b. Department of Botany, Faculty of Science, Charles University, Benátská 2, 128 00, Prague, Czech Republic
Many interesting relationships between DNA content and infraspecific environmental range variables have been described (Grime and Mowforth, 1982; Bennett, 1987; Price, 1988; Poggio and Naranjo, 1990; Bromham et al., 2015). All these reports suggest that plant genome size may be predictable, adaptive, and of evolutionary significance (Poggio et al., 1998; Soltis et al., 2007; Te Beest et al., 2012). Increasing relative DNA content by polyploidization has proven to be an important mechanism of speciation in angiosperms and is a key driver of diversity (De Bodt et al., 2005; Soltis et al., 2009; Tank et al., 2015; Soltis and Soltis, 2016; Landis et al., 2018). Autopolyploid collections are usually treated as multiple copies of a single cytotype within a species, while allopolyploids arise via hybridization between species and are typically viewed as different species. Disentangling the cytological characterization of species across their entire geographic range is important to understand patterns of cytotype formation, establishment and migration, and to uncover previously undetected cytotype variation that can yield better estimates of diversification (Soltis et al., 2007, 2010; Severns and Liston, 2008; Laport and Ng, 2017).
Drylands phylogeography theory in the Western Hemisphere was developed mostly in North American deserts, but South American deserts were a key centre for the emergence of Neotropical lineages and have been understudied. Some of these lineages later migrated northwards and diversified in the Northern Hemisphere. Moreover, arid land vegetation evolved in the temperate Neotropics since the Oligocene (Burnham and Graham, 1999), encompassing heterogeneous climate and topography differing from their current distribution. The genus Larrea Cav. (Zygophyllaceae), commonly known as creosote bush, is amphitropically distributed, with species separated spatially by areas that are not bridged by shared vegetation type or climatic regimes. Ancestral Larrea species is thought to have been South American, and reached North America through long distance dispersal and species have diverged allopatrically since the dispersal events (Barbour, 1969; Raven, 1972; Lia et al., 2001). The genus exhibits both intra-and interspecific variation in DNA content (Hunziker et al., 1972; Laport et al., 2012), and grows in arid and semiarid regions of North and South America. Phylogeographic studies suggest establishment of polyploid populations of the North American species accompanying its rapid spread (Laport et al., 2012).
Due to the large latitudinal span of the genus Larrea (20–35° N in Mexico and United States, and 22–46° S in Monte Desert of Argentina, plus scattered sites in Chile, Bolivia and Peru), environmental selective pressures should differentially impact species inhabiting different portions of the climatic range. In general, Larrea straddles the 200-mm annual isohyet (Ezcurra et al., 1991), but among North American deserts, precipitation seasonality varies from summer rains, regularly distributed monthly rains to mainly winter rainfall, coupled with an increasing aridity gradient from east to west (Chihuahua desert-> Sonora desert-> Mojave desert, respectively) (Yang, 1970). Meanwhile, although mean annual precipitation is relatively constant in the Monte Desert, its distribution changes, in a latitudinal fashion, from monsoon-type summer rains in northern Argentina to a winter rainfall peak near the Patagonian Andes, and to a more regular monthly distribution along the southern Atlantic coast (Paruelo et al., 1998; Roig et al., 2009). These environmental characteristics provide a good model for investigating correlations between genome size and climatic variables, which can shed some light on drylands phylogeography in the Americas.
Our goals are to investigate the adaptive potential of DNA content within the genus Larrea and among four Larrea species, by relating variation in relative DNA content to climatic variables throughout their collective distributions in North and South America. We ask the following specific questions: (1) Are there differences in relative DNA content measured by flow cytometry and relative monoploid genome value between species of the genus Larrea across their collective distributions? (2) Are there differences in climatic ranges among Larrea species? (3) Are climatic variables related to relative DNA content variation in the genus Larrea? We predict that relative DNA content will be higher in harsher (i.e. drier and hotter) climatic conditions, since changes in DNA content may influence an organism's ecological tolerance or increase ecological amplitude or facilitate the acquisition of new genetic functions. Finally, we revisit the idea of integrating estimates of DNA content, measured using modern techniques of flow cytometry, with fine scale, functionally relevant climatic variables, and discuss the evolutionary importance of hybridization and polyploidization within the genus as potential drivers of diversification.
2. Materials and methods 2.1. Study systemWe sampled four out of five Larrea species throughout their respective ranges, leaving out Larrea ameghinoi Speg., which is an extremely rare species, restricted to few localities in Chubut and Rio Negro provinces in Argentina. The only North American species, Larrea tridentata (DC) Coville has multiple cytotypes, whose ploidy level increase across an aridity gradient; with diploid individuals (2n = 26) occurring in the Chihuahua Desert, tetraploids (2n = 4x = 52) in the Sonora Desert and hexaploids (2n = 6x = 78) in the Mojave Desert. Although the taxonomic status of autopolyploids and their influence in biodiversity levels remains a subject of active debate in the literature (Soltis et al., 2007); the different L. tridentata cytotypes have historically been considered as representing a single species (Yang, 1970; Hunziker et al., 1972; Laport et al., 2013). We follow this precedent in our study and consider ploidal variation as representing intra-specific DNA content variation. Interestingly, the 4x and 6x cytotypes are found interdigitated in the field in Sonora and Mojave deserts (Laport and Ramsey, 2015). The South American L. nitida Cav., and L. divaricata Cav., are diploids (2n = 26), while L. cuneifolia Cav. is an allotetraploid (2n = 4x = 52) (Hunziker et al., 1972) (Fig. 1).
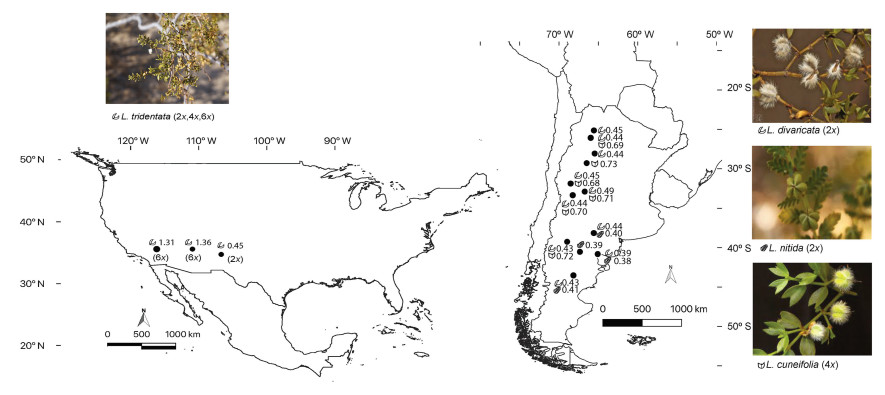
|
| Fig. 1 Map of sampled localities of four Larrea species: L. divaricata, L. cuneifolia, L. nitida, L. tridentata in deserts of North and South America. Including average local relative DNA content ratio (RCR) and sketches of leaf morphology. |
The genus Larrea is more speciose in the Monte Desert phytogeographic region in Argentina, which resembles the North American warm deserts, being a heterogeneous mosaic of landscapes, from open sand dune habitats to shrublands dominated by creosote bush. In South America, 12 localities were sampled, representing three species, L. cuneifolia, L. divaricata, and L. nitida, totalling 106 individuals collected. Meanwhile, 14 individuals of L. tridentata were sampled from three localities, representing the three North American deserts (Chihuahua, Sonora and Mojave Deserts). Two to five individuals per locality were sampled and kept in silica gel until flow cytometry analysis (Table 1). Voucher specimens are deposited in the BCRU herbarium (Bariloche, Argentina).
| Localities | LAT | LON | ELEV m a.s.l. | LC (29) | LD (46) | LN (22) | LT (14) | BIO1 | BIO14 | Solar Radiation | |||||||
| RCR | Cx | RCR | Cx | RCR | Cx | RCR | Cx | ||||||||||
| Los Cardones | −25.140 | −65.576 | 1912 | 0.445 | 0.225 | 16 | 2 | 16340.33 | |||||||||
| Cafayate | −26.073 | −65.978 | 1609 | 0.686 | 0.172 | 0.450 | 0.223 | 17 | 0 | 18324.58 | |||||||
| Catamarca | −28.065 | −65.475 | 427 | 0.437 | 0.218 | 20 | 7 | 16290.50 | |||||||||
| La Rioja | −29.284 | −66.491 | 377 | 0.728 | 0.182 | 20 | 4 | 17804.00 | |||||||||
| San Juan | −31.878 | −68.525 | 588 | 0.681 | 0.142 | 0.448 | 0.224 | 18 | 3 | 17949.75 | |||||||
| Mendoza | −33.362 | −68.266 | 607 | 0.702 | 0.140 | 0.442 | 0.221 | 16 | 4 | 17683.92 | |||||||
| San Luis | −32.900 | −66.741 | 766 | 0.714 | 0.143 | 0.429 | 0.215 | 18 | 8 | 17659.00 | |||||||
| Lihuel Calel | −38.136 | −65.592 | 236 | 0.438 | 0.219 | 0.401 | 0.201 | 15 | 13 | 16440.25 | |||||||
| Neuquén | −39.243 | −68.974 | 697 | 0.720 | 0.144 | 0.431 | 0.216 | 0.495 | 0.248 | 12 | 6 | 16137.08 | |||||
| Río Negro | −40.520 | −67.353 | 504 | 0.394 | 0.197 | 13 | 9 | 15804.00 | |||||||||
| Las Grutas | −40, 811 | −65, 094 | 22 | 0.392 | 0.196 | 0.383 | 0.192 | 15 | 15 | 15749.25 | |||||||
| Madryn | −43.515 | −68.181 | 304 | 0.429 | 0.214 | 0.407 | 0.204 | 12 | 9 | 13668.58 | |||||||
| NM, Chihuahua | 34.325 | −106.704 | 1066 | 0.445 | 0.223 | 13 | 9 | 19545.00 | |||||||||
| Tucson, Sonora | 32.221 | −110.925 | 1525 | 1.360 | 0.227 | 21 | 5 | 19331.33 | |||||||||
| Baker, Mojave | 35.185 | −116.172 | 915 | 1.315 | 0.219 | 18 | 1 | 19687.50 | |||||||||
| Total mean (SD) | 0.702 (0.044) | 0.175 (0.011) | 0.433 (0.027) | 0.217 (0.014) | 0.397 (0.013) | 0.198 (0.007) | 0.444 (0.003) 1.335 (0.049) |
0.222 (0.005) | |||||||||
We measured relative nuclear DNA content by flow cytometry using the AT-selective fluorochrome 4′, 6′-diamidino-2 phenylindole (DAPI) assay following a simplified two-step protocol (Doležel et al., 2007). We used Solanum pseudocapsicum L. (2C = 2.61 pg) (Temsch et al., 2010) as primary internal standard, but in some cases peaks of standard and sample overlapped. Therefore, in those cases we employed Bellis perennis L. (2C = 3.38 pg) (Schönswetter et al., 2007) and we derived genome size of Bellis directly from simultaneous analysis with Solanum. We further express relative DNA content values as ratios to Solanum. We chopped leaf tissue together with the standard using a sharp razor blade in a Petri dish containing Otto I buffer (0.1 M citric acid, 0.5% Tween 20). We filtered the suspension and incubated for 5 min at room temperature. After incubation, we added 1 ml of Otto II buffer (0.4 M Na2HPO4 12H2O), along with 2-mercaptoethanol (2 μg/ml) and DAPI (4 μg/ml). All samples were analysed with a Partec CyFlow Space instrument (Partec GmbH, Münster, Germany), equipped with a UV LED chip as the excitation source. Flow cytometry histograms were evaluated using the Partec FloMax software (Partec GmbH). Measurements with coefficient of variation (CV) values, for standard and sample peaks, below 5% were accepted. The extent of the total variation in intraspecific relative DNA content index was calculated as a percentage of the difference between its highest and lowest values, and was expressed as % of the minimum. Also the monoploid genome value (Cx) relative equivalent was calculated by dividing the relative DNA content of each sample by the cytotype ploidy level to test the role of polyploidy in the diversification of the genus.
2.4. Climatic variablesFor broad characterization of the variable climatic conditions of both North and South American deserts, we looked for available occurrence data of focal species in seven data reservoirs including herbaria (MO, NY, ARIZ, COLO, SI) and online repositories (GBIF: gbif.org, Mincyt: datos.mincyt.gob.ar/#/biologicos). We then excluded repeated locations or doubtful coordinates. This curated occurrence data set (399 localities), including our sampled localities (15 sites), represents registered GPS-locations for each occurring specie, including latitude, longitude and elevation. For these 399 records we extracted local climate variables from WorldClim 1.4, a set of 19 global bioclimatic variables with a spatial resolution of 30 arc seconds (Hijmans et al., 2005), (http://www.worldclim.org/) (Supplementary material S3). For further analyses we winnowed down the dataset to a reduced set of temperature and precipitation proxies that are functionally relevant in arid environments keeping uncorrelated bioclimatic variables. The climatic matrix included the following variables: BIO1: Annual mean temperature, and BIO14: Precipitation of the driest month and solar radiation. Solar radiation data for South American localities was generated with the ClimateSA v.1.0 software package, available at tinyurl.com/ClimateSA (Hamann et al., 2013), and for North American localities with ClimateNA available at climatena.ca (Wang et al., 2016). Solar radiation is particularly relevant since it has been suggested to have influenced the evolution of leaf orientation for Larrea species (Ezcurra et al., 1991). We used this matrix to characterize the climatic envelope for each species, and a subset of the climatic values for the 15 sampled localities were correlated with relative DNA content data.
2.5. Statistical analysesWe tested for differences among species in relative DNA content ratio (RCR) and monoploid genome value (Cx), via Kruskal–Wallis one-way analysis of variance on ranks models and Tukey's post-hoc tests. To study the degree of ecological divergence among species, we used boxplots for the climatic variables for each species to graphically depict observed differences among species. As many of the variables demonstrated a non-normal distribution, we used Kruskal–Wallis tests, followed by a nonparametric post-hoc test, to check the significance of differences between species, using all pairwise multiple comparison procedure (Dunn's Method). To investigate the relationship between climate and relative DNA content, we performed Pearson's correlations and linear mixed effect models, using species identity as random factor. Statistical analyses were carried out in R 3.4.4 (R Development Core Team, 2017), using above mentioned functions and, when appropriate, packages nlme and ggplot for graphic representation of the results.
3. Results 3.1. Flow cytometryIn this study we sampled for flow cytometry two diploid species, one species comprising diploids, autotretraploids and autohexaploids, and one allotetraploid species within the genus Larrea (Table 1, Supplementary material S1). As expected, L. tridentata showed different relative DNA content ratios (RCR) that represent different cytotypes. Samples from New Mexico (Chihuahua desert), have a diploid cytotype, and showed an average RCR (=sample: standard fluorescence ratio (±s.d.)) of 0.445 ± 0.004; while the other two North American populations showed greater RCR, as they are known to be polyploids: Tucson (6x, Sonora desert) yields RCR of 1.360 ± 0.013, and Baker (6x, Mojave Desert) yields RCR of 1.315 (Table 1, Supplementary material S1). Diploid L. tridentata and the South American diploid L. divaricata have similar RCR and relative monoploid genome values (Cx) (Table 1, Supplementary material S1). L. divaricata has a mean RCR of 0.433 ± 0.027, the total variation for this species was 38.98% of the minimum value, with minimum RCR in the South (Rio Negro, 0.392 ± 0.005) and maximum in the North (Cafayate, 0.450 ± 0.008). L. cuneifolia is the South American species with highest RCR and lower Cx as it is an allotetraploid. Across its geographic distribution, this species has an average RCR of 0.702 ± 0.044 and Cx of 0.175 ± 0.011 (measured with B. perennis, and recalculated for S. pseudocapsicum standard), total variation for this species was 37.36% of the minimum value. L. nitida is the diploid species with the lowest RCR (0.397 ± 0.013), that varied from 0.383 ± 0.004 in southeast Atlantic coast (Las Grutas) to 0.407 ± 0.004 in the South (Madryn), with total genome size variation of 12.35% of the minimum value (Table 1, Supplementary material S1).
3.2. Climatic envelopesData based on the 399 records matrix obtained from online repositories and sampled localities shows that Larrea species grow at statistically different temperature and precipitation ranges (Fig. 2, Supplementary material S2, S3). L. divaricata and L. cuneifolia grow under similar conditions in terms of mean annual temperature (15.01 and 15.33 ℃), while L. nitida grows at lower temperatures (11.80 ℃) and L. tridentata in North America grows at much higher temperatures (18.22 ℃) (Fig. 2). In terms of precipitation, L. divaricata (8.36 mm) has a wider range than the other species. L. cuneifolia (7.09 mm) differs from L. nitida and L. tridentata, the latter is the most different since it grows in sites with very low precipitation levels (4.2 mm). While L. nitida grows in higher precipitation sites (9.8 mm) (Fig. 2). In terms of mean solar radiation L. divaricata and L. cuneifolia grow under similar conditions (16, 624 and 17, 550 W/m2), while L. nitida grows at lower radiation (15, 415 W/m2), and L. tridentata grows at much higher solar radiation (19, 521 W/m2) (Fig. 2).
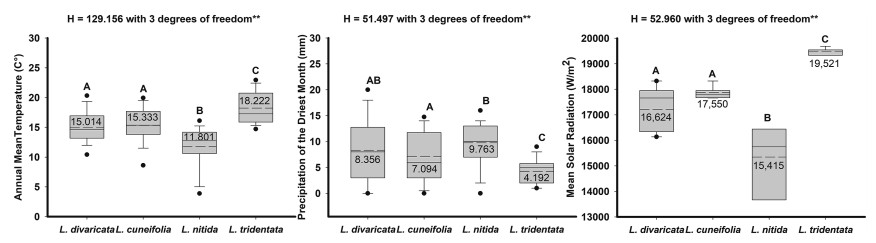
|
| Fig. 2 Box plot of bioclimatic variables extracted from WorldClim using occurrence records of focal species obtained from herbaria, online repositories and our field collections: Annual mean temperature (BIO1), Precipitation of the driest month (BIO14), and Mean solar radiation in four Larrea species. Central complete and dashed lines in the square are the mean and median values, respectively, the box represents the standard deviation, and whiskers are 95% confidence intervals. Including results of Kruskal–Wallis One Way Analysis of Variance on Ranks, different letters represent All Pairwise Multiple Comparison Procedures (Dunn's Method). ∗∗Statistically significant models P ≤ 0.001 (details in). |
When the four studied species are analysed together there is a general pattern suggesting that DNA content increases in warmer and drier climates, with higher solar radiation (Fig. 3, Table 2, Supplementary material S2). All three examined variables are significantly correlated with the relative DNA content for the genus, although correlation is around 0.5. Moreover, within each species some environmental variables showed apparent covariation with relative DNA content ratio (RCR), but they were not significantly correlated, probably due to the low sample size (Table 2). In L. cuneifolia, the South American allotetraploid, RCR decreases with elevation and increases with summer precipitation (southward), with higher DNA content in sites with lower precipitation all year round, i.e. less precipitation seasonality. With higher solar radiation RCR in L. cuneifolia decreases. In L. divaricata RCR increases northward where summer precipitation decreases (i.e. less precipitation → higher DNA content). RCR in L. nitida increases with elevation and decreases with summer temperature and precipitation, and, as with L. cuneifolia, increases when precipitation has more seasonality, i.e. restricted to certain season of the year. The North American species, L. tridentata, show lower RCR eastward and it decreases with summer precipitation, but increases with summer temperature, and precipitation seasonality (Table 2).
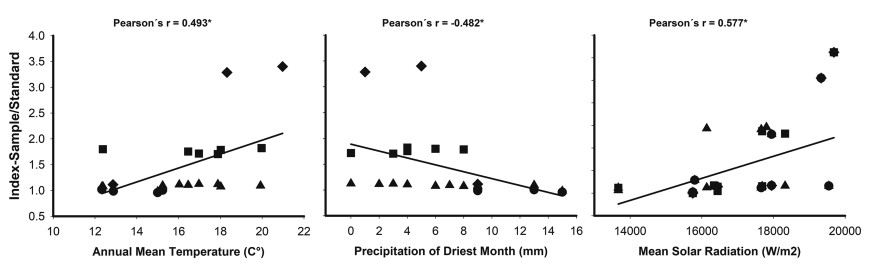
|
| Fig. 3 Pearson correlations between bioclimatic variables (Annual mean temperature (BIO1), Precipitation of the driest month (BIO14), Mean solar radiation) and Relative DNA content ratio (RCR) for four Larrea species. ∗Statistically significant correlations P ≤ 0.05. Triangles up: Larrea divaricata, squares: L. cuneifolia, circles: L. nitida, romboids: L. tridentata. |
| Latitude | Longitude | Elevation | BIO1 | BIO14 | MSR | |
| LCRCR | −0.389 | 0.055 | −0.528 | −0.033 | 0.645 | −0.543 |
| LDRCR | 0.645a | −0.255 | 0.613 | 0.311 | −0.793a | 0.438 |
| LNRCR | 0.016 | −0.511 | 0.905 | −0.333 | −0.228 | 0.108 |
| LTRCR | −0.055 | −0.919 | 0.100 | 0.872 | −0.942 | 0.068 |
| All species | 0.593a | −0.619a | 0.322 | 0.493a | −0.482a | 0.577a |
| a Statistically significant correlations P ≤ 0.05. | ||||||
Our results are consistent with the hypothesis that selective climatic pressures in the genus Larrea might have facilitated repeated whole genome duplication events in North America (Hunziker et al., 1972; Lia et al., 2001; Laport et al., 2013). In South America, reticulate evolution as allopolyploidization by hybridization could have arisen in an environmentally-dependent way that has resulted in the observed correlations between climate and genome size. Studying the degree of relative DNA content changes among the four more abundant species of Larrea throughout their distributions in North and South American deserts, we found differences in relative DNA content (RCR) and monoploid genome value among species. The small variance observed in their relative DNA content could be potentially helpful to identify species by their genome size. We acknowledge that relative genome size values were calculated based on DAPI staining AT bases. Data on interspecific geographic AT content variation would be necessary to account for this bias. However, the AT bases variation would slightly affect the general patterns observed in the present study.
All studied species showed a correlation between relative DNA content and key temperature and precipitation gradients. L nitida has the lowest DNA content among the diploid species and also the lowest variation; along with differential morphological characters; enough so that it is classified in another section of the genus (Hunziker and Comas, 2002). L. cuneifolia, as an allotetraploid shows intermediate values of relative DNA content, between hexaploid L. tridentata and the diploid species. The genomic composition of L. cuneifolia is still unknown, but it has been suggested to have originated from diploid genomes (Lia et al., 2001), whose relative genome sizes should be added together to get comparative theoretic allotetraploid size. On the other extreme, the three populations of L. tridentata sampled for this study (New Mexico-Chihuahua, Baker-Mojave and Tucson-Sonora), have the highest DNA content, and are inferred to contain diploid and hexaploid cytotypes, respectively. We only sampled 6x individuals in Tucson (Sonora desert), which is not a surprise since Laport et al. (2012) reported several broadly interdigitated populations of tetraploids and hexaploids throughout central Arizona.
Temperature, precipitation and solar radiation have been found to be influential predictors of plant distribution; as higher DNA content tend to occur in harsher climates (Stebbins, 1985; Poggio et al., 2014; Rice et al., 2019). Due to the large and disrupt latitudinal span of the genus Larrea (22–46° S in South American Monte Desert, plus scattered sites in Bolivia and Peru, and 20–35° N in North America), environmental selective pressures are expected to differentially affect species inhabiting different portions of the climatic range. Consistent with this hypothesis, in Larrea, climatic variables are correlated with relative DNA content as well. We found that South American Larrea species are more likely to have higher DNA content in sites with lower annual precipitation, higher solar radiation, and temperature. L. cuneifolia, the only polyploid species in South America, has the same climatic breadth as the diploid L. divaricata, but they differ in how their RCR relates with climate. The South American allotetraploid shows no correlation between DNA content and environment. Unexpectedly, RCR in L. cuneifolia decreases with solar radiation, probably as a result of the interaction of this climatic variable and elevation. Meanwhile, significant levels of variation in DNA content are observed along the distribution of L. divaricata with latitude and precipitation of the driest month. The correlation between genome size and latitude suggests adaptation to varying climatic conditions at different latitudes, favouring plants possessing certain genome sizes if they are able to tolerate local environmental conditions that might constrain their fitness (Ohri et al., 1986; Bottini et al., 2000; Leong-Škorničková et al., 2007; Zhang et al., 2019). The observed pattern might also respond to genetic variation of local population's genomes in combination with DNA amount. Further studies on genetic variation should be conducted over the range of the species to better understand the selection forces acting on the different species. L. nitida also partially shares its geographic range with the other South American Larrea species, although, it is found only in colder and more humid environments, being a species with high habitat specificity. DNA content in L. nitida increases with elevation and decreases with increasing temperature and precipitation. In the North American species (L. tridentata) RCR, along with ploidy level, is positively correlated with temperature and negatively correlated with precipitation (i.e., individuals from drier sites with higher temperatures have larger genomes) in agreement with global patterns reported by Rice et al. (2019).
The reduced set of temperature and precipitation proxies selected here, annual mean temperature, precipitation of the driest month, and solar radiation, differs for South and North American Larrea congeners. In South America, DNA content is negatively correlated with precipitation and positively correlated with temperature. Larrea species are hypothesized to have responded to heterogeneous environments through diversification into different species during the Neogene (Lia et al., 2001). Along with this diversification, alloploidization arose by hybridization at least once. The allotetraploid, L. cuneifolia, might have had an advantage at the time of its origin, although today its range overlaps with L. divaricata. The harsh environment of a climatic refugia may have triggered the sympatric diversification via hybridization between L. divaricata and other congeneric species that formed L. cuneifolia. In contrast, L. tridentata, is hypothesized to have evolved three ploidal races via autoploidization in the North American deserts during the Quaternary (Lia et al., 2001), potentially as a response to selective pressures due to climatic variation as the species reached North America from South America. This strategy, i.e. whole genome duplication, has been proposed to play a major role in rapid plant evolution and as an important determinant of invasiveness facilitating the establishment in novel environments (Te Beest et al., 2012).
Our results show that both South American and North American Larrea occupy a wide range of climatic conditions, but there are differences in climatic ranges among species of the genus. We found that L. nitida with its small genome size and low DNA content variation, tolerates lower temperatures and low levels of solar radiation, while growing in localities with more precipitation. L. divaricata and L. cuneifolia have relatively high DNA content variability along their respective distributions and show high tolerance for a range of temperatures and precipitations. L. cuneifolia's climatic envelope is characterized by lower precipitation and higher solar radiation than the other species. The literature suggests that polyploids might tolerate harsher environmental conditions compared to diploid progenitors due to their greater potential fitness (Maceira et al., 1993; Petit and Thompson, 1999; Zhang et al., 2019). L. tridentata is exposed to more severe climatic conditions than the South American species; temperatures in North American deserts are higher and precipitation much lower than those found in South America. The climatic envelope for L. tridentata is characterized by high values of temperature and solar radiation along with low precipitation, harsh conditions that might have been selective drivers for polyploidization. L. divaricata and L. tridentata are sister taxa (Lia et al., 2001) and L. divaricata has a similar monoploid genome value (Cx) and relative nuclear DNA content (RCR) to diploid L. tridentata. This might indicate that the extra DNA in L. tridentata tetraploids and hexaploids is not random, suggesting selection for polyploidization, as postulated for Hippeastrum spp. (Poggio et al., 2014). These duplicated genomes can undergo divergent evolution for new functions, giving polyploids ecological benefits that contribute to their success in nature (Soltis and Soltis, 2000; Comai, 2005). Also, as changes in DNA content can influence an organism's ecological amplitude or tolerance, have an important role in gene regulation, or the acquisition of new genetic functions, genome duplication in L. tridentata may have allowed the species to survive in harsher environments (i.e. drier and hotter). Different species respond in a different fashion to the same climatic conditions. However, differences in DNA content can influence the ecological breadth of the species. L. tridentata has three cytotypes and grows under much higher temperature than the South American species and in localities with very low precipitation. The ploidal volatility of L. tridentata might have provided adaptive advantages in those extreme environments. Energy availability, determined by temperature, light, and water, has been proposed as the primary driver of patterns of species diversity (Rohde, 1992; Hawkins et al., 2003; Willis et al., 2009; Flenley, 2011). Consistent with this hypothesis, temperature has been identified as a correlate of molecular evolutionary rate in plants (Davies et al., 2004), as has water availability (Goldie et al., 2010). So, it is reasonable to expect that plants at drier, hotter and with higher solar radiation environments (i.e. harsher environments) that undergo processes of increasing DNA content may also increase ecological amplitude or acquisition of new genetic functions.
5. ConclusionsIn this study we shed light on how DNA content variation in South American desert vegetation taxa played an important role in structuring Western Hemisphere dryland plant communities. Pointing out how South American deserts have been understudied. Correlation between environmental factors and relative DNA content reveal a potentially adaptive role for intraspecific genome size variation. In some cases, repeated whole genome duplication events in plant species might have a selective advantage in harsh environments where climate pressures are high. On the other hand, reticulate evolution as allopolyploidization via hybridization might arise climate-dependently showing particular climate – genome size signals. Increasing DNA content and/or ploidy level, have played a major role in plant evolution, conferring plants a genetically advantageous character, i.e. giving them the ability to adapt to a wider range of climatic conditions.
Author contributionsC.P.S. designed the study and managed the project. M.T. and C.P.S. collected samples. R.V.R. prepared DNA samples and performed cytogenetic analyses along with R.U. and T.U. Data analyses were performed by R.V.R., C.P.S. and R.U. C.P.S. and R.V.R. wrote the manuscript. M.T. and T.U. modified and improved the manuscript. All authors read and approved the final manuscript.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgementsThis work was supported by Argentine National Found for Science and Technology, under Grant PICT 2014-3478-BID and 2019-0149 BID; and Council of Science and Technology-CONICET under Grant PIP-0712. R.V.R., M.T., and C.P.S. are members of CONICET. Grant Agency of the Czech Republic 17-12420S. We thank Peter Smouse and two anonymous reviewers for helpful comments that improved the quality of our manuscript. Thanks also to Thomas Kitzberger, Julieta Betinelli and Daiana Jaume, Carolina Calviño, Laura Zalazar, who collaborated in field work, sample processing, and downloading climatic data respectively.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.11.006 .
Barbour, M.G., 1969. Patterns of genetic similarity between Larrea divaricata of North and South America. Am. Midl. Nat., 81: 54-67. DOI:10.2307/2423651 |
Bennett, M.D., 1987. Variation in genomic form in plants and its ecological implications. New Phytol., 106: 177-200. |
Bottini, M., Greizerstein, E., Aulicino, M.B., et al., 2000. Relationships among genome size, environmental conditions and geographical distribution in natural populations of NW Patagonian species of Berberis L.(Berberidaceae). Ann. Bot., 86: 565-573. DOI:10.1006/anbo.2000.1218 |
Bromham, L., Hua, X., Lanfear, R., et al., 2015. Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am. Nat., 185: 507-524. DOI:10.1086/680052 |
Burnham, R.J., Graham, A., 1999. The history of neotropical vegetation: new developments and status. Ann. Mo. Bot. Gard., 86: 546-589. DOI:10.2307/2666185 |
Comai, L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet., 6: 836-846. DOI:10.1038/nrg1711 |
Davies, T.J., Savolainen, V., Chase, M.W., et al., 2004. Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. Lond. B Biol. Sci., 271: 2195-2200. DOI:10.1098/rspb.2004.2849 |
De Bodt, S., Maere, S., Van de Peer, Y., 2005. Genome duplication and the origin of angiosperms. Trends Ecol. Evol., 20: 591-597. DOI:10.1016/j.tree.2005.07.008 |
Doležel, J., Greilhuber, J., Suda, J., 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc., 2: 2233. DOI:10.1038/nprot.2007.310 |
Ezcurra, E., Montana, C., Arizaga, S., 1991. Architecture, light interception, and distribution of Larrea species in the Monte Desert, Argentina. Ecology, 72: 23-34. DOI:10.2307/1938899 |
Flenley, J.R., 2011. Ultraviolet insolation and the tropical rainforest: altitudinal variations, Quaternary and recent change, extinctions, and the evolution of biodiversity. In: Bush, M., Flenley, J., Gosling, W. (Eds.), Tropical Rainforest Responses to Climatic Change. Springer Praxis Books. Springer, Berlin, Heidelberg, pp. 241e258.
|
Goldie, X., Gillman, L., Crisp, M., et al., 2010. Evolutionary speed limited by water in arid Australia. Proc. R. Soc. Lond. B Biol. Sci., 277: 2645-2653. DOI:10.1098/rspb.2010.0439 |
Grime, J., Mowforth, M., 1982. Variation in genome size—an ecological interpretation. Nature, 299: 151-153. DOI:10.1038/299151a0 |
Hamann, A., Wang, T., Spittlehouse, D.L., et al., 2013. A comprehensive, high-resolution database of historical and projected climate surfaces for western North America. Bull. Am. Meteorol. Soc., 94: 1307-1309. DOI:10.1175/BAMS-D-12-00145.1 |
Hawkins, B.A., Field, R., Cornell, H.V., et al., 2003. Energy, water, and broad-scale geographic patterns of species richness. Ecology, 84: 3105-3117. DOI:10.1890/03-8006 |
Hijmans, R., Cameron, S., Parra, J., et al., 2005. WorldClim. University of California, Berkeley. Version 1.3.
|
Hunziker, J., Palacios, R., De Valesi, A.G., et al., 1972. Species disjunctions in Larrea: evidence from morphology, cytogenetics, phenolic compounds, and seed albumins. Ann. Mo. Bot. Gard., 59: 224-233. DOI:10.2307/2394755 |
Hunziker, J.H., Comas, C., 2002. Larrea interspecific hybrids revisited (Zygophyllaceae). Darwiniana, 40: 33-38. |
Landis, J.B., Soltis, D.E., Li, Z., et al., 2018. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot., 105: 348-363. DOI:10.1002/ajb2.1060 |
Laport, R.G., Hatem, L., Minckley, R.L., et al., 2013. Ecological niche modeling implicates climatic adaptation, competitive exclusion, and niche conservatism among Larrea tridentata cytotypes in North American deserts. J. Torrey Bot. Soc., 140: 349-363. DOI:10.3159/TORREY-D-13-00009.1 |
Laport, R.G., Minckley, R.L., Ramsey, J., 2012. Phylogeny and cytogeography of the North American creosote bush (Larrea tridentata, Zygophyllaceae). Syst. Bot., 37: 153-164. DOI:10.1600/036364412X616738 |
Laport, R.G., Ng, J., 2017. Out of one, many: the biodiversity considerations of polyploidy. Am. J. Bot., 104: 1119-1121. DOI:10.3732/ajb.1700190 |
Laport, R.G., Ramsey, J., 2015. Morphometric analysis of the North American creosote bush (Larrea tridentata, Zygophyllaceae) and the microspatial distribution of its chromosome races. Plant Syst. Evol., 301: 1581-1599. DOI:10.1007/s00606-014-1179-5 |
Leong-Škorničková, J., Šída, O., Jarolímová, V., et al., 2007. Chromosome numbers and genome size variation in Indian species of Curcuma (Zingiberaceae). Ann. Bot., 100: 505-526. DOI:10.1093/aob/mcm144 |
Lia, V.V., Confalonieri, V.A., Comas, C.I., et al., 2001. Molecular phylogeny of Larrea and its allies (Zygophyllaceae): reticulate evolution and the probable time of creosote bush arrival to North America. Mol. Phylogenet. Evol., 21: 309-320. DOI:10.1006/mpev.2001.1025 |
Maceira, N.O., Jacquard, P., Lumaret, R., 1993. Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytol., 124: 321-328. DOI:10.1111/j.1469-8137.1993.tb03822.x |
Ohri, D., Kumar, A., Pal, M., 1986. Correlations between 2C DNA values and habit in Cassia (Leguminosae: Caesalpinioideae). Plant Syst. Evol., 153: 223-227. DOI:10.1007/BF00983689 |
Paruelo, J.M., Beltrán, A., Jobbágy, E., et al., 1998. The climate of Patagonia: general patterns and controls on biotic processes. Ecol. Austral, 8: 85-101. |
Petit, C., Thompson, J.D., 1999. Species diversity and ecological range in relation to ploidy level in the flora of the Pyrenees. Evol. Ecol., 13: 45-65. DOI:10.1023/A:1006534130327 |
Poggio, L., Naranjo, C., 1990. Contenido de ADN y evolucion en plantas superiores, vol. 5. Academia Nacional Ciencias Exactas Fisicas y Naturales, Buenos Aires, Argentina, Monografia, pp. 27e37
|
Poggio, L., Realini, M.F., Fourastié, M.F., et al., Genome downsizing and karyotype constancy in diploid and polyploid congeners: a model of genome size variation. AoB Plants. |
Poggio, L., Rosato, M., Chiavarino, A.M., et al., 1998. Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Ann. Bot., 82: 107-115. DOI:10.1006/anbo.1998.0757 |
Price, H.J., 1988. DNA content variation among higher plants. Ann. Mo. Bot. Gard., 75: 1248-1257. DOI:10.2307/2399283 |
Raven, P.H., 1972. Plant species disjunctions: a summary. Ann. Mo. Bot. Gard., 59: 234-246. DOI:10.2307/2394756 |
Rice, A., Šmarda, P., Novosolov, M., et al., 2019. The global biogeography of polyploid plants. Nat. Ecol. Evol., 3: 265-273. DOI:10.1038/s41559-018-0787-9 |
Rohde, K., 1992. Latitudinal gradients in species diversity: the search for the primary cause. Oikos, 65: 514-527. DOI:10.2307/3545569 |
Roig, F., Roig-Juñent, S., Corbalán, V., 2009. Biogeography of the Monte Desert. J. Arid Environ., 73: 164-172. DOI:10.1016/j.jaridenv.2008.07.016 |
Schönswetter, P., Suda, J., Popp, M., et al., 2007. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Mol. Phylogenet. Evol., 42: 92-103. DOI:10.1016/j.ympev.2006.06.016 |
Severns, P.M., Liston, A., 2008. Intraspecific chromosome number variation: a neglected threat to the conservation of rare plants. Conserv. Biol., 22: 1641-1647. DOI:10.1111/j.1523-1739.2008.01058.x |
Soltis, D.E., Albert, V.A., Leebens Mack, J., et al., 2009. Polyploidy and angiosperm diversification. Am. J. Bot., 96: 336-348. DOI:10.3732/ajb.0800079 |
Soltis, D.E., Buggs, R.J., Doyle, J.J., et al., 2010. What we still don't know about polyploidy. Taxon, 59: 1387-1403. DOI:10.1002/tax.595006 |
Soltis, D.E., Soltis, P.S., Schemske, D.W., et al., 2007. Autopolyploidy in angiosperms: have we grossly underestimated the number of species?. Taxon, 56: 13-30. DOI:10.2307/25065732 |
Soltis, P.S., Soltis, D.E., 2000. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. U.S.A., 97: 7051-7057. DOI:10.1073/pnas.97.13.7051 |
Soltis, P.S., Soltis, D.E., 2016. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol., 30: 159-165. DOI:10.1016/j.pbi.2016.03.015 |
Stebbins, G.L., 1985. Polyploidy, hybridization, and the invasion of new habitats. Ann. Mo. Bot. Gard., 72: 824-832. DOI:10.2307/2399224 |
Tank, D.C., Eastman, J.M., Pennell, M.W., et al., 2015. Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol., 207: 454-467. DOI:10.1111/nph.13491 |
Te Beest, M., Le Roux, J.J., Richardson, D.M., et al., 2012. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot., 109: 19-45. DOI:10.1093/aob/mcr277 |
Temsch, E.M., Temsch, W., Ehrendorfer-Schratt, L., et al., 2010. Heavy metal pollution, selection, and genome size: the species of the Žerjav study revisited with flow cytometry. J. Bot.: 1-11. DOI:10.1155/2010/596542 |
Wang, T., Hamann, A., Spittlehouse, D., et al., 2016. Locally downscaled and spatially customizable climate data for historical and future periods for North America. PLoS One, 11: e0156720. DOI:10.1371/journal.pone.0156720 |
Willis, K.J., Bennett, K.D., Birks, H.J.B., 2009. Variability in thermal and UV-B energy fluxes through time and their influence on plant diversity and speciation. J. Biogeogr., 36: 1630-1644. DOI:10.1111/j.1365-2699.2009.02102.x |
Yang, T.W., 1970. Major chromosome races of Larrea divaricata in North America. J. Ariz. Acad. Sci., 6: 41-45. DOI:10.2307/40022846 |
Zhang, K., Wang, X., Cheng, F., 2019. Plant polyploidy: origin, evolution, and its influence on crop domestication. Hortic. Plant J., 5: 231-239. DOI:10.1016/j.hpj.2019.11.003 |



