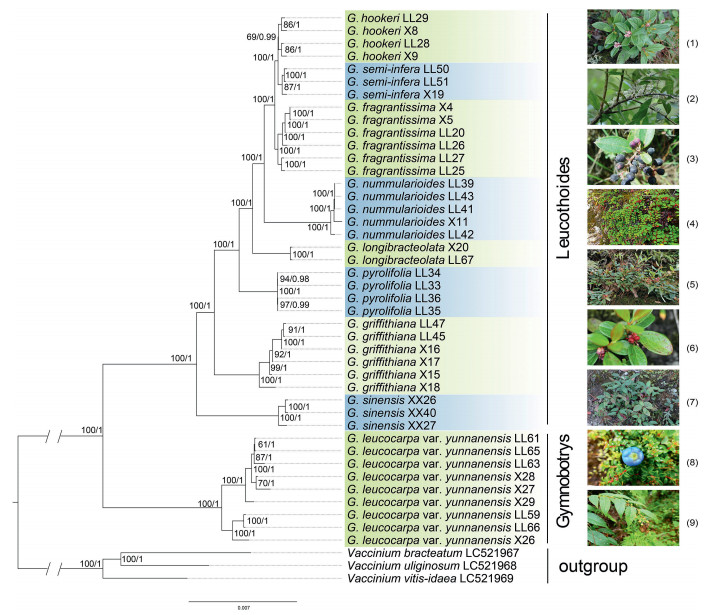
|
| Fig. 5 A phylogenetic tree based on 42 plastome sequences of nine Chinese medicinal wintergreens (Gaultheria). Likelihood bootstrap support values (BS) and Bayesian posterior probability values (PP) are shown next to the nodes. Images on the right side show the plants of the nine species: (1) G. hookeri, (2) G. semi-infera, (3) G. fragrantissima, (4) G. nummularioides, (5) G. longibracteolata, (6) G. pyrolifolia, (7) G. griffithiana, (8) G. sinensis, and (9) G. leucocarpa var. yunnanensis. |
We selected seven DNA regions (incl., infA, ndhF, psaI-trnM, rps3-rpl16, trnQ-psaA, trnV-rps12, and ycf3-trnK) with higher Pi values in the sliding window analyses, which were expected to have high potential for species delimitation in Chinese medicinal wintergreens (Fig. 6). We evaluated the fitness of these sequences as DNA barcoding regions by reconstructing ML phylogenetic trees (Figs. S1–S7). The results showed that the trnV-rps12 region could discriminate all nine species successfully (BS ≥ 89). The trnQ-psaA region could discriminate eight out of nine species (BS ≥ 85), except for Gaultheria hookeri. The ndhF region could discriminate seven out of nine species (BS ≥ 96), except for G. fragrantissima and G. hookeri. The ycf3-trnK and psaI-trnM region each could discriminate six out of nine species; specifically, the ycf3-trnK region could identify G. griffithiana, G. leucocarpa var. yunnanensis, G. longibracteolata, G. nummularioides, G. pyrolifolia, and G. sinensis (BS ≥ 95). The psaI-trnM region had high resolution among G. griffithiana, G. leucocarpa var. yunnanensis, G. longibracteolata, G. nummularioides, G. pyrolifolia and G. sinensis (BS = 100). The rps3-rpl16 region could discriminate five out of nine species: G. griffithiana, G. leucocarpa var. yunnanensis, G. nummularioides, G. pyrolifolia, and G. sinensis (BS ≥ 93). The infA region could discriminate only four out of nine species, i.e., G. fragrantissima, G. leucocarpa var. yunnanensis, G. griffithiana, and G. sinensis (BS ≥ 93).

|
| Fig. 6 Sliding window analysis of the complete plastomes of nine Chinese medicinal wintergreens (Gaultheria). X-axis: position of the window midpoint, Y-axis: nucleotide diversity within each window (window length: 600 bp, step size: 100 bp). Genic regions with Pi values > 0.2 are marked; region names with asterisk superscripts denote recommended DNA barcodes, while other regions are considered unsuitable for DNA barcodes due to the lack of either species discrimination or PCR amplification feasibility. |
The gene content of plastomes of land plants is generally considered to be conserved (Kumar et al., 2016), and the loss or duplication of different genes has been found in a variety of plants (Mohanta et al., 2020). It has been inferred that genes lost in plastomes may have moved to the nuclear genome or the function of these genes has been replaced by nuclear-encoded genes (Bryant et al., 2011; Savage et al., 2013; Mohanta et al., 2020). We identified several protein-coding gene losses and duplications in the Gaultheria plastomes. Both the ycf1 and ycf2 genes were absent in all studied Gaultheria samples. These two genes usually have elevated substitution rates and may have undergone pseudogenization in some land plant lineages (Oliver et al., 2010; Wolf et al., 2011). ycf1 has also been found missing in grasses and some parasitic plants, and independent losses of the ycf2 gene have occurred in multiple angiosperm groups, including Poaceae, Podostemaceae, Geraniaceae, and non-photosynthetic Ericaceae (Huang et al., 2010; Jin et al., 2020a). However, the mechanism for the loss of these two genes in the plastome is still not well understood.
In contrast to gene losses, gene duplications, attributed to repeat regions or IR expansions, were also detected in Gaultheria plastomes (incl., ndhF, rpl23, rps14, and trnfM). The ndhF gene was duplicated in all the samples of G. leucocarpa var. yunnanensis due to IR expansion. The chloroplast NAD(P)H dehydrogenase (ndh) complex is involved in photosystem I (PSI) cyclic electron transport and chlororespiration (Peng et al., 2011), and their mutation rates are high and sensitive to environmental conditions and stress (Silva et al., 2016). G. leucocarpa var. yunnanensis has a widespread distribution in southern China, while other wintergreens inhabit the Himalaya-Hengduan regions. This difference in distribution implies that the duplication of ndhF gene is related to environmental adaptation in this species. In addition, plastomes with duplicated ndh genes usually exhibit extensive rearrangements and a higher frequency of pseudogenes (Martin et al., 1996; Casano et al., 2001; Paredes and Quiles, 2013). Consistent with this phenomenon, we identified an inversion in the LSC of some samples of G. leucocarpa var. yunnanensis.
It is rare that duplication of plastid genes does not involve movement of IR boundaries (Raubeson and Jansen, 2005; Bock, 2007; Guisinger et al., 2011). In fact, only a few instances have been documented, for example, the duplication of clpP gene in Silene L. and Lychnis L. (Erixon and Oxelman, 2008), the duplication of psbJ and trnI-CAU in Trachelium Tourn. ex L. (Haberle et al., 2008), and the duplication of rpl23 and three tRNA genes in Jasminum L. (Lee et al., 2006). In this study, we identified duplication of three genes (i.e., rpl23, rps14 and trnfM) in Gaultheria plastomes that were independent of IR boundary shifts. The long repeats located in the LSC (between psaB and trnG) and both IR regions (between rrn16 and trnH) that covered rps14 and trnfM genes seemingly generated the three copies of these two genes. Additionally, we identified a fourth copy of trnfM in the LSC region adjacent to atpI, but with a slightly divergent sequence. By checking against other plastomes within Ericales, we inferred that the rps14 and trnfM genes between psaB and trnG are likely the original copies. Similarly, the rpl23 located in the LSC region (between trnI and rpl2) was not identical to the other two copies in the IR regions (between trnH and rps16); in this case, the LSC copy is likely the original copy.
Variation in plastome structure most typically involves expansion or contraction of the IRs into or out of adjacent single-copy regions (Ravi et al., 2008; Downie et al., 2015; Ye et al., 2018). The trnN-GUU gene is considered the ancestral IR/SSC endpoint that has been retained in most land plants, and the trnV-GAC gene represents the ancestral IR/LSC endpoint among most land plants (Zhu et al., 2016). Plastome IR expansions have been found in different plant groups, including Geraniaceae (Guisinger et al., 2011), Euphorbiaceae (Li et al., 2017), Solanaceae (Amiryousefi et al., 2018), Bignoniaceae (Thode and Lohmann, 2019) and Leguminosae (Bai et al., 2021). All plastomes of Gaultheria fragrantissima, G. griffithiana, G. hookeri, G. longibracteolata, G. nummularioides, G. pyrolifolia, G. semi-infera and G. sinensis expanded toward the SSC region from trnN to rpl32, and all samples of G. leucocarpa var. yunnanensis expanded toward the SSC region from trnN to ndhF. Different mechanisms have been proposed to explain IR expansions, such as gene conversion or double-strand DNA breaks (Goulding et al., 1996; Wang et al., 2008). In addition, some studies point out that the contraction and expansion of IR regions is a major contributor of plastomes size variation (Wang et al., 2018). For example, the IR regions of species from the genera Acorus and Magnolia are not expanded, and their plastome sizes are between 150 and 160 kb (Zhu et al., 2016), whereas the plastome size of Chinese medicinal wintergreens is between 176 and 182 kb. Our study validated that expansion of the IR region contributes to the increased genome size of Gaultheria species.
Similar to IR boundary shifts, inversions also represent essential mechanisms for plastome rearrangements, which contribute to the structural diversification of plant plastomes (Wicke et al., 2011; Jansen and Ruhlman, 2012). In nearly all photosynthetic land plants, plastomes are highly conserved in structural organization and gene arrangement (Jansen and Ruhlman, 2012; Zhu et al., 2016; Ye et al., 2018). Despite the commonly held view that plastomes have a conserved structure and sequence, they have been found to exhibit considerable variation in an increasing number of taxa (Wang et al., 2018; Ye et al., 2018). In our study, we found that there was one inversion in the LSC regions of six samples (LL59, LL66, X26, X27, X28 and X29) within Gaultheria leucocarpa var. yunnanensis, which disrupts the canonical order of the plastome genes. In addition, except for the inversion in the above samples in G. leucocarpa var. yunnanensis, we found that there were apparent multiple rearrangement events between the Gaultheria plastomes and those from the other five families (Lecythidaceae, Pentaphylacaceae, Primulaceae, Styracaceae and Theaceae) in Ericales. Our study provides new evidence of the variation between Ericales plastomes.
4.2. Abundant and variable repeat sequencesThe long repeats of Gaultheria plastomes were characterized in terms of the number, length and distribution. Previous studies indicated that long repeats mainly occur in non-coding DNA sequences (Raubeson and Jansen, 2005). However, we detected long repeats in coding regions of Gaultheria plastomes (e.g., infA, ndhA, ndhF, ndhI, petD, rpl22, rpl23, rpl32, rpoA, rpoC1, rps14, rps3, trnfM, and trnI). The most abundant sizes of long repeats among angiosperms are on average smaller than 50 bp (Raubeson and Jansen, 2005). With the increase in the number of plastomes sequenced, some longer repetitive sequences (approximately 100 bp) have also been identified within some taxa. For example, the lengths of repeats range from 30 to 90 bp in Orchidaceae (Dong et al., 2018), 91 to 132 bp in Poaceae (Zhang et al., 2011), and 30 to 307 bp in Polypodiaceae (Liu et al., 2021). Generally, the number of repeats in the plastome is less than 50; for example, there are approximately 30 long repeats in Cymbidium (Yang et al., 2013), and 30 long repeats in Debregeasia (Wang et al., 2020). However, the longest long-repeat of each Gaultheria sample ranged from 706 bp to 1722 bp, and the number of long repeats was up to 259, which is significantly more abundant and variable than for many previously published plastomes.
Previous studies have shown that long repeats play a role in sequence rearrangement and variation in plastomes (Cavalier-Smith, 2002; Asano et al., 2004; Timme et al., 2007; Yang et al., 2013), and that large numbers of repeats can damage the stability of the plastome structure, for example, by producing inversions (Alexandre and Normand, 2010). Approximately 25 long repeats were located in the psbB-psbJ and trnE-petA regions of Gaultheria plastomes; thus, we speculate that this is one of the reasons for the 14-kb inversion in some samples of G. leucocarpa var. yunnanensis. Previous studies suggest that overall repeat content is positively correlated with the degree of genomic rearrangements (Wu et al., 2021b). This could be one of the reasons for the rearrangement of the plastomes in Ericaceae compared to other families within Ericales.
4.3. Phylogenetic relationships and delimitation of Chinese wintergreen speciesPrevious phylogenetic studies using a multiple-gene approach and international DNA barcoding (ITS, Waxy, Lfy, matK, rbcL, rpl16, trnH-psbA, trnL-trnF and trnS-trnG) failed to resolve the phylogenetic relationships among Chinese wintergreen species (Lu et al., 2010, 2019b; Ren et al., 2011). In this study, using plastomes in which one of the IR copies had been removed to reduce redundancy, we resolved the phylogenetic relationships among all nine Chinese medicinal wintergreens.
Previous phylogenetic analysis recovered three major clades in Chinese Gaultheria, i.e., the Leucothoides clade, the Trichophyllae clade, and the Gymnobotrys clade, with the Leucothoides clade sister to the Trichophyllae clade (Lu et al., 2019b). In this and other analyses, G. leucocarpa var. yunnanensis, which diverged earlier than other studied species, was resolved as a member within the Gymnobotrys clade; G. fragrantissima, G. griffithiana, G. hookeri, G. longibracteolata, G. nummularioides, G. pyrolifolia and G. semi-infera were placed within the Leucothoides clade, but with poorly supported infraspecific relationships, and G. sinensis was placed in the Trichophyllae clade (Lu et al., 2010, 2019a; Zhang et al., 2017). Our research supports this phylogeny, with G. sinensis sister to the grade comprising G. fragrantissima, G. semi-infera, G. griffithiana, G. hookeri, G. nummularioides and G. longibracteolata. We found that G. nummularioides had a sister relationship with the grade of G. hookeri, G. semi-infera and G. fragrantissima, and the clade comprising G. hookeri, G. semi-infera and G. fragrantissima was the most derived, which is consistent with the results of Lu et al. (2010). However, the relationship among the latter three species was poorly resolved in previous research (Lu et al., 2010; Fritsch et al., 2011), whereas our study suggests that each of the three species form a monophyletic lineage. In contrast to previous studies, we succeeded in clarifying the phylogenetic relationships among these nine species within the genus Gaultheria.
4.4. Candidate DNA barcodesChinese medicinal wintergreens are an important commodity in China, and the lack of genomic resources for Gaultheria has been the main obstacle to taxonomical and genetic analysis, as well as identification and conservation. Common barcodes are not effective for these species (Lu et al., 2010; Ren et al., 2011); although plastomes have been successfully used as a plant super-barcode to distinguish these species, plastome super-barcodes are cost-prohibitive and require complex bioinformatics processing (Shen et al., 2019). Short DNA barcodes, or markers, may provide an effective and economical alternative for identification of target plants (Chase and Fay, 2009; Chen et al., 2010; Shen et al., 2019). Fortunately, the mutation events in plastomes are not randomly distributed within the sequence and are concentrated in certain 'hotspot' regions; therefore, specific barcodes or markers can be exploited from the plastome (Dong et al., 2012). In this study, we proposed eleven regions with high nucleotide variability (Pi) values that might have a high potential for species delimitation in Chinese medicinal wintergreens. These mutational hotspots have the potential to resolve taxonomic issues in the genus and to be used in the future as barcodes for species identification. An ideal DNA barcode should be easily retrievable with a universal primer pair, an appropriately short sequence length to facilitate DNA extraction and amplification (Shen et al., 2019). The intergenic regions psbB-psbJ and trnfM-rrn16 possess high nucleotide variability (Pi) values, however, their PCR success rates were rather low (8.3% and 0 for each, respectively; tested with six samples of each wintergreen by using four primer pairs); thus, these two regions were excluded for the consideration of DNA barcodes. The rpoA-rpoB region was too long (approximately 6500 bp) for Sanger sequencing. Therefore, we finally selected seven regions (infA, ndhF, psaI-trnM, rps3-rpl16, trnQ-psaA, trnV-rps12 and ycf3-trnK) that have the potential to develop into DNA barcodes. Our results show that the trnV-rps12 region provided the best resolution for identifying all nine species of Chinese medicinal wintergreens. It was followed by trnQ-psaA region, which identified eight out of nine species. The infA gene provides the least resolution, which allowed the identification of only four out of nine species. In addition, eight divergence hotspot regions, including rpl36-infA, trnF (GAA)-ndhJ, trnD (GUC)-psbM, trnL (UAA)-trnF (GAA), trnT (UGU)-trnL (UAA), rpl23b, rps3b and petN-trnC (GCA), were recommended as candidate DNA barcodes for species identification of the Trichophyllae clade (Zhang et al., 2017). In summary, different sets of DNA barcodes were detected for delimitation of species from the Trichophyllae clade and the Leucothoides clade.
5. ConclusionsOur research showed that the complete plastome could be used as a plant super-barcode to distinguish closely related medicinal wintergreens successfully, and that even specific molecular markers can be screened based on plastome sequences to distinguish the species. In addition, several unique characteristics of plastomes in Gaultheria, such as the loss of genes, rare gene duplication events, inversions (within G. leucocarpa var. yunnanensis), multiple rearrangement events, and remarkably long repeats, reveal new perspectives for understanding the dynamics of angiosperm plastomes. Furthermore, reticulate hybridization or species radiation that may have occurred during the evolutionary history of Gaultheria was not revealed by plastome sequence data, and further studies with increased taxa sampling and nuclear DNA sequences are necessary.
Author contributionsLL and XYD conceived the work. YLX performed the experiments and analyses. HHS contributed materials/analysis tools. YLX and XYD wrote the paper. XYD and LL revised the paper. All authors approved the final paper.
Declaration of interestsThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsWe thank the Germplasm Bank of Wild Species at the Kunming Institute of Botany for skillful laboratory assistance; we are grateful to Liang-Xin Gao and Zheng-Yu Zuo for the help of DNA extraction, PCR, and Sanger sequencing works to validate some repeat regions. This research was co-supported by the National Natural Science Foundation of China (31960080, 41671052 and 42175139), the Reserve Talents of Young and Middle-aged Academic and Technical Leaders in Yunnan province (202005AC160020), and the Program Innovative Research Team in Science and Technology in Yunnan Province (202005AE160004).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.06.002.
Ahmed, I., Matthews, P.J., Biggs, P.J., et al., 2013. Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol. Ecol. Resour., 13: 929-937. DOI:10.1111/1755-0998.12128 |
Alexandre, M., Normand, B., 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol., 186: 299-317. DOI:10.1111/j.1469-8137.2010.03195.x |
Amiryousefi, A., Hyvönen, J., Poczai, P., 2018. The chloroplast genome sequence of bittersweet (Solanum dulcamara): plastid genome structure evolution in Solanaceae. PLoS One, 13: e0196069. DOI:10.1371/journal.pone.0196069 |
Aruna, P., Murugan, K., Priya, A., et al., 2014. Larvicidal, pupicidal and repellent activities of Gaultheria Oil (Plantae: Ericaceae) against the filarial vector, Culex quinquefasciatus (Insecta: Diptera: Culicidae). J. Entomol. Zool. Stud., 2: 290-294. |
Asano, T., Tsudzuki, T., Takahashi, S., et al., 2004. Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res., 11: 93-99. DOI:10.1093/dnares/11.2.93 |
Bai, H.R., Oyebanji, O., Zhang, R., et al., 2021. Plastid phylogenomic insights into the evolution of subfamily Dialioideae (Leguminosae). Plant Divers., 43: 27-34. DOI:10.1016/j.pld.2020.06.008 |
Barrett, C.F., Baker, W.J., Comer, J.R., et al., 2016. Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol., 209: 855-870. DOI:10.1111/nph.13617 |
Beier, S., Thiel, T., Münch, T., et al., 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics, 33: 2583-2585. DOI:10.1093/bioinformatics/btx198 |
Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27: 573-580. DOI:10.1093/nar/27.2.573 |
Bock, R., 2007. Structure, function, and inheritance of plastid genomes. Cell and Molecular Biology of Plastids. Springer, Berlin, pp. 29-63.
|
Bryant, N., Lloyd, J., Sweeney, C., et al., 2011. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol., 155: 1678-1689. DOI:10.1104/pp.110.168120 |
Casano, L.M., Martın, M., Sabater, B., 2001. Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol, 125: 1450-1458. DOI:10.1104/pp.125.3.1450 |
Cavalier-Smith, T., 2002. Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr. Biol., 12: R62-R64. DOI:10.1016/S0960-9822(01)00675-3 |
Chase, M.W., Fay, M.F., 2009. Barcoding of plants and fungi. Science, 325: 682-683. DOI:10.1126/science.1176906 |
Chen, S., Yao, H., Han, J., et al., 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One, 5: e8613. DOI:10.1371/journal.pone.0008613 |
Cheng, Y., Miao, J.H., Ma, L.Y., et al., 2009. Study advances on chemical constituents and bioactivities from plants of genus Gaultheria. Lishizhen Med. Mater. Med. Res., 20: 3. |
Daniell, H., Lin, C.S., Yu, M., et al., 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol., 17: 1-29. DOI:10.1186/s13059-015-0866-z |
Darling, A.C., Mau, B., Blattner, F.R., et al., 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res., 14: 1394-1403. DOI:10.1101/gr.2289704 |
Dong, W., Liu, J., Yu, J., et al., 2012. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One, 7: e35071. DOI:10.1371/journal.pone.0035071 |
Dong, W.L., Wang, R.N., Zhang, N.Y., et al., 2018. Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci., 19: 716. DOI:10.3390/ijms19030716 |
Downie, S.R., Jansen, R.K., Botany, S.J., 2015. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot., 40: 336-351. DOI:10.1600/036364415X686620 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Erixon, P., Oxelman, B., 2008. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS One, 3: e1386. DOI:10.1371/journal.pone.0001386 |
Fajardo, D., Senalik, D., Ames, M., et al., 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet. Genomes, 9: 489-498. DOI:10.1007/s11295-012-0573-9 |
Fritsch, P.W., Lu, L., 2020. A taxonomic revision of Gaultheria series Trichophyllae (Ericaceae). J. Bot. Res. Instit. Texas, 14: 289-341. DOI:10.17348/jbrit.v14.i2.1010 |
Fritsch, P.W., Lu, L., Bush, C.M., et al., 2011. Phylogenetic analysis of the wintergreen group (Ericaceae) based on six genic regions. Syst. Bot., 36: 990-1003. DOI:10.1600/036364411X604994 |
Fu, C.N., Mo, Z.Q., Yang, J.B., et al., 2022. Testing genome skimming for species discrimination in the large and taxonomically difficult genus Rhododendron. Mol. Ecol. Resour., 22: 404-414. DOI:10.1111/1755-0998.13479 |
Goulding, S.E., Wolfe, K., Olmstead, R., et al., 1996. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet., 252: 195-206. DOI:10.1007/BF02173220 |
Guisinger, M.M., Kuehl, J.V., Boore, J.L., et al., 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol., 28: 583-600. DOI:10.1093/molbev/msq229 |
Haberle, R.C., Fourcade, H.M., Boore, J.L., et al., 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol., 66: 350-361. DOI:10.1007/s00239-008-9086-4 |
Hu, Y.F., Li, X., Li, Z.Z., et al., 2020. Research progress on chemical constituents, pharmacological activities and quality control of Gaultheria leucocarpa var. yunnanensis. Chin. Trad. Pat. Med., 42: 162-167. DOI:10.3847/1538-4357/abb1c3 |
Huang, J.L., Sun, G.L., Zhang, D.M., 2010. Molecular evolution and phylogeny of the angiosperm ycf2 gene. J. Syst. Evol., 48: 240-248. DOI:10.1111/j.1759-6831.2010.00080.x |
Jansen, R.K., Ruhlman, T.A., 2012. Plastid genomes of seed plants. Genomics of Chloroplasts and Mitochondria. Springer, Dordrecht, pp. 103-126.
|
Jin, D.M., Jin, J.J., Yi, T.S., 2020a. Plastome structural conservation and evolution in the clusioid clade of Malpighiales. Sci. Rep., 10: 1-6. DOI:10.1038/s41598-019-56847-4 |
Jin, J.J., Yu, W.B., Yang, J.B., et al., 2020b. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 1-31. DOI:10.1117/1.oe.59.1.016110 |
Katoh, K., Standley, D.M.J., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Kron, K., Fritsch, P., Lu, L., et al., 2020. New combinations and new and resurrected names in Gaultheria (Ericaceae). Gard Bull, 72: 299-317. DOI:10.26492/gbs72(2).2020-13 |
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Kurtz, S., Choudhuri, J.V., Ohlebusch, E., et al., 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res., 29: 4633-4642. DOI:10.1093/nar/29.22.4633 |
Lanfear, R., Frandsen, P.B., Wright, A.M., et al., 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol., 34: 772-773. |
Lee, S.B., Kaittanis, C., Jansen, R.K., et al., 2006. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics, 7: 1-12. DOI:10.3348/jkrs.2006.54.1.1 |
Li, H., Guo, Q., Li, Q., et al., 2020. Long-reads reveal that Rhododendron delavayi plastid genome contains extensive repeat sequences, and recombination exists among plastid genomes of photosynthetic Ericaceae. PeerJ, 8: e9048. DOI:10.7717/peerj.9048 |
Li, Z., Long, H., Zhang, L., et al., 2017. The complete chloroplast genome sequence of tung tree (Vernicia fordii): organization and phylogenetic relationships with other angiosperms. Sci. Rep., 7: 1-11. DOI:10.1038/s41598-016-0028-x |
Librado, P., Rozas, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Liu, Q., Li, X., Li, M., et al., 2020. Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny. BMC Plant Biol., 20: 1-20. DOI:10.1186/s12870-019-2170-7 |
Liu, S., Wang, Z., Su, Y., et al., 2021. Comparative genomic analysis of Polypodiaceae chloroplasts reveals fine structural features and dynamic insertion sequences. BMC Plant Biol., 21: 1-15. DOI:10.1186/s12870-020-02777-7 |
Liu, W.R., Qiao, W.L., Liu, Z.Z., et al., 2013. Gaultheria: phytochemical and pharmacological characteristics. Molecules, 18: 12071-12108. DOI:10.3390/molecules181012071 |
Lu, L., Fritsch, P.W., Bush, C.M., et al., 2019a. Allopolyploidy in the Wintergreen Group of tribe Gaultherieae (Ericaceae) inferred from low-copy nuclear genes. Nord. J. Bot., 37: e02077. DOI:10.1111/njb.02077 |
Lu, L., Fritsch, P.W., Cruz, B.C., et al., 2010. Reticulate evolution, cryptic species, and character convergence in the core East Asian clade of Gaultheria (Ericaceae). Mol. Phylogenet. Evol., 57: 364-379. DOI:10.1016/j.ympev.2010.06.002 |
Lu, L., Fritsch, P.W., Matzke, N.J., et al., 2019b. Why is fruit colour so variable? Phylogenetic analyses reveal relationships between fruit-colour evolution, biogeography and diversification. Glob. Ecol. Biogeogr., 28: 891-903. DOI:10.1111/geb.12900 |
Luo, B.S., Gu, R.H., Kennelly, E.J., et al., 2018. Gaultheria ethnobotany and bioactivity: blueberry relatives with anti-inflammatory, antioxidant, and anticancer constituents. Curr. Med. Chem., 25: 5168-5176. |
Luo, B.S., Kastrat, E., Morcol, T., et al., 2021. Gaultheria longibracteolata, an alternative source of wintergreen oil. Food Chem., 342: 128244. DOI:10.1016/j.foodchem.2020.128244 |
Lv, S.Y., Ye, X.Y., Li, Z.H., et al., 2022. Testing complete plastomes and nuclear ribosomal DNA sequences for species identification in a taxonomically difficult bamboo genus Fargesia. Plant Divers. DOI:10.1016/j.pld.2022.04.002 |
Ma, X., Zhao, L., Han, Z., et al., 2002. Comparison of the contents of lignans glycosides in 5 medicinal plants of Gaultheria by HPLC. J. Plant Resour. Environ., 11: 2. |
Ma, X.J., Zhao, L., Du, C.F., et al., 2001. Advances in studies on Gaultheria leucocarpa var. yunnanensis and medicinal plants of Gaultheria L. Chin. Trad. Herb. Drugs, 32: 5. |
Martin, M., Casano, L.M., Sabater, B., 1996. Identification of the product of ndhA gene as a thylakoid protein synthesized in response to photooxidative treatment. Plant Cell Physiol., 37: 293-298. DOI:10.1093/oxfordjournals.pcp.a028945 |
Mohanta, T.K., Mishra, A.K., Khan, A., et al., 2020. Gene loss and evolution of the plastome. Genes, 11: 1133. DOI:10.3390/genes11101133 |
Moore, M.J., Dhingra, A., Soltis, P.S., et al., 2006. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol., 6: 1-13. DOI:10.1186/1471-2229-6-1 |
Mukhopadhyay, M., Bantawa, P., Mondal, T.K., et al., 2016. Biological and phylogenetic advancements of Gaultheria fragrantissima: economically important oil bearing medicinal plant. Ind. Crop. Prod., 81: 91-99. DOI:10.1016/j.indcrop.2015.11.042 |
Mwanzia, V.M., He, D.X., Gichira, A.W., et al., 2020. The complete plastome sequences of five Aponogeton species (Aponogetonaceae): insights into the structural organization and mutational hotspots. Plant Divers., 42: 334-342. DOI:10.1016/j.pld.2020.02.002 |
Nguyen, L.T., Schmidt, H.A., Von Haeseler, A., et al., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32: 268-274. DOI:10.1093/molbev/msu300 |
Nikolić, M., Marković, T., Mojović, M., et al., 2013. Chemical composition and biological activity of Gaultheria procumbens L. essential oil. Ind. Crop. Prod., 49: 561-567. DOI:10.1016/j.indcrop.2013.06.002 |
Oliver, M.J., Murdock, A.G., Mishler, B.D., et al., 2010. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics, 11: 1-8. DOI:10.1186/1471-2164-11-1 |
Paredes, M., Quiles, M.J., 2013. Stimulation of chlororespiration by drought under heat and high illumination in Rosa meillandina. J. Plant Physiol., 170: 165-171. DOI:10.1016/j.jplph.2012.09.010 |
Peng, L., Yamamoto, H., Shikanai, T., 2011. Structure and biogenesis of the chloroplast NAD (P)H dehydrogenase complex. Biochim. Biophys. Acta, Bioenerg, 1807: 945-953. DOI:10.1016/j.bbabio.2010.10.015 |
Raubeson, L.A., Jansen, R.K., 2005. Chloroplast genomes of plants. Plant Diversity and Evolution: genotypic and phenotypic variation in higher plants. CABI, Cambridge, pp. 45-68.
|
Ravi, V., Khurana, J., Tyagi, A., et al., 2008. An update on chloroplast genomes. Plant Syst. Evol., 271: 101-122. DOI:10.1007/s00606-007-0608-0 |
Ren, H., Lu, L., Wang, H., et al., 2011. DNA barcoding of Gaultheria L. in China (Ericaceae: Vaccinioideae). J. Syst. Evol., 49: 411-424. DOI:10.1111/j.1759-6831.2011.00152.x |
Ronquist, F., Teslenko, M., Van Der Mark, P., et al., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Savage, L.J., Imre, K.M., Hall, D.A., et al., 2013. Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS One, 8: e73291. DOI:10.1371/journal.pone.0073291 |
Shen, Z., Lu, T., Zhang, Z., et al., 2019. Authentication of traditional Chinese medicinal herb "Gusuibu" by DNA-based molecular methods. Ind. Crop. Prod., 141: 111756. DOI:10.1016/j.indcrop.2019.111756 |
Silva, S.R., Diaz, Y.C., Penha, H.A., et al., 2016. The chloroplast genome of Utricularia reniformis sheds light on the evolution of the ndh gene complex of terrestrial carnivorous plants from the Lentibulariaceae family. PLoS One, 11: e0165176. DOI:10.1371/journal.pone.0165176 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Thode, V.A., Lohmann, L.G., 2019. Comparative chloroplast genomics at low taxonomic levels: a case study using Amphilophium (Bignonieae, Bignoniaceae). Front. Plant Sci., 10: 796. DOI:10.3389/fpls.2019.00796 |
Timme, R.E., Kuehl, J.V., Boore, J.L., et al., 2007. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am. J. Bot., 94: 302-312. DOI:10.3732/ajb.94.3.302 |
Wang, R.J., Cheng, C.L., Chang, C.C., et al., 2008. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol., 8: 1-14. DOI:10.1186/1471-2148-8-1 |
Wang, R.N., Milne, R.I., Du, X.Y., et al., 2020. Characteristics and mutational hotspots of plastomes in Debregeasia (Urticaceae). Front. Genet., 11: 729. DOI:10.3389/fgene.2020.00729 |
Wang, Y., Wang, S., Liu, Y., et al., 2021. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics, 22: 1-12. |
Wang, Y.H., Wicke, S., Wang, H., et al., 2018. Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae). Front. Plant Sci., 9: 138. DOI:10.3389/fpls.2018.00138 |
Wick, R.R., Schultz, M.B., Zobel, J., et al., 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics, 31: 3350-3352. DOI:10.1093/bioinformatics/btv383 |
Wicke, S., Schneeweiss, G.M., Depamphilis, C.W., et al., 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol., 76: 273-297. DOI:10.1007/s11103-011-9762-4 |
Williams, A.V., Miller, J.T., Small, I., et al., 2016. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol. Phylogenet. Evol., 96: 1-8. DOI:10.1016/j.ympev.2015.11.021 |
Wolf, P.G., Der, J.P., Duffy, A.M., et al., 2011. The evolution of chloroplast genes and genomes in ferns. Plant Mol. Biol., 76: 251-261. DOI:10.1007/s11103-010-9706-4 |
Wu, H., Ma, P.F., Li, H.T., et al., 2021a. Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae). Plant Divers, 43: 15-26. DOI:10.1016/j.pld.2020.07.004 |
Wu, S., Chen, J., Li, Y., et al., 2021b. Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol., 21: 1-16. DOI:10.1186/s12870-020-02777-7 |
Yang, J.B., Tang, M., Li, H.T., et al., 2013. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol., 13: 1-12. DOI:10.1186/1471-2148-13-1 |
Ye, W.Q., Yap, Z.Y., Li, P., et al., 2018. Plastome organization, genome-based phylogeny and evolution of plastid genes in Podophylloideae (Berberidaceae). Mol. Phylogenet. Evol., 127: 978-987. DOI:10.1016/j.ympev.2018.07.001 |
Zhang, M.Y., Fritsch, P.W., Ma, P.F., et al., 2017. Plastid phylogenomics and adaptive evolution of Gaultheria series Trichophyllae (Ericaceae), a clade from sky islands of the Himalaya-Hengduan Mountains. Mol. Phylogenet. Evol., 110: 7-18. DOI:10.1016/j.ympev.2017.01.015 |
Zhang, Y., Li, J., He, F., et al., 2020. Chemical constituents of Gaultheria leucocarpa var. yunnanensis (Franch.) T.Z. Hsu & R.C. Fang. Centr. South Pharm., 18: 1800-1802. |
Zhang, Y.J., Ma, P.F., Li, D.Z., 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One, 6: e20596. DOI:10.1371/journal.pone.0020596 |
Zhu, A., Guo, W., Gupta, S., et al., 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol., 209: 1747-1756. DOI:10.1111/nph.13743 |



