b. Center of Plant Ecology, Core Botanical Gardens, Chinese Academy of Sciences, Mengla, Yunnan 666303, China;
c. Department of Biology, Duke University, Box 90338, Durham, NC 27708, USA;
d. University of Chinese Academy of Sciences, Beijing 100049, China
Polyploidy, or whole-genome duplication (WGD), contributes substantially to speciation and genomic evolution (Otto, 2007; Van de Peer et al., 2017). However, due to high extinction rates (Mayrose et al., 2011), a reduced efficacy of selection (Stebbins, 1971), or the possibility of returning to a diploidized state (Li et al., 2021), few polyploids succeed during evolution (Arrigo and Barker, 2012; Soltis et al., 2014; Levin, 2019). Accordingly, it is not clear how polyploids persist in nature (Fawcett and Van de Peer, 2010; Madlung, 2013). Polyploid success may be evaluated by the longevity, diversification, or adaptation of derived lineages (Fig. 1; Otto, 2007; Hegarty and Hiscock, 2008; Fawcett and Van de Peer, 2010; te Beest et al., 2012; Van de Peer et al., 2017). Success based on longevity emphasizes the survival of polyploidy-derived lineages over tens of millions of years (Fawcett and Van de Peer, 2010). Ancient waves of WGD events during the history of plant evolution highlight the success of polyploid lineages from this perspective (Zhang et al., 2020). Success based on diversification reflects the ability of polyploid lineages to diversify at the species level or above (Stebbins, 1971; Otto, 2007; Kellogg, 2016). Recent work has revealed that polyploidy may promote the diversification of plant lineages, such as Allium or Brassicaceae (Han et al., 2020; Román-Palacios et al., 2020). Finally, success based on adaptation (referred to here as the polyploid adaptation hypothesis) asserts that successful polyploids are capable of consistently colonizing or adapting to new environments after speciation (Hegarty and Hiscock, 2008; te Beest et al., 2012; Van de Peer et al., 2017), irrespective of a short-term advantage in initial establishment. Surprisingly, there is still little evidence for this hypothesis (but see Ramsey, 2011; Selmecki et al., 2015; Wu et al., 2020). The polyploid adaptation hypothesis can be tested at the population level or on contemporary timescales, making it feasible to detect the microevolutionary impacts of polyploid success.

|
| Fig. 1 Overview of hypotheses for evaluating polyploid success. There are three hypotheses for evaluating polyploid success: (1) the diversification hypothesis, in which polyploids show a high rate of species diversification (e.g., the number of species per time interval); (2) the longevity hypothesis, which predicts that polyploids persist for long periods; and (3) the adaptation hypothesis, characterized by significant niche divergence from diploid progenitors or ancestral polyploid population. Species are indicated by circles along time scales from ancestral t1 to recent t2, with the pie size proportional to ploidy level (where smaller pies represent diploidy and larger pies represent polyploidy); the yellow star indicates a whole-genome duplication event. Colors indicate different polyploid species, with shading indicating their corresponding niches. |
Multiple factors may contribute to adaptation in polyploids, including the initial genomic changes mediated by the speciation mode (e.g., allo- or autopolyploid: polyploidization with or without hybridization) or changes in key traits, ecological amplitudes, or genomic plasticity (Soltis and Soltis, 2000; Comai, 2005; Leitch and Leitch, 2008; te Beest et al., 2012). For example, allopolyploids may attain initial success by fixed-heterozygosity or preadapted ecological tolerances inherited from progenitors (Barker et al., 2016). To achieve establishment and avoid extinction induced by minority cytotype exclusion, polyploids are likely to be asexual, selfing, or perennial (Van Drunen and Husband, 2019; Spoelhof et al., 2020), or to show rapid ecological differentiation (Baniaga et al., 2020; Huynh et al., 2020). However, the speciation mode or associated phenotypic changes may only result in the establishment of a nascent polyploid population (Bomblies, 2020). Genomic modifications, such as unidirectional introgression from diploids during secondary contact (Han et al., 2015; Arnold et al., 2016), are more likely to improve fitness in local populations (Schmickl and Yant, 2021), without influencing allopatric populations. Therefore, to evaluate the evolutionary fates of polyploids, it is important to quantify medium- to long-term adaptation (Van de Peer et al., 2017), either across populations or in broader natural contexts.
Polyploids experiencing extreme climates may provide ideal opportunities to test their evolutionary fates, because the advantages of polyploidy might be only revealed under such environmental stimuli (Van de Peer et al., 2021), for example, extreme salt stress (Chao et al., 2013) or cold and dark conditions (Wu et al., 2020). Polyploids may also benefit from reduced competition in habitats with lower species diversity (Rice et al., 2019), such as denuded or barren areas created by Pleistocene glaciation in high-altitude temperate mountains (Wen et al., 2014).
Located in southwest China, the biodiversity hotspot of the Hengduan Mountains (HDM) provides a natural laboratory for studying the evolution of polyploids (Nie et al., 2005; Wang et al., 2017). The two phytogeographical areas in the HDM (southern and northern) show distinct differences in environments and species assemblages (Zhang et al., 2009). The terrain and vegetation in the southern HDM are more diverse than those in the north (An et al., 2011; Zhao et al., 2020), making the southern HDM a regional biodiversity center or refugium (Yu et al., 2019; Zhang et al., 2021).
Here, we tested the polyploid adaptation hypothesis using natural populations of two polyploid yellowcresses in the HDM, the endemic Rorippa elata and the widespread Rorippa palustris, by speciation modeling, comparative phylogeography, and a transplant experiment. Under the polyploid adaptation hypothesis, we expect to observe adaptation to new environment during the colonization of the HDM. R. elata and R. palustris represent a tractable experimental system to examine adaptive responses of polyploids during colonization in the HDM. First, R. elata and R. palustris are the only two Rorippa species distributed in both parts of the HDM (Al-Shehbaz, 2016). Second, both species are polyploids that have successfully colonized altitudinal ranges from 2000 to 4600 m, significantly higher than the ranges of other Rorippa species in Eurasia (Fig. S1). Third, R. elata plants have evolved a set of putative adaptive traits for high altitudes (Figs. 2 and S2), such as an enlarged plant body, showy flowers, high germination rates, perenniality, clonality, and a mixed mating system (Hu et al., 2021). By contrast, R. palustris plants are relatively weedier, with annual and short-lived life-histories, self-compatibility, and plentiful fruits or seeds (Jonsell, 1968; Les, 2018). In this study, we aim to decipher the polyploid speciation mode and phylogeographic history for R. elata and R. palustris, and then evaluate whether the two polyploids spread via adaptation in the HDM. Our study will better our understanding of the contribution of adaptation to polyploid success.
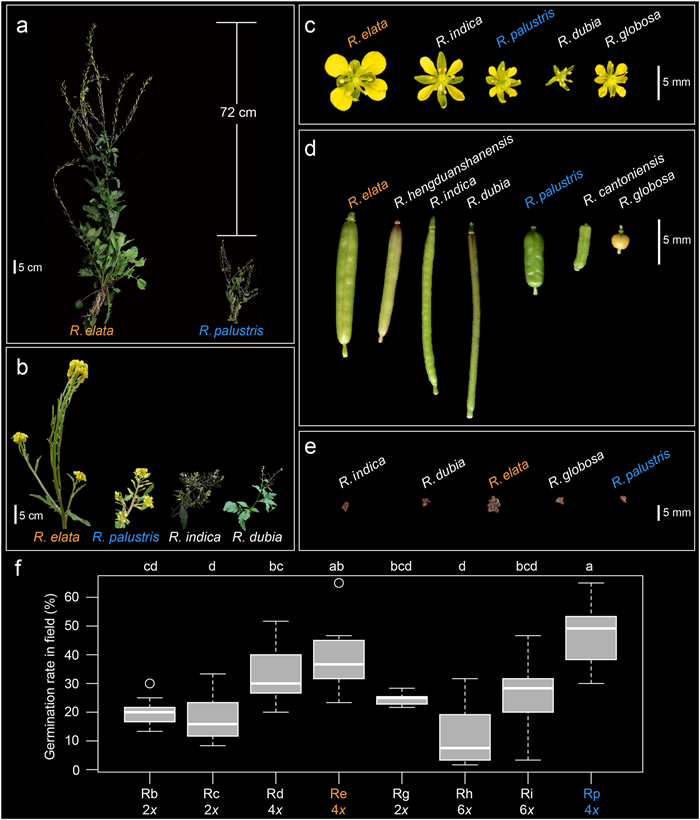
|
| Fig. 2 Trait variation among Rorippa species in East Asia. (a) Whole plants of sympatric Rorippa elata and R. palustris, with difference in plant height (72 cm) shown. Comparisons of Rorippa species inflorescence (b), flower (c), fruit (d), 10-seeds (e) and germination rates (f). R. elata is labeled in orange. R. palustris is labeled in blue. Scale bars in (a) and (b) are shown as 5 cm; in (c), (d) and (e) scale bars are shown as 5 mm. Photos of (a) and (b) were taken from field. Photos of (c), (d) and (e) were taken from the growth chamber. Germination rates, which were collected from SABG common garden, are indicated as box-and-whisker plots with the median (horizontal line), 25th and 75th percentiles (bottom and top of the box), and limits of the 95% confidence intervals (lower and upper whiskers). Letters above boxes indicate results of multiple comparisons based on least significance difference post hoc test, and ploidy levels (revealed by the number before the basic chromosome number x) are shown below species abbreviations (Rb, R. barbareifolia; Rc, R. cantoniensis; Rd, R. dubia; Re, R. elata; Rg, R. globosa; Rh, R. hengduanshanensis; Ri, R. indica; Rp, R. palustris). |
Fieldwork was performed in southwest China in Yunnan, Sichuan, Qinghai, Gansu, and Tibet (Xizang) between 2017 and 2019. A total of 346 Rorippa elata individuals were sampled from 46 populations, covering most of the native range (Table S1; Fig. S3). For each population, we sampled 4–17 individuals (mean = 7.5) that were separated from each other by at least 10 m. In addition, samples were analyzed from 46 populations of R. palustris plus related Rorippa species (R. globosa, R. dubia, R. indica, and the newly identified R. hengduanshanensis (Zheng et al., 2021)), as well as Nasturtium officinale and Barbarea intermedia as outgroups (Table S1; Fig. S3). Given the selfing character of R. palustris (Jonsell, 1968), only one individual was sampled from most populations. Seeds or healthy leaves were collected from each sampled individual. In total, 27 and 18 populations of R. elata, and 21 and 17 populations of R. palustris were located in the southern and northern HDM, respectively.
To investigate the polyploid speciation histories of Rorippa elata and R. palustris, 10 samples from seven Rorippa diploids (2n = 2x = 16), covering nearly all continents were selected as subgenomic representatives (Bleeker et al., 2002; Table S1), including Rorippa austriaca (2 samples), R. cantoniensis (1), R. globosa (1), and R. islandica (2) from Eurasia; and R. curvisiliqua (1), R. sessiliflora (1), and R. sinuata (2) from North America. Two samples of Nasturtium officinale were used as outgroups.
2.2. Flow cytometryGenome sizes of germinated seedling samples were estimated by flow cytometry (FCM) as described previously (Doležel et al., 2007), using a BD FACSVerse Flow Cytometer (USA) with Capsella rubella (haploid genome size = 219 Mb) as an internal standard (Slotte et al., 2013). For each sample, at least 5000 nuclei were collected, with a CV threshold for gated FCM peaks of < 5%. The ploidy level was calibrated using the ratio of genome sizes between the tested sample and diploid Rorippa islandica (2n = 2x = 16; haploid genome size = 245 Mb) (Koenig and Weigel, 2015). Chromosome numbers for representative samples with small, middle, or large genome sizes were counted according to the method of Han et al. (2015). In total, the genome sizes of 186 R. elata samples from 92 populations and 62 R. palustris samples from 38 populations were estimated by FCM (Fig. S4).
2.3. Sequence dataDNA was extracted using a modified CTAB method (Doyle and Doyle, 1987) from either fresh leaves of seedlings germinated in a growth chamber or silica gel-dried materials obtained through fieldwork. For phylogeographic reconstruction, four of ten plastid markers, psbC-trnS, trnG-trnM, trnL, and trnL-trnF, were tested and filtered for amplification efficiency and nucleotide polymorphism across 72 randomly selected Rorippa elata samples. In addition, one ITS marker was included. PCR primers were designed based on previous work (White et al., 1990; Taberlet et al., 1991; Stanford et al., 2000; Nakayama et al., 2014) (Table S2). PCR products were sequenced directly using an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA).
To infer the speciation mode, we cloned and sequenced 12 low-copy nuclear loci among 14 candidates, including ACO1, CHS, CIP7, CRD1, FTSZ1-1, HY4, MCM5, MLH1, SMC2, SYP61, AT3G50910, and AT5G52810 (Table S2). PCR primers and procedures were designed and performed according to previous work (Koch et al., 2000; Stockenhuber et al., 2015; Cai and Ma, 2016). PCR products were cloned into the pEASY-T1 Cloning Vector System (TransGen, Beijing, China) and proceeded to Sanger sequencing. Sequences for 8–16 clones were collected for each locus and sample. For error-free sequences, raw clonal sequencing data were processed, and haplotypes or genotypes were then inferred using PURC and Fluidigm2PURC scripts in Python v.2.7 (Rothfels et al., 2017; Blischak et al., 2018). A total of 12 regimes with different clustering thresholds or UCHIME settings were implemented in two sets (Fig. S5). Nucleotide sequences were aligned and edited using Geneious Prime v.2019.0.3 (Kearse et al., 2012).
The sequences can be accessed by GenBank IDs: MW031315-MW031737, and MW120371-MW122062.
2.4. Speciation modelingPhylogenetic methods were used to infer the speciation mode (autopolyploidy vs. allopolyploidy) for Rorippa elata and R. palustris according to a proposed pipeline for polyploid phylogenetic studies (Rothfels, 2021). First, single-gene trees for these two species and seven diploid progenitors were constructed for all 12 nuclear loci using BEAST v.1.8.4 (Drummond et al., 2012), as described previously (Han et al., 2020). Based on the topologies of these single-gene trees, subgenomic origins of PURC-inferred homoeologs in R. elata or R. palustris were classified as g1 and g2 for each locus, according to their phylogenetic positions within the backbone built for seven diploid Rorippa species (Fig. S6). Second, a concatenated sequence matrix was generated for each species based on the classified homoeologs and was used to construct a Bayesian species tree in BEAST v.1.8.4. Third, using the gene-tree reconciliation algorithm implemented in GRAMPA (Thomas et al., 2017), speciation modes for polyploid R. elata or R. palustris were distinguished (e.g., autopolyploidy, allopolyploidy, or no polyploidy). The species tree was used to generate two multi-labeled trees (MUL-trees) with g1 or g2 tips dropped. Based on each single-gene tree, 1000 new gene trees were generated with simulated gene gains or losses in JPrIME (https://github.com/arvestad/jprime). A matrix of 12, 000 simulated gene trees was used in the two sets of GRAMPA runs for g1 or g2. The MUL-tree with the minimum parsimony score was selected as the best-fitted species tree. The speciation mode was identified according to the topology of the selected MUL-tree.
2.5. Phylogeography and ecological niche modelingTo evaluate evolutionary relationships among Rorippa haplotypes, both reticulate and bifurcating trees were generated. Haplotypes of concatenated plastid sequences or ITS genotypes were generated separately using DnaSP v.6.10.04 (Rozas et al., 2017), with consideration of indels. The geographic distribution of plastid haplotypes or ITS genotypes was checked for the 72 R. elata samples used during marker screening tests (Fig. S7). A dated phylogenetic tree was generated for Rorippa plastid haplotypes and outgroups (Barbarea intermedia and Nasturtium officinale in the tribe Cardamineae) using BEAST v.1.8.4 (Drummond et al., 2012), with secondary calibration at the crown of the tribe Cardamineae according to the 95% highest posterior density (95% HPD: 10.2–18.6 Myr, million years), based on the previous estimation in Guo et al. (2017). Additionally, a phylogenetic network was constructed using TCS v.1.21 and visualized using tcsBU (Clement et al., 2000; Santos et al., 2015). The maximum connection unit for TCS inference was defined as the 95% limit (= 20 mutation steps), and indels were treated as the fifth state. Estimators of genetic diversity, including the haplotypic diversity Hd and the nucleotide diversity π, were evaluated for sites without indels using DnaSP v.6.10.04.
To evaluate the geographic pattern of haplotypic variation, the distribution and frequency of haplotypes were mapped for each population. The ancestral geographic ranges were reconstructed along the dated BEAST tree without outgroups using BioGeoBEARS v.1.1.2 implemented in R v.4.0.2 (Matzke, 2018). The best-fitted biogeographic model was tested under six scenarios (Table S3). Two bioregions, southern HDM (S) and northern HDM (N), were assigned to haplotypes according to the Flora of the Pan-Himalaya (Al-Shehbaz, 2016).
Bioclimatic variables obtained from WorldClim (Fick and Hijmans, 2017) and Oscillayers (Gamisch, 2019) were used to model potential ecological niches of R. elata and R. palustris using MaxEnt v.3.4.3 (Phillips et al., 2006). Five bioclimatic variables with low correlations (absolute Pearson's correlation coefficient R < 0.70; Fig. S8) within each species were selected, including mean diurnal temperature range (bio2), isothermality (bio3), mean temperature of coldest quarter (bio11), precipitation of wettest month (bio13), and precipitation seasonality (bio15). Based on multiple field collections, 119 and 102 geo-referenced occurrence records (pairwise geographic distance > 300 m) for R. elata and the HDM R. palustris were used, respectively (Table S4). The ecological niches were first modeled across three periods, the last interglacial (LIG: 0.14–0.12 Ma, million years ago), the last glacial maxima (LGM: 0.021–0.018 Ma), and the present, with bioclimatic data obtained from WorldClim, at a spatial resolution of 30 arc-seconds. To accurately reflect niche evolution of R. elata and R. palustris across historical periods in the HDM, ecological niche modeling (ENM) was then performed using bioclimatic data extracted from Oscillayers, ranging from 0.46 to 0.01 Ma with 10 Kyr (thousand years) intervals, at a spatial resolution of 2.5 arc-minutes. For each ENM panel per species per time period, the range size was estimated as the number of grids at different probabilities of modeled niches (e.g., ≥0.5, medium; ≥ 0.7, high; and ≥0.9, full) using DIVA-GIS v.7.5.0.0 (Hijmans et al., 2012). The latitudinal borders were recorded at the southern margin (SM) or northern margin (NM) for each species across the HDM.
2.6. Common garden experimentA field common garden was constructed in Shangri-La, Yunnan, a potential site of glacial refugium for Rorippa elata. The site was located in the Shangri-La Alpine Botanical Garden (SABG), at an latitude of 27.908°N, longitude of 99.64°E, and altitude of 3304 m (Hu et al., 2021).
For Rorippa elata, 633 seedlings were germinated from seeds of 150 accessions and 77 populations. They were then transplanted into the SABG on April 29, 2019, after 8 weeks of propagation in a growth chamber under long-day (LD) conditions (16 hours light and 8 hours dark) at 20 ℃ and 60% humidity. Plants were randomized into 5 × 5 plots (75 × 75 cm2) using custom R code (https://github.com/Ting-Shen/Rorippa-polyploids-in-HDM), with 15 cm as the interval between individuals. At the end of August 2019, approximately 91% of plants (576) survived, excluding 17 living individuals that were virtually destroyed by rodents in one plot. The main data sets with enough replicates (≥10) per population were collected from 29 populations across the HDM in 2020 (with no observed rodent activity). In each population, the mean number of individuals was 13.9, ranging from 10 to 22 plants per population. Several potential fitness-related traits (FRTs) were measured in 2020 (Table S5), including the survival rate, flowering time, number and length of primary sprouts or secondary inflorescences, total number of fruits, fruit length, and number of ovules per fruit.
All traits were categorized into three groups: survival, plant architecture, and reproduction (Table S5). First, two survival rates (SR) were recorded, the overwinter survival rate based on the survival or mortality of plants in May 2020 (SR_01) and the juvenile survival rate at the end of the growing season, collected in late August of 2020 (SR_02). Second, multiple traits related to plant architecture were measured at the end of the growing season, including the total number (TN_), mean length (ML_), or mean basal diameter (MD_) of primary sprouts (PS) or fruiting primary sprouts (PSF) and secondary inflorescences (SI). Plant height (PH) was quantified based on the longest primary sprout. Third, reproductive characters were recorded, including the flowering time (FT), total number of fruits (TN_F), mean fruit length (ML_F), and mean number of ovules per fruit (MN_O). In particular, for MN_O, about 2160 fruits were counted for 44 populations in the common garden, with an average of 50 fruits (30–105 fruits) per population. Fitness for each accession was estimated based on the rate of survival to reproduction age and the total number of seeds produced, including three components (SR_02) × (TN_F) × (MN_O).
To test whether local Rorippa polyploid species (R. elata and R. palustris) were better adapted to the environment of the HDM, we quantified germination rates for R. elata, R. palustris, and other lowland diploids or polyploids in the SABG common garden. Eight Rorippa species were sampled, including three diploids, Rorippa barbareifolia, R. cantoniensis, and R. globosa; three tetraploids, R. dubia, R. elata, and R. palustris; and two hexaploids, R. hengduanshanensis and R. indica. For each species, 10 accessions (except seven for R. globosa and nine for R. indica) were selected from collections in or around the HDM, with six replicates for each accession and 10 seeds for each replicate. All replicates were randomized into 5 × 10 plots. Seeds were stratified in 0.1% agar and 3.3 mg L−1 gibberellin in the dark at 4 ℃ for 10 days and then sowed into soil (nutrient soil to vermiculite = 1:3) directly. The number of germinated seedlings in each replicate plot were counted at 18 days after sowing (18DAS).
2.7. Growth chamber experimentWe investigated flowering responses to vernalization treatment for 27 randomly selected Rorippa palustris accessions from the HDM. Because R. elata cannot flower even under long-term vernalization treatment (e.g., 2 months), we did not include these plants in the growth chamber experiment. Seeds were surface-sterilized and germinated on half-strength MS media and then stratified in the dark at 4 ℃ for 7 days. After 10 days of germination under LD conditions at 20 ℃, seedlings were transplanted into soil (nutrient soil to vermiculite = 1:3) and grown in the growth chamber with LD conditions, at 20 ℃ and 60% humidity. For the vernalization treatment, 7-day-old seedlings were moved to 4 ℃ and short-day (SD) conditions (8 hours light and 16 hours dark) for 4 weeks and then grown under 20 ℃ and LD conditions. Three to six biological replicates were used for each accession. Individual plants were randomized to control for position effects. Bolting time (BT) was recorded as the time to the first sprout or elongated inflorescence from rosettes. Flowering time (FT) was based on the date of the first opened flower. To evaluate vernalization sensitivity, the differences in BT or FT between non-vernalization and vernalization treatments were calculated for each accession.
2.8. Statistical analysisThe effects of source latitude or altitude or their interaction on traits, as well as the relationship between traits and geographic parameters or fitness components, were evaluated by analysis of variance (ANOVA) and a correlation analysis. Variables were treated as putative proxies for selective pressures (e.g., latitude) or variables under selection (e.g., FRTs). To test combined effects, traits belonging to the same functional category were treated as one multivariate response variable (Table S5); the joint influence of source latitude or altitude was then analyzed by a multivariate analysis of variance (MANOVA). The relationships between the single trait variable and source latitude or altitude or estimated fitness were evaluated by Pearson's correlation coefficients (R) across populations. Traits significantly related to fitness components (e.g., SR_02, TN_F, or MN_O) or fitness itself were classified as FRTs. The fitness function was quantified based on the Pearson's correlation coefficient R between the trait and fitness component, and was used to quantify how selection would act on quantitative FRTs. In addition, to test whether the above relationships (e.g., between source latitudes and FRTs) are obscured by the effects of other conditioning processes (e.g., source altitudes), coplots conditioned on additional variables were generated when exploring the focal relationship. Multiple comparisons with least square means (lsmean) were performed to test differences in germination rates among Rorippa species, with Bonferroni adjustment of P-values using the lsmeans function in the 'lsmeans' package. All statistical analyses were conducted in R v.4.0.2 (R Core Team, 2018) according to descriptions in the R Book (Crawley, 2013).
3. Results 3.1. Speciation modeThe estimated mean (±SD) genome sizes were 562.431 ± 11.219 Mb for Rorippa elata (Fig. S4; range, 541–587 Mb) and 545.750 ± 10.779 Mb (514–568 Mb) for R. palustris. In comparison with diploid R. islandica (245 Mb; 2n = 2x = 16), samples of both R. elata and R. palustris were identified as tetraploids with 2n = 4x = 32, with further verification by chromosome counting (Fig. S4).
We inferred the hybrid speciation history for tetraploid Rorippa elata and R. palustris. First, after two sets of PURC runs under six analysis regimes, the number of inferred homoeologs at each locus decreased to two across regimes (Fig. S5). Second, according to phylogenetic topologies of 12 single-gene trees (Fig. S6), one homoeolog of R. elata or R. palustris clustered with diploid R. globosa across all gene trees and the other homoeolog of R. elata diverged from all other Rorippa species, except at MLH1. The other homoeolog of R. palustris clustered with diploid R. islandica. These two kinds of homoeologs were distinguished as subgenome g1 or g2. Third, based on two GRAMPA runs, the MUL-trees with the lowest parsimony score supported a mode of allotetraploid speciation for both tetraploids (Fig. 3a-b). According to the Bayesian species tree built on concatenated sequence data, both R. elata and R. palustris showed hybrid origins within the Eurasian clade (Fig. 3c-d). Based on the phylogenetic relationships, one ancestral progenitor of R. elata or R. palustris was inferred as closely related to R. globosa and the other progenitor was identified as an unknown species, possibly extinct or unsampled for R. elata and closely related to R. islandica for R. palustris.
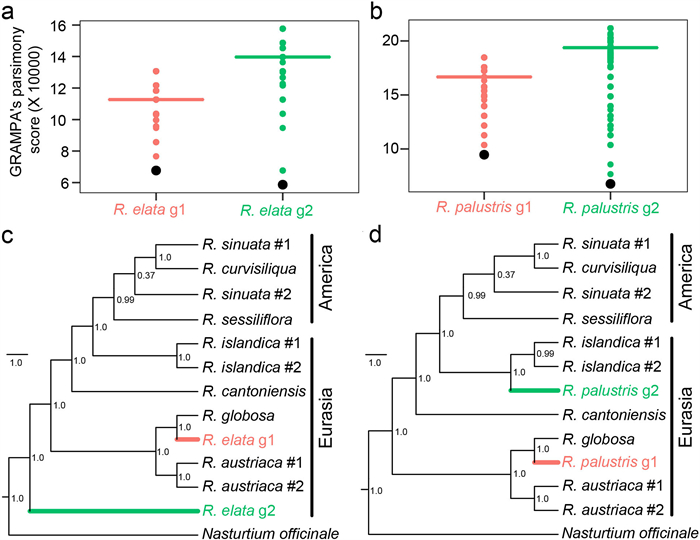
|
| Fig. 3 Speciation modeling. (a–b) Distribution of GRAMPA parsimony scores for simulated (colored dots) and observed data (black dots) for each subgenome of Rorippa elata (a) and R. palustris (b), where horizontal lines indicate the mean levels. (c–d) Multi-labeled (MUL) Bayesian trees showing the hybrid origin of R. elata (c) and HDM R. palustris (d), with outgroups and representative diploid species native to Eurasia or America. Posterior probabilities are shown near the nodes. The scale bars represent branch lengths. |
Haplotypic or genetic diversity varied among species as well as geographic regions. Based on the investigated plastid and ITS nucleotides, 17 plastid haplotypes and 14 ITS genotypes were phased for Rorippa and related plants when including indels (Figs. 4, S9 and S10). Seven plastid haplotypes and two ITS genotypes were unique to R. elata, and one plastid haplotype and one ITS genotype were unique to R. palustris in the HDM, with four plastid haplotypes (Hap_2, 3, 6, and 14) and three ITS genotypes (Gen_3, 4, and 11) shared between plants in the HDM and Himalayas (HIM; Fig. 4, S10). For populations in the HDM, R. palustris had a higher overall plastid haplotypic and nucleotide diversity (Hd = 0.257, π = 0.00080) than did R. elata (0.158 and 0.00052; Table S6). At the regional scale, populations of R. palustris from the southern HDM harbored more genetic variation (Hd = 0.325, π = 0.00131) than did populations from the northern HDM (0.166 and 0.00012). For R. elata, genetic diversity was similar in the southern and northern HDM populations (Hd = 0.162 vs. 0.155, π = 0.00054 vs. 0.00051).
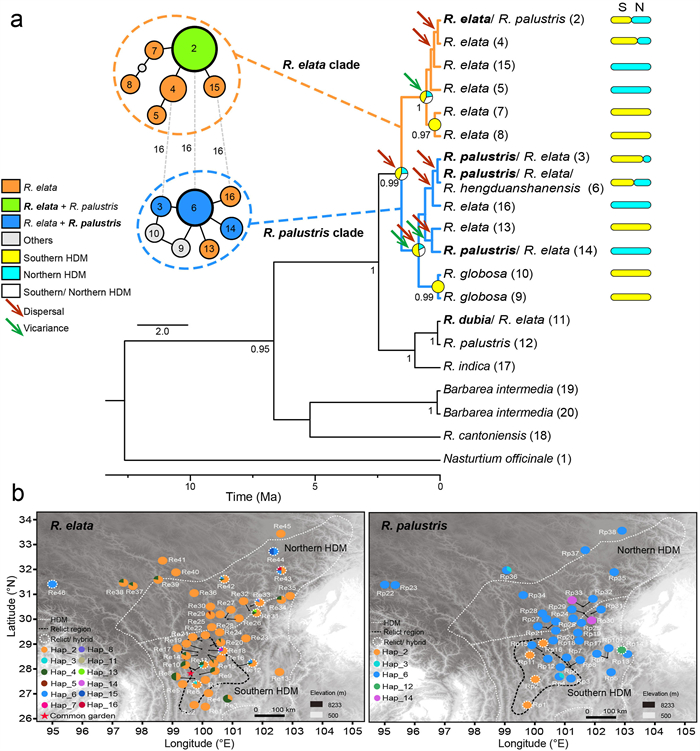
|
| Fig. 4 Haplotype network, phylogeny, and distribution of Rorippa species in the Hengduan Mountains. (a) Plastid haplotypic network and phylogeny of Rorippa species in the HDM. In the network, pie colors represent the species or clade (details in left legend), pie size is proportional to sample size, haplotype codes are labeled in each pie (grey indicates unsampled haplotypes), and mutation steps are indicated as numbers along dashed grey lines. Haplotypes with 'Others' include species other than Rorippa elata and R. palustris. In the phylogeny, values of posterior probabilities above 0.9 are labeled near branches; species names with core or dominant haplotypes are shown in bold; and haplotype codes are shown in parentheses, with a scale bar representing branch length. The distribution regions for haplotypes are colored in green (S: southern HDM), yellow (N: northern HDM), or white (southern and northern HDM), with occurring proportions shown by bar length. Colored pies at the main nodes represent the frequencies of ancestral regions inferred by BioGeoBEARS, with arrows in red showing dispersal events and in green showing vicariance events. Ma, million years ago. (b) Geographic distribution of plastid haplotypes for Rorippa elata (left) and Rorippa palustris (right). Background grey-to-black gradients indicate elevation scale (m), with legends at the bottom-right. Haplotypes are coded by different colors and numbers. Potential relict or hybrid haplotypes are labeled by dashed white circles. Dashed white lines outline the region or subregion of the HDM. Dashed black lines enclose regions accumulated by multiple relict haplotypes. The red star shows the location of the common garden in Shangri-La Alpine Botanical Garden (SABG). The leftmost populations on the maps (including Re46, Rp22, and Rp23) were collected from Tibet (Xizang); see for exact GPS information. |
Patterns of genetic variation were inferred through the construction of a haplotypic or genotypic network and phylogeny across Rorippa populations in the HDM. The plastid haplotypic network showed two main genetic clades, including one clade dominated by R. elata (named the R. elata clade) and another dominated by R. palustris or other Rorippa species (named the R. palustris clade; Fig. 4a). The core haplotype of the R. elata clade, labeled Hap_2, was shared by 84% of R. elata individuals. The core haplotype of the R. palustris clade, Hap_6, was shared by 78% of R. palustris individuals and 16 R. elata individuals. Six haplotypes of R. elata (Hap_3, 6, 11, 13, 14, and 16) and two haplotypes of R. palustris (Hap_2 and 12) were not within these main clades (estimated by pairwise p-distances; Fig. S9). These haplotypes accounted for only 6% of R. elata individuals or 17% of R. palustris individuals, indicating that they are rare haplotypes or relicts with extremely high pairwise divergence by the criteria of Alonso-Blanco et al. (2016). However, some of these haplotypes may also result from hybridization with related species, especially haplotypes shared among multiple species, such as Hap_3, 6, and 14 for R. elata or Hap_2 for R. palustris.
The two genetic clades were also well-supported (posterior = 0.99) in the dated BEAST tree (Fig. 4a). The R. elata clade and R. palustris clade originated c. 0.55 Ma (95% HPD: 1.30–0.14 Ma) and c. 0.86 Ma (1.94–0.22 Ma), respectively, with a crown age of c. 1.55 Ma (3.25–0.51 Ma). The core clade of R. elata (excluding Hap_5, 7, and 8) covered 92% of R. elata samples, with a crown age of c. 0.37 Ma (0.89–0.04 Ma).
3.4. Phylogeography and niche dynamicsAncestral geographic ranges were reconstructed for Rorippa plants in the HDM along the well-supported plastid haplotypic tree (Fig. 4a). The southern HDM was the most likely ancestral area for both the R. elata clade and R. palustris clade in a BioGeoBEARS analysis. Divergence between the R. elata and R. palustris clades was mediated by an ancient dispersal event from the southern to southern or northern HDM. Multiple dispersal (two for R. elata and four for R. palustris) or vicariance events (one for R. elata and two for R. palustris) were also identified within each clade (Fig. 4a).
The geographic distribution of haplotypic variation revealed different population structures across species or regions (Figs. 4 and S10). For R. elata, no clear phylogeographic structure was observed among populations. Plastid haplotype 2 (Hap_2) was the most frequent type across populations, with a mean frequency per population of up to 91% (Fig. 4b). Only four populations were dominated by other haplotypes (i.e., the summed percentage of other haplotypes exceeded 50% per population), including two populations located in the southern HDM (Re3 and Re8), one in the northern HDM (Re44), and one in HIM (Re46). However, populations with relictual haplotypes were unevenly distributed. Among 10 populations harboring relicts or more than three haplotypes (Mao et al., 2021), three were found in the southern HDM, particularly in a region below the 29°N latitudinal line (referred to as the relict region, hereafter) along the Muli, Jianchuan, Shangri-La, and Daocheng counties, clockwise from east to west. For R. palustris, the core haplotype (Hap_6) was mainly found in the northern HDM (Fig. 4b), whereas most of the relict haplotypes of R. palustris were located in a relict region that fully overlapped with the relict region for R. elata. Based on the above phylogeographic results, the southern HDM would be a stable ancestral area for either R. elata or the HDM R. palustris.
Temporal dynamics of suitable niche ranges were reconstructed from 0.46 Ma to the present by ENM (Figs. 5 and S11-S14). Irrespective of the probability levels of modeled niches (e.g., 0.5, 0.7, or 0.9), suitable niches for R. elata or R. palustris populations expanded northward recurrently, approximately along the main rivers during interglacial periods (e.g., 0.14–0.12 Ma, LIG), and withdrew to south of the 29°N latitudinal line during glacial periods (e.g., 0.021–0.018 Ma, LGM; Figs. 5 and S11-S14). During the past 0.46 myr, the main ranges of R. elata and R. palustris expanded northward at least six times (counted as the number of peaks above the 29°N latitudinal line), with the largest expansion around the LIG (c. 0.12 Ma) and the latest one at present (Figs. 5 and S13-S14). The ranges with high habitat suitability (≥0.9) were stably localized in the southern HDM (e.g., below the 29°N latitudinal line; Fig. S14), overlapping with areas inhabited by relict haplotypes of either R. elata or R. palustris (Figs. 4b and S10b). Geologically, niche ranges during the LGM were mainly limited by the reconstructed fallen-snowline and the expansion of glaciers in the northern HDM (Fig. S12), in which areas below the lower snowline altitudes (≤4600 m) and glacier intervals were suitable for R. elata or R. palustris plants. By contrast, present niche ranges shifted to a higher altitude snowline (≤5200 m) and were restricted to the wet glacier forefields at the northern rim of the maritime-type glaciers (type I; Fig. S12). Overall, we demonstrated a clear pattern of recurrent northward movement by populations of R. elata or R. palustris in response to glacial dynamics in the HDM.

|
| Fig. 5 Niche evolution of Rorippa elata and R. palustris in the Hengduan Mountains. (a) Temporal changes in range size estimated by the number of suitable niche grids (probability ≥70%) for R. elata (orange line) and R. palustris (blue line) in HDM. (b–c) The northern margin (NM; black line) and southern margin (SM; grey line) of latitudes for R. elata (b) or R. palustris (c) in HDM across timelines. The dashed lines in (b–c) indicate the 29°N latitudinal line. Ma, million years ago. |
Among Rorippa species in Eurasia, plants of R. elata inhabited areas with the highest altitudes (mean elevation = 3589 ± 612.096 m, P < 0.0001; Fig. S1), together with the HDM R. palustris (3192 ± 823.859 m in HDM vs. 387 ± 67.171 m in other places). Among Rorippa species found in sympatry in East Asia, tetraploids had significantly higher 18DAS germination rates (LSD test, lsmean = 3.96, df = 71) than did those of either diploids (2.02, P < 0.0001) or hexaploids (1.87, P < 0.0001; Fig. 2f) in the field common garden of SABG. Of the eight species tested, HDM R. palustris showed the highest 18DAS germination rate (lsmean = 4.73; Fig. 2f), followed by R. elata (3.90), R. dubia (3.25), R. indica (2.50), R. globosa (2.45), R. barbareifolia (2.03), R. cantoniensis (1.72), and R. hengduanshanensis (1.17). The germination rate of HDM R. palustris differed significantly from those of other species (df = 66, P ≤ 0.030) except R. elata (df = 66, P = 1.000). Accessions of R. elata had significantly higher germination rates than those of other Rorippa species (df = 66, P ≤ 0.002) except R. palustris, R. dubia, R. indica, and R. globosa (df = 66, P ≥ 0.069). The 2-year common garden experiment further revealed clinal variation in multiple FRTs in R. elata along source latitudes or altitudes (Table 1; Fig. S15).
| Category | Sub-category | Trait | Latitude | Altitude | Latitude × Altitude | |||||
| F statistic | P | F statistic | P | F statistic | P | |||||
| Survival rate | Overwinter | SR_01 | 10.532 | 0.003** | 12.601 | 0.002** | 0.069 | 0.795 | ||
| End of growing period | SR_02 | 11.738 | 0.002** | 5.049 | 0.034* | 0.009 | 0.923 | |||
| Total | – | 7.042 | 0.004** | 6.154 | 0.007** | 0.069 | 0.934 | |||
| Plant architecture | Primary sprout (PS) | PH | 5.553 | 0.027* | 0.281 | 0.601 | 0.211 | 0.651 | ||
| TN_PS | 6.619 | 0.017* | 3.127 | 0.090 | 0.022 | 0.882 | ||||
| TN_PSF | 9.696 | 0.005** | 1.727 | 0.201 | 0.025 | 0.875 | ||||
| Fre_PSF | 21.177 | 0.000*** | 2.816 | 0.106 | 1.481 | 0.235 | ||||
| MD_PS | 0.022 | 0.882 | 0.000 | 0.983 | 0.034 | 0.856 | ||||
| Total | 5.660 | 0.002** | 1.119 | 0.382 | 0.346 | 0.879 | ||||
| Secondary inflorescence (SI) | TN_SI | 0.035 | 0.853 | 1.755 | 0.199 | 1.520 | 0.231 | |||
| TN_SIF | 1.266 | 0.273 | 0.006 | 0.940 | 3.292 | 0.083 | ||||
| TL_SI | 5.042 | 0.035* | 0.580 | 0.455 | 1.738 | 0.201 | ||||
| ML_SI | 6.832 | 0.016* | 0.857 | 0.365 | 0.702 | 0.411 | ||||
| Total | 2.674 | 0.064 | 0.715 | 0.592 | 5.235 | 0.005** | ||||
| Total | – | 6.101 | 0.010* | 0.963 | 0.513 | 1.024 | 0.481 | |||
| Reproduction (Rorippa elata) | Flower | FT | 9.913 | 0.006** | 19.184 | 0.000*** | 6.555 | 0.021* | ||
| Fruit | TN_F | 0.255 | 0.621 | 0.000 | 0.994 | 0.258 | 0.618 | |||
| ML_F | 0.010 | 0.921 | 0.614 | 0.445 | 0.529 | 0.478 | ||||
| Ovule | MN_O | 4.874 | 0.042* | 0.912 | 0.354 | 0.753 | 0.398 | |||
| Total | – | 3.150 | 0.051 | 6.765 | 0.004** | 1.628 | 0.226 | |||
| Reproduction (R. palustris) | Non-vernalization | BT | 4.215 | 0.052 | 4.669 | 0.041* | 0.299 | 0.590 | ||
| FT | 8.418 | 0.008** | 2.722 | 0.113 | 1.563 | 0.224 | ||||
| Total | 6.167 | 0.007** | 2.927 | 0.075 | 2.562 | 0.100 | ||||
| Vernalization | BT | 0.885 | 0.357 | 2.752 | 0.111 | 0.181 | 0.675 | |||
| FT | 0.241 | 0.628 | 2.535 | 0.125 | 0.839 | 0.369 | ||||
| Total | 1.312 | 0.290 | 1.566 | 0.231 | 1.159 | 0.332 | ||||
| Vernalization sensitivity | – | 18.112 | < 0.001*** | 2.063 | 0.157 | 1.381 | 0.246 | |||
| *, P < 0.05; **, P < 0.01; ***, P < 0.001. Abbreviations: SR, survival rate; PH, plant height; TN, total number; PS, primary sprout; PSF, primary sprout with fruit; Fre, frequency; MD, mean diameter; SI, secondary inflorescence; SIF, secondary inflorescence with fruit; TL, total length; ML, mean length; FT, flowering time; F, fruit; MN, mean number; O, ovule; Total, treating the group of response variables as one multivariate response; BT, bolting time; FT, flowering time. -, not available. | ||||||||||
The estimated fitness varied from 279 ± 68.525 to 6922 ± 84.709 (estimated as the survival rate to reproduction age plus the total seed produced) across populations of Rorippa elata (Fig. S15), with the lowest value for a population from the southern HDM (27.853°N, 102.478°E) and the highest value for a population from the northern HDM (31.541°N, 99.978°E). Fitness function analysis suggested that plant architecture traits, such as plant height (Pearson's correlation coefficient R = 0.45, P = 0.032; Fig. S15) and the total number of primary sprouts or secondary inflorescence (e.g., TN_PS/PSF/SI; R ≥ 0.59, P ≤ 0.007), and reproductive traits, such as flowering time (R = 0.60, P = 0.006) and mean length of fruit (ML_F, R = 0.40, P = 0.037), were non-randomly related to fitness or its components. Therefore, these traits are candidate FRTs, functioning as indicators of the strength of the phenotypic response to selection.
3.5.2. SurvivalMore than 91% of Rorippa elata plants survived to the end of August 2019 in the common garden of SABG; however, only 63% resprouted in May 2020. Significant effects were detected for both source latitudes and altitudes on survival rate variation (SR_01/02; F ≥ 5.049, P ≤ 0.034; Table 1) but not for the interaction (P ≥ 0.795), particularly at the lowest (2047–3200 m, mainly located in the southern HDM) or highest levels (3560–4309 m, mainly located in the northern HDM) of the conditioned altitudinal variable (Fig. S16). The calculated mean overwinter germination rates (SR_01) per population were positively correlated with source latitudes (Fig. S15; R = 0.47, P = 0.011) but not altitudes (P = 0.96). A significant positive relationship was also observed between source latitudes and survival rates recorded in August 2020 (SR_02; R = 0.53, P = 0.003). Collectively, R. elata populations in the high-latitude northern HDM had overwintering germination rates of 68% ± 19% and juvenile survival rate of 58% ± 13%, compared with values of 55% ± 18% and 49% ± 19% for populations in the southern HDM.
3.5.3. Plant architectureVariation in primary sprout or secondary inflorescence traits of Rorippa elata differed with respect to geographic variables. We detected significant effects of source latitude on most FRTs of primary sprouts (PS, including PH, TN_PS/PSF, and Fre_PSF; F ≥ 5.553, df = 1, P ≤ 0.027) other than mean basal diameter (MD_PS; P = 0.882), and a significant interaction effect of source latitude and altitude on combined variation in secondary inflorescences (SI; F = 5.235, P = 0.005; Table 1). There were no significant effects of source altitude on primary sprout or secondary inflorescence traits or their combined multivariate trait (P ≥ 0.09; Table 1). In particular, the total number of fruiting sprouts (TN_PSF) decreased across both source latitudes and altitudes (R = −0.45, P = 0.015; R = −0.51, P = 0.004; Fig. S15). Also, plant height was negatively correlated with source latitudes and altitudes (PH; R = −0.43, P = 0.022; R = −0.38, P = 0.044; Fig. S15). No significant latitudinal or altitudinal correlations were detected with the mean basal diameter of sprouts (MD_PS; absolute value of R ≤ 0.10, P ≥ 0.610) or the total number of secondary inflorescences (TN_SI; absolute value of R ≤ 0.23, P ≥ 0.170). However, the mean length of secondary inflorescences (ML_SI) clearly increased across source latitudes and altitudes (R = 0.47, P = 0.014; R = 0.46, P = 0.019). Taken together, the plant architecture of R. elata at high latitudes or altitudes was more compact than the more diffuse growth form at low latitudes or altitudes, characterized by shorter stems, fewer sprouts, and longer branches.
3.5.4. ReproductionBoth source latitude and altitude significantly influenced flowering time variation for Rorippa elata (FT; F = 9.913, P = 0.006; F = 19.184, P < 0.001; Table 1) but not fruit number or length or ovule number per fruit. Genotypes from higher latitudes or altitudes flowered earlier than those from low latitudes or altitudes (R = −0.42, P = 0.024; R = −0.57, P = 0.001; Fig. S15). The total number of fruits or mean fruit length did not differ across source latitudes or altitudes (TN_F and ML_F; absolute value of R ≤ 0.13, P ≥ 0.410). Statistically significant negative correlations were found between the mean number of ovules per fruit and latitude or altitude (MN_O; R = −0.42, P = 0.004; R = −0.34, P = 0.025; Fig. S15). Overall, high-latitude or altitude populations of R. elata showed early flowering and produced fewer ovules compared with populations at low latitudes or altitudes.
3.5.5. Flowering timeFor Rorippa palustris, variation in both bolting time and flowering time was significantly influenced by or correlated with source latitude and altitude under non-vernalizing conditions (Table 1). The observed changes in bolting time were negatively related to altitude (R = −0.49, P = 0.009; Fig. S17). Similarly, flowering time decreased significantly from lower to higher latitudes (R = −0.49, P = 0.010) or altitudes (R = −0.45, P = 0.020). However, these patterns were not observed for plants in the vernalization treatment (P ≥ 0.100). Furthermore, the calculated vernalization sensitivity was also negatively related to source latitudes (R = −0.52, P = 0.005) or altitudes (R = −0.42, P = 0.031; Fig. S18).
4. DiscussionThe success of polyploids can be attributed to a number of non-mutually exclusive factors, including their contributions to the longevity or diversification of descendent lineages or to long-term adaptability after speciation (Fig. 1). Here, we tested the polyploid adaptation hypothesis, which predicts that successful polyploids have an advantage with respect to persistence and adaptation to new environments. We found evidence for the recurrent post-glacial colonization of HDM from the south to north by two allotetraploids, R. elata and R. palustris, with adaptive alterations in fitness-related traits across latitudinal gradients.
4.1. Hybrid origins of tetraploid Rorippa elata and Rorippa palustris in the mid-PleistocenePolyploid success is thought to be influenced by the mode of speciation, under the assumption that allopolyploids have an evolutionary advantage (Otto, 2007; Barker et al., 2016). We inferred hybrid origins for both Rorippa elata and HDM R. palustris (Figs. 3, S5 and S6) during the mid-Pleistocene around c. 0.55 Ma and c. 0.86 Ma, respectively, in the southern HDM (Fig. 4). The estimated time of origin is younger than those for other hybrid species in the HDM, such as Picea purpurea (1.30 Ma) (Sun et al., 2014) or Ostryopsis intermedia (1.80 Ma) (Wang et al., 2021). This period corresponded with the Mid–Pleistocene Transition (1.25–0.70 Ma), with ice–age cycles shifting from symmetric 41 Kyr to asymmetric 100 Kyr, as well as an increased severity of glaciations (Chalk et al., 2017; Sun et al., 2021). These climatic fluctuations resulted in stepwise alterations in habitat connectivity, which may in turn have influenced gene flow among populations and facilitated hybrid speciation (Muellner-Riehl, 2019; Rahbek et al., 2019).
A hybrid origin could facilitate the establishment of polyploids by the co-option of preadapted genes or traits from progenitors (Comai, 2005; Otto, 2007; Wu et al., 2022). For example, it has been proposed that the establishment and spread of wild allopolyploid Aegilops wheats was favored by the eco-genetic additivity of their progenitors (Huynh et al., 2020). Similarly, such eco-genetic additivity may have promoted the success of several widespread weeds, such as the allotetraploid Capsella bursa-pastoris (Han et al., 2015) and Cardamine flexuosa (Mandáková et al., 2014), as well as the allohexaploid Echinochloa crus-galli (Ye et al., 2020), thus supporting the role of allopolyploidization in plant colonization (Stebbins, 1985). Our results indicate that allopolyploidy may have facilitated the initial establishment and later success of Rorippa elata and R. palustris, contrary to previous arguments that polyploidy had a minor role in promoting species diversity in the HDM (Nie et al., 2005; Wang et al., 2017; but see Mao et al., 2021).
4.2. Northward colonization shaped by glacial dynamics in the late-PleistoceneNeopolyploids are common in deglaciated areas (Brochmann et al., 2004; Novikova et al., 2018), reflecting a biogeographical response of plants to the Quaternary climatic oscillations. Here, we found that Pleistocene glaciation shaped the phylogeography of Rorippa polyploids in the HDM, with recurrent northward expansion during interglacial periods and repeated southward withdrawals during glacial periods (Fig. 4, Fig. 5 and S11-S14).
The initial expansion of the core clade of Rorippa elata (covered 92% of R. elata samples, with the crown age of c. 0.37 Ma) followed weak summer monsoons in the HDM (c. 0.41 Ma) (An et al., 2011; Zhao et al., 2020) and was influenced by glacial dynamics (Figs. 5 and S11-S14). The larger interglacial expansions of R. elata may have been driven by the reduced precipitation and drought stresses associated with weakening summer monsoons in either the southern (c. 0.40 to 0.13 Ma, in Heqing Basin, 26.562°N, 100.171°E; An et al., 2011) or northern (c. 0.62 or 0.41 Ma to present, in Zoige Basin, 33.969°N, 102.331°E; Zhao et al., 2020) HDM. In agreement with paleoclimatic or ancient DNA evidence (Li et al., 1996; Liu et al., 2021), the niche ranges of R. elata or R. palustris were largely shaped by shifts in snowline altitudes (Fig. S12). For example, the northern niche margins for both species were limited by the snowline altitudes ranging from 4600 m during the LGM (c. 0.021 Ma) to 5200 m at present (Fig. S12), accompanied by the availability of ice-free and newly-opened lands in the northern HDM (c. 0.014 Ma, in Lake Naleng, 31.100°N, 99.750°E; Liu et al., 2021). The climatic region defined by maritime-type glaciers also may have played an important role in limiting the northern margins of R. elata and R. palustris (Fig. S12), supporting the influence of glacial dynamics in shaping the geography of alpine plants in the HDM (Sun et al., 2017).
In summary, we propose that the current distributions of Rorippa elata or R. palustris in the HDM was molded by the increasing altitude of the snowline and the expansion of glacier forefields during glacial retreats (Wallis et al., 2016; Cauvy-Fraunié and Dangles, 2019), as well as the reduced summer precipitation driven by weak monsoons since c. 0.41 Ma (An et al., 2011; Zhao et al., 2020). The denuded habitats created during the late-Pleistocene were stressful but lacking interspecific competition (Liu et al., 2021), which may have provided a chance for polyploids like R. elata or R. palustris to conquer. However, the causal relationship and the underlying adaptive basis for those inferences have yet to be clearly demonstrated (Mao et al., 2021).
4.3. Clinal adaptation across latitudinal gradientsThe stress response is an important determinant for the success of polyploidy (Van de Peer et al., 2021) in recent or geologic eras (Chao et al., 2013; Wu et al., 2020), although unambiguous data supporting this conjecture are scarce (Ramsey, 2011; but see Čertner et al., 2019). In the HDM, species richness decreases significantly from south to north (Zhang et al., 2009), corresponding with the higher prevalence of species with hybrid origins in the south (Yang et al., 2019; Wang et al., 2021). Only a few hybrids are known to have spread northwards or expanded their ranges across the HDM (Wu et al., 2022), such as populations of Rorippa elata and R. palustris (Figs. 2, S1 and S2). How these Rorippa allotetraploids have adapted better than diploids across the HDM is an intriguing question that awaits further investigation. Our results agree with the polyploid adaptation hypothesis (Fig. 1) (Hegarty and Hiscock, 2008; Van de Peer et al., 2017), which stipulates that successful Rorippa polyploids may spread via adaptive responses to the environments in the HDM.
We tested the hypothesis in Rorippa polyploids through common garden and growth chamber experiments. Seedling survival rates were higher for Rorippa polyploids than for diploids (Fig. 2), implying an evolutionary advantage under the high-altitude environments of the HDM. For R. elata and R. palustris, clinal variation in fitness-related traits was found along latitudinal and altitudinal gradients, suggesting that adaptive changes accompanied their range expansions (Table 1; Figs. S15-S18). In general, R. elata plants at high latitudes or altitudes are biennials or short-lived perennials (e.g., high overwinter resprouting rate) with early flowering, which could confer an increased tolerance to seasonal climatic changes, such as a short growing period (Molau, 1993; Folk et al., 2020). Similarly, the annual R. palustris from the northern HDM highlands showed early flowering and low vernalization sensitivity (Figs. S17 and S18), consistent with findings in the annual model plant Arabidopsis thaliana (Stinchcombe et al., 2005).
For plants in cold or dry conditions, compact architectures may be advantageous to conserve warmth, save water, or resist wind (Körner, 2020). In contrast to lowland plants with more diffuse architectures, the highland Rorippa elata evolved a relatively compact architecture, characterized by a shorter height, reduced sprout numbers, and longer branches (Table 1; Fig. S15). A trade-off between survival and reproduction was also found in R. elata populations. Along latitudes or altitudes, survival rates increased as ovule numbers decreased (Fig. S15), perhaps reflecting a general K-selection strategy for plants inhabiting cold, density-dependent environments (Gadgil and Solbrig, 1972).
Latitudinal or altitudinal gradients may covary with other climatic or geographic factors, such as diurnal amplitude, seasonal precipitation, or rate of snow melt. In addition to shaping the ecological niches of Rorippa polyploids in the HDM, these factors may modulate the sensitivity of plants to local environments undergoing climate change. Plant responses to these variables require further investigation. In addition, the correlation-based adaptive potential estimated at one site (e.g., SABG) does not fully capture the performance of populations in other places (e.g., the northern HDM). Additional work is needed to evaluate adaptation in Rorippa polyploids across their ranges in the HDM. Further studies should also examine the specific genomic regions or genes responsible for the observed adaptive changes, along with more exact demographic inferences for Rorippa polyploids in the HDM.
5. ConclusionWe found that Rorippa elata, which is endemic to the HDM, and the widely distributed R. palustris, are allotetraploids with mid-Pleistocene origins. Both species have repeatedly colonized the HDM from south to north by taking advantage of the availability of new niches created by glacial retreats. Following colonization of the northern HDM, clinal variation in several fitness-related traits was found along a latitudinal gradient. These findings for both species support the polyploid adaptation hypothesis (Fig. 1) and suggest that polyploids may have spread via adaptation in response to environmental changes during the Pleistocene (Novikova et al., 2018). In conclusion, our results link the evolutionary success of polyploids to both intrinsic (e.g., adaptive clinal responses in fitness-related traits) and extrinsic (e.g., glacial dynamics) factors during medium-to long-term evolution, contributing to our understanding of the significance of polyploidy from the perspective of adaptation.
Author contributionsT-S.H. and Y–W.X. conceived the project. T-S.H. designed the experiments, collected the data, and performed analyses. Z-Y.H. and Z-Q.D. performed the common garden work. Q-J.Z. did the cytological experiments and phylogenetic analysis. J.L. explained the ecological results. T-S.H. drafted the manuscript. T.M-O. revised the manuscript and edited the language. All authors reviewed the final manuscript.
Declaration of competing interestThere are no conflicts of interest to declare.
AcknowledgementsThis work was supported by the National Natural Science Foundation of China (31800177, 32170224, and U1802242), and the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31000000). T-S.H. is also supported by the Youth Innovation Promotion Association CAS (2020391), China Scholarship Council (201804910061), and CAS Light of West China Program. We acknowledge Ya-Long Guo at the Institute of Botany CAS for providing Capsella rubella seeds and revising the manuscript. We also thank members of the Paleoecology group, and Biogeography and Ecology group at the Xishuangbanna Tropical Botanical Garden CAS, as well as Zhendong Fang and Lin Liu at the Shangri-La Alpine Botanical Garden who helped with valuable discussions on the transplanting experiment. We thank the anonymous reviewers for helpful comments which greatly improved the manuscript.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.02.002.
Al-Shehbaz, I.A. 2016. Brassicaceae. In: Hong D-Y ed. Flora of the Pan-Himalaya. Cambridge University Press, Cambridge, UK. pp. 275-288.
|
Alonso-Blanco, C., Andrade, J., Becker, C., et al., 2016. 1, 135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell, 166: 481-491. DOI:10.1016/j.cell.2016.05.063 |
An, Z., Clemens, S.C., Shen, J., et al., 2011. Glacial-interglacial Indian summer monsoon dynamics. Science, 333: 719-723. DOI:10.1126/science.1203752 |
Arnold, B.J., Lahner, B., DaCosta, J.M., et al., 2016. Borrowed alleles and convergence in serpentine adaptation. Proc. Natl. Acad. Sci. U.S.A., 113: 8320-8325. DOI:10.1073/pnas.1600405113 |
Arrigo, N., Barker, M.S., 2012. Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol., 15: 140-146. DOI:10.1016/j.pbi.2012.03.010 |
Baniaga, A.E., Marx, H.E., Arrigo, N., et al., 2020. Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives. Ecol. Lett., 23: 68-78. DOI:10.1111/ele.13402 |
Barker, M.S., Arrigo, N., Baniaga, A.E., et al., 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytol., 210: 391-398. DOI:10.1111/nph.13698 |
Bleeker, W., Weber-Sparenberg, C., Hurka, H., 2002. Chloroplast DNA variation and biogeography in the genus Rorippa scop. (Brassicaceae). Plant Biol., 4: 104-111. DOI:10.1055/s-2002-20442 |
Blischak, P.D., Latvis, M., Morales-Briones, D.F., et al., 2018. Fluidigm2PURC: automated processing and haplotype inference for double-barcoded PCR amplicons. Appl. Plant Sci., 6: 1-6. |
Bomblies, K., 2020. When everything changes at once: finding a new normal after genome duplication. Proc. Royal Soc. B., 287: 1-14. |
Brochmann, C., Brysting, A.K., Alsos, I.G., et al., 2004. Polyploidy in arctic plants. Biol. J. Linn. Soc. Lond., 82: 521-536. DOI:10.1111/j.1095-8312.2004.00337.x |
Cai, L., Ma, H., 2016. Using nuclear genes to reconstruct angiosperm phylogeny at the species level: a case study with Brassicaceae species. J. Syst. Evol., 54: 438-452. DOI:10.1111/jse.12204 |
Cauvy-Fraunié, S., Dangles, O., 2019. A global synthesis of biodiversity responses to glacier retreat. Nat. Ecol. Evol., 3: 1675-1685. DOI:10.1038/s41559-019-1042-8 |
Čertner, M., Sudová, R., Weiser, M., et al., 2019. Ploidy-altered phenotype interacts with local environment and may enhance polyploid establishment in Knautia serpentinicola (Caprifoliaceae). New Phytol., 221: 1117-1127. DOI:10.1111/nph.15426 |
Chalk, T.B., Hain, M.P., Foster, G.L., et al., 2017. Causes of ice age intensification across the Mid-Pleistocene Transition. Proc. Natl. Acad. Sci. U.S.A., 114: 13114-13119. DOI:10.1073/pnas.1702143114 |
Chao, D. -Y., Dilkes, B., Luo, H., et al., 2013. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science, 341: 658-659. DOI:10.1126/science.1240561 |
Clement, M., Posada, D., Crandall, K.A., 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol., 9: 1657-1659. DOI:10.1046/j.1365-294x.2000.01020.x |
Comai, L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet., 6: 836-846. DOI:10.1038/nrg1711 |
Crawley, M.J. 2013. The R Book (2nd). John Wiley & Sons.
|
Doležel, J., Greilhuber, J., Suda, J., 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc., 2: 2233-2244. DOI:10.1038/nprot.2007.310 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Drummond, A.J., Suchard, M.A., Xie, D., et al., 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol., 29: 1969-1973. DOI:10.1093/molbev/mss075 |
Fawcett, J.A., Van de Peer, Y., 2010. Angiosperm polyploids and their road to evolutionary success. Trends Ecol. Evol., 2: 13-21. |
Fick, S.E., Hijmans, R.J., 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol., 37: 4302-4315. DOI:10.1002/joc.5086 |
Folk, R.A., Siniscalchi, C.M., Soltis, D.E., 2020. Angiosperms at the edge: extremity, diversity, and phylogeny. Plant Cell Environ., 43: 2871-2893. DOI:10.1111/pce.13887 |
Gadgil, M., Solbrig, O.T., 1972. The concept of r- and K-selection: evidence from wild flowers and some theoretical considerations. Am. Nat., 106: 14-31. DOI:10.1086/282748 |
Gamisch, A., 2019. Oscillayers: a dataset for the study of climatic oscillations over Plio-Pleistocene time-scales at high spatial-temporal resolution. Global Ecol. Biogeogr., 28: 1552-1560. DOI:10.1111/geb.12979 |
Guo, X., Liu, J., Hao, G., et al., 2017. Plastome phylogeny and early diversification of Brassicaceae. BMC Genom., 18: 1-9. DOI:10.1186/s12864-016-3406-7 |
Han, T. -S., Wu, Q., Hou, X. -H., et al., 2015. Frequent introgressions from diploid species contribute to the adaptation of the tetraploid Shepherd's purse (Capsella bursa-pastoris). Mol. Plant, 8: 427-438. DOI:10.1016/j.molp.2014.11.016 |
Han, T. -S., Zheng, Q. -J., Onstein, R.E., et al., 2020. Polyploidy promotes species diversification of Allium through ecological shifts. New Phytol., 225: 571-583. DOI:10.1111/nph.16098 |
Hegarty, M.J., Hiscock, S.J., 2008. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol., 18: R435-R444. DOI:10.1016/j.cub.2008.03.043 |
Hijmans, R.J., Guarino, L., Mathur, P. 2012. DIVA-GIS Version 7.5.
|
Hu, Z., Zheng, Q., Mu, Q., et al., 2021. The mating system and reproductive assurance of Rorippa elata (Brassicaceae) across latitude. Biodivers. Sci., 29: 712-721. DOI:10.17520/biods.2021056 |
Huynh, S., Broennimann, O., Guisan, A., et al., 2020. Eco-genetic additivity of diploids in allopolyploid wild wheats. Ecol. Lett., 23: 663-673. DOI:10.1111/ele.13466 |
Jonsell, B., 1968. Studies in the North-West European Species of Rorippa S. Str.. Uppsala, Sweden: Acta Universitatis Upsaliensis: 1-221. |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Kellogg, E.A., 2016. Has the connection between polyploidy and diversification actually been tested?. Curr. Opin. Plant Biol., 30: 25-32. DOI:10.1016/j.pbi.2016.01.002 |
Koch, M.A., Haubold, B., Mitchell-Olds, T., 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol., 17: 1483-1498. DOI:10.1093/oxfordjournals.molbev.a026248 |
Koenig, D., Weigel, D., 2015. Beyond the thale: comparative genomics and genetics of Arabidopsis relatives. Nat. Rev. Genet., 16: 285-298. DOI:10.1038/nrg3883 |
Korner, C. 2020. Plant adaptations to alpine environments. In: Goldstein MI, DellaSala DA eds. Encyclopedia of the World's Biomes. Elsevier, Oxford. pp. 355-361.
|
Leitch, A., Leitch, I., 2008. Genomic plasticity and the diversity of polyploid plants. Science, 320: 481-483. DOI:10.1126/science.1153585 |
Les, D.H. 2018. Core Eudicots: Dicotyledons IV: "Rosid" Tricolpates: Malvid Rosids (Eurosids II): Order 9: Brassicales: Family 2: Brassicaceae: 7. Rorippa. Aquatic Dicotyledons of North America: Ecology, Life History, and Systematics. CRC Press. pp. 398-406.
|
Levin, D.A., 2019. Why polyploid exceptionalism is not accompanied by reduced extinction rates. Plant Syst. Evol., 305: 1-11. DOI:10.1007/s00606-018-1552-x |
Li, J., Feng, Z., Zhou, S. 1996. The vestiges of Quaternary glaciation in the Hengduan Mountains region. In: Li J, Su Z eds. Glacier in the Hengduan Mountains. Science Press, Beijing, China. pp. 157-173.
|
Li, Z., McKibben, M.T.W., Finch, G.S., et al., 2021. Patterns and processes of diploidization in land plants. Annu. Rev. Plant Biol., 72: 387-410. DOI:10.1146/annurev-arplant-050718-100344 |
Liu, S., Kruse, S., Scherler, D., et al., 2021. Sedimentary ancient DNA reveals a threat of warming-induced alpine habitat loss to Tibetan Plateau plant diversity. Nat. Commun., 12: 1-9. DOI:10.1038/s41467-020-20314-w |
Madlung, A., 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity, 110: 99-104. DOI:10.1038/hdy.2012.79 |
Mandáková, T., Marhold, K., Lysak, M.A., 2014. The widespread crucifer species Cardamine flexuosa is an allotetraploid with a conserved subgenomic structure. New Phytol., 201: 982-992. DOI:10.1111/nph.12567 |
Mao, K. -S., Wang, Y., Liu, J. -Q., 2021. Evolutionary origin of species diversity on the Qinghai–Tibet plateau. J. Syst. Evol., 42: 1142-1158. DOI:10.1111/jse.12809 |
Matzke, N.J. 2018. BioGeoBEARS: BioGeography with Bayesian (And Likelihood) Evolutionary Analysis with R Scripts. Version 1.1.1.
|
Mayrose, I., Zhan, S.H., Rothfels, C.J., et al., 2011. Recently formed polyploid plants diversify at lower rates. Science, 333: 1257-1257. DOI:10.1126/science.1207205 |
Molau, U., 1993. Relationships between flowering phenology and life history strategies in tundra plants. Arctic Antarct. Alpine Res., 25: 391-402. DOI:10.2307/1551922 |
Muellner-Riehl, A.N., 2019. Mountains as evolutionary arenas: patterns, emerging approaches, paradigm shifts, and their implications for plant phylogeographic research in the Tibeto-Himalayan region. Front. Plant Sci., 10: 1-18. DOI:10.3389/fpls.2019.00001 |
Nakayama, H., Fukushima, K., Fukuda, T., et al., 2014. Molecular phylogeny determined using chloroplast DNA inferred a new phylogenetic relationship of Rorippa aquatica (Eaton) EJ Palmer & Steyermark (Brassicaceae)—lake cress. Am. J. Plant Sci., 5: 48-54. DOI:10.4236/ajps.2014.51008 |
Nie, Z. -L., Wen, J., Gu, Z. -J., et al., 2005. Polyploidy in the flora of the Hengduan Mountains hotspot, southwestern China. Ann. Mo. Bot. Gard., 92: 275-306. |
Novikova, P.Y., Hohmann, N., Van de Peer, Y., 2018. Polyploid Arabidopsis species originated around recent glaciation maxima. Curr. Opin. Plant Biol., 42: 8-15. DOI:10.1016/j.pbi.2018.01.005 |
Otto, S.P., 2007. The evolutionary consequences of polyploidy. Cell, 131: 452-462. DOI:10.1016/j.cell.2007.10.022 |
Phillips, S.J., Anderson, R.P., Schapire, R.E., 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model., 190: 231-259. DOI:10.1504/IJGENVI.2006.010156 |
R Core Team. 2018. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria.
|
Rahbek, C., Borregaard, M.K., Antonelli, A., et al., 2019. Building mountain biodiversity: geological and evolutionary processes. Science, 365: 1114-1119. DOI:10.1126/science.aax0151 |
Ramsey, J., 2011. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. U.S.A., 108: 7096-7101. DOI:10.1073/pnas.1016631108 |
Rice, A., Šmarda, P., Novosolov, M., et al., 2019. The global biogeography of polyploid plants. Nat. Ecol. Evol., 3: 265-273. DOI:10.1038/s41559-018-0787-9 |
Román-Palacios, C., Molina-Henao, Y.F., Barker, M.S., 2020. Polyploids increase overall diversity despite higher turnover than diploids in the Brassicaceae. Proc. Royal Soc. B., 287: 1-9. |
Rothfels, C.J., 2021. Polyploid phylogenetics. New Phytol., 230: 66-72. DOI:10.1111/nph.17105 |
Rothfels, C.J., Pryer, K.M., Li, F.W., 2017. Next-generation polyploid phylogenetics: rapid resolution of hybrid polyploid complexes using PacBio single-molecule sequencing. New Phytol., 213: 413-429. DOI:10.1111/nph.14111 |
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J.C., et al., 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol., 34: 3299-3302. DOI:10.1093/molbev/msx248 |
Santos, A.M., Cabezas, M.P., Tavares, A.I., et al., 2015. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics, 32: 627-628. |
Schmickl, R., Yant, L., 2021. Adaptive introgression: how polyploidy reshapes gene flow landscapes. New Phytol., 230: 457-461. DOI:10.1111/nph.17204 |
Selmecki, A.M., Maruvka, Y.E., Richmond, P.A., et al., 2015. Polyploidy can drive rapid adaptation in yeast. Nature, 519: 349-352. DOI:10.1038/nature14187 |
Slotte, T., Hazzouri, K.M., Ågren, J.A., et al., 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet., 45: 831-835. DOI:10.1038/ng.2669 |
Soltis, D.E., Segovia-Salcedo, M.C., Jordon-Thaden, I., et al., 2014. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytol., 202: 1105-1117. DOI:10.1111/nph.12756 |
Soltis, P.S., Soltis, D.E., 2000. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. U.S.A., 97: 7051-7057. DOI:10.1073/pnas.97.13.7051 |
Spoelhof, J.P., Keeffe, R., McDaniel, S.F., 2020. Does reproductive assurance explain the incidence of polyploidy in plants and animals?. New Phytol., 227: 14-21. DOI:10.1111/nph.16396 |
Stanford, A.M., Harden, R., Parks, C.R., 2000. Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS sequence data. Am. J. Bot., 87: 872-882. DOI:10.2307/2656895 |
Stebbins, G.L., 1971. Chromosomal Evolution in Higher Plants. London, UK: Edward Arnold.
|
Stebbins, G.L., 1985. Polyploidy, hybridization, and the invasion of new habitats. Ann. Mo. Bot. Gard., 72: 824-832. DOI:10.2307/2399224 |
Stinchcombe, J.R., Caicedo, A.L., Hopkins, R., et al., 2005. Vernalization sensitivity in Arabidopsis thaliana (Brassicaceae): the effects of latitude and FLC variation. Am. J. Bot., 92: 1701-1707. DOI:10.3732/ajb.92.10.1701 |
Stockenhuber, R., Zoller, S., Shimizu-Inatsugi, R., et al., 2015. Efficient detection of novel nuclear markers for Brassicaceae by transcriptome sequencing. PLoS One, 10: 1-19. |
Sun, H., Zhang, J., Deng, T., et al., 2017. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers., 39: 161-166. |
Sun, Y., Abbott, R.J., Li, L., et al., 2014. Evolutionary history of Purple cone spruce (Picea purpurea) in the Qinghai–Tibet Plateau: homoploid hybrid origin and Pleistocene expansion. Mol. Ecol., 23: 343-359. DOI:10.1111/mec.12599 |
Sun, Y., McManus, J.F., Clemens, S.C., et al., 2021. Persistent orbital influence on millennial climate variability through the Pleistocene. Nat. Geosci., 14: 812-818. DOI:10.1038/s41561-021-00794-1 |
Taberlet, P., Gielly, L., Pautou, G., et al., 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol., 17: 1105-1109. DOI:10.1007/BF00037152 |
te Beest, M., Le Roux, J.J., Richardson, D.M., et al., 2012. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot., 109: 19-45. DOI:10.1093/aob/mcr277 |
Thomas, G.W.C., Ather, S.H., Hahn, M.W., 2017. Gene-tree reconciliation with MUL-trees to resolve polyploidy events. Syst. Biol., 66: 1007-1018. DOI:10.1093/sysbio/syx044 |
Van de Peer, Y., Ashman, T.-L., Soltis, P.S., et al., 2021. Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell, 33: 11-26. DOI:10.1093/plcell/koaa015 |
Van de Peer, Y., Mizrachi, E., Marchal, K., 2017. The evolutionary significance of polyploidy. Nat. Rev. Genet., 18: 411-424. DOI:10.1038/nrg.2017.26 |
Van Drunen, W.E., Husband, B.C., 2019. Evolutionary associations between polyploidy, clonal reproduction, and perenniality in the angiosperms. New Phytol., 224: 1266-1277. DOI:10.1111/nph.15999 |
Wallis, G.P., Waters, J.M., Upton, P., et al., 2016. Transverse alpine speciation driven by glaciation. Trends Ecol. Evol., 31: 916-926. |
Wang, J.-J., Peng, Z.-B., Sun, H., et al., 2017. Cytogeographic patterns of angiosperms flora of the Qinghai-Tibet plateau and Hengduan mountains. Biodivers. Sci., 25: 218-225. DOI:10.11646/phytotaxa.295.3.2 |
Wang, Z., Jiang, Y., Bi, H., et al., 2021. Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol. Plant, 14: 208-222. |
Wen, J., Zhang, J., Nie, Z.-L., et al., 2014. Evolutionary diversifications of plants on the Qinghai-Tibetan plateau. Front. Genet., 5: 1-16. |
White, T.J., Bruns, T., Lee, S., et al., . 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Sninsky JJ, Gelfand DH, White TJ, Innis MA eds. PCR Protocols: a Guide to Methods and Applications. Academic Press, San Diego, California, USA. pp. 315-322.
|
Wu, S., Han, B., Jiao, Y., 2020. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant, 13: 59-71. |
Wu, S., Wang, Y., Wang, Z., et al., 2022. Species divergence with gene flow and hybrid speciation on the Qinghai-Tibet Plateau. New Phytol., 234: 392-404. DOI:10.1111/nph.17956 |
Yang, R., Folk, R., Zhang, N., et al., 2019. Homoploid hybridization of plants in the Hengduan mountains region. Ecol. Evol., 9: 8399-8410. DOI:10.1002/ece3.5393 |
Ye, C.-Y., Wu, D., Mao, L., et al., 2020. The genomes of the allohexaploid Echinochloa crus-galli and its progenitors provide insights into polyploidization-driven adaptation. Mol. Plant, 13: 1298-1310. |
Yu, H., Favre, A., Sui, X., et al., 2019. Mapping the genetic patterns of plants in the region of the Qinghai–Tibet Plateau: implications for conservation strategies. Divers. Distrib., 25: 310-324. DOI:10.1111/ddi.12847 |
Zhang, D.C., Boufford, D.E., Ree, R.H., et al., 2009. The 29°N latitudinal line: an important division in the Hengduan Mountains, a biodiversity hotspot in southwest China. Nord. J. Bot., 27: 405-412. |
Zhang, L., Wu, S., Chang, X., et al., 2020. The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant Cell Environ., 43: 2847-2856. DOI:10.1111/pce.13898 |
Zhang, Y., Qian, L., Spalink, D., et al., 2021. Spatial phylogenetics of two topographic extremes of the Hengduan Mountains in southwestern China and its implications for biodiversity conservation. Plant Divers., 43: 181-191. |
Zhao, Y., Tzedakis, P.C., Li, Q., et al., 2020. Evolution of vegetation and climate variability on the Tibetan Plateau over the past 1.74 million years. Sci. Adv., 6: 1-12. |
Zheng, Q.-J., Yu, C.-C., Xing, Y.-W., et al., 2021. A new Rorippa species (Brassicaceae), R. hengduanshanensis, from the Hengduan mountains in China. Phytotaxa, 480: 210-222. DOI:10.11646/phytotaxa.480.3.1 |



