b. Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, National Engineering Laboratory for Tree Breeding, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing 100083, China
Dove tree (Davidia involucrata Baill.) is a member of the family Davidiaceae (Qi et al., 2009). The plant is a deciduous tree that is exclusively found in China's southwest region (Su and Zhang, 1999; Li et al., 2002). The dormancy stage of D. involucrata seeds is extremely long, typically lasting between 3 and 4 years, and it is this property that makes this species an excellent model for understanding the mechanisms of seed dormancy. Cloning the genes involved in seed dormancy and investigating their functions has been very effective in several plant species (He et al., 2004; Shimozu et al., 2017; Sun et al., 2008; Su and Zhang, 1999).
Agronomic, horticultural, ecological, and therapeutic properties of this endangered tree species have been demonstrated (He et al., 2004; Qi et al., 2009; Sun et al., 2008; Su and Zhang, 1999; Tang et al., 2017). Geographically, Davidia involucrata populations are scarce and dispersed at high elevations in southern and south-central China, as a result of the extremely severe ecotype requirement (Su and Zhang, 1999; Wahid et al., 2007). High temperatures are often detrimental to the growth and development of D. involucrata, as the species is best suited to cool conditions. In particular, this species is projected to be very vulnerable to future rapid climate warming (Tang et al., 2017). In order to maintain the genetic resources, Chinese researchers have been investigating the introduction of Davidia and ex situ cultivation of this species since 1979 (Gang et al., 2004). Nonetheless, such studies are not progressing smoothly since high-temperature stress during the summer months is the most significant factor inhibiting the efficient proliferation of D. involucrata outside of its natural habitats (Li et al., 2016a). D. involucrata has been the subject of numerous investigations, most of which have concentrated on physiological or biochemical changes (Tang et al., 2017; Zhang et al., 1995). Currently, little is known about the molecular regulatory mechanisms that D. involucrata uses to respond to dormancy in the seeds.
In seed dormancy, an intact, healthy seed that is put under favorable conditions cannot germinate. As the range of conditions under which seeds complete germination expands, the level of dormancy steadily decreases. The range of environmental conditions under which seeds germinate and mature increases as the dormancy of the seed is gradually removed (Debeaujon et al., 2000; Hilhorst, 1995; Mares et al., 2021). In most cases, three phases are involved: the first phase is the imbibition phase, which is characterized by the rapid uptake of water by the seed; the second phase is the lag phase, which represents a metabolic reactivation; and the third phase occurs when a tiny portion of the embryo pushes out of the seed coat. The process of germination starts with the quiescent seed soaking up water, and it concludes with the radicle protruding and the embryonic axis lengthening (Kucera et al., 2005; Song and Zhu, 2019).
To obtain energy and the necessary building blocks, metabolism is initiated to drive seed germination (Park and Seo, 2015; Li et al., 2016b; Mesihovic et al., 2016). A previous study found that while dormant seeds are physiologically indistinguishable from non-dormant seeds, metabolic processes and protein metabolism may be important in initiating seed germination (Han and Yang, 2015). A distinct amount of 25 proteins was found to accumulate in the dormant and non-dormant Arabidopsis seeds after being immersed in water for only one day (Cornelius et al., 2011; Debeaujon et al., 2000). Additionally, approximately one out of eight non-dormant seed proteins are involved in energy metabolism. The amount of ABA down-regulates 90% of Arabidopsis seed proteins; most of these proteins are used for energy and protein metabolism (Yang et al., 2013). Seed dormancy is invoked if a block is present in metabolic pathways (Kucera et al., 2005; Li et al., 2021a; Song and Zhu, 2019). At the global level, it is possible to detect differentials in the levels of several metabolites (Song and Zhu, 2019). A previous study has found that seed dormancy in a wide range of herbaceous plants is associated with metabolic pathways (Song and Zhu, 2019). Some excellent examples are a repressed sucrose metabolism downregulated in imbibed dormant seeds (Das et al., 2017), energy metabolism downregulated in imbibed dormant seeds (Gao et al., 2012), lipid metabolism downregulated in imbibed dormant seeds (Cadman et al., 2006), and amino acid metabolism downregulated in imbibed dormant seeds (Hance and Bevington, 1992; Noland and Brad Murphy, 1986). It has been demonstrated that the ability of imbibed dormant seeds to manufacture protein (Arc et al., 2012; Cadman et al., 2006) and generate adenosine triphosphate (ATP) decreases once the seeds are transferred to conditions conducive for germination (Szczotka et al., 2003). However, two studies reported that imbibed dormant seeds of Picea glauca (Downie and Bewley, 2000) and Julans regia (Ali Reza Einali, 2007) were unable to inactivate the synthesis of sucrose and amino acids relative to wet cold seeds. To date, metabolic alterations during germination have only been reported in seeds that are non-dormant.
In addition to enhancing soil moisture, seed stratification has been shown to significantly increase germination and break seed dormancy (Hance and Bevington, 1992; Song and Zhu, 2016). A recent cold seed stratification study in Arabidopsis established that FHY3 interacts with phytochrome B to regulates seed dormancy and germination (Liu et al., 2021a). Again, Arc et al. (2012) reported that cold stratification and exogenous nitrates yielded similar functional proteome adjustments during Arabidopsis seed dormancy release. The present study explored targeted metabolome profiles of moist sand stratified seeds of D. involucrata, and putative responsive metabolites and pathways were discovered. To our knowledge, this is the first time a metabolome profiling of stratified D. involucrata seeds have been performed under dormancy conditions. These findings provide first insights into the mechanisms by which D. involucrata adapts to protracted dormancy, and they will serve as useful information for future research on the improvement of germination in Davidia involucrata.
2. Materials and methods 2.1. Plant materials and sample preparationDavidia involucrata seeds were collected in 2018 from the natural population of Dove tree (50 years plants) in Mulinzi National Nature Reserve, Hubei Province (109°42′6.6″ east longitude, 29°38′53.2″ north latitude). The phenotypes of seed dormancy and germination were determined as previously described (Jiang et al., 2016). Seeds were harvested from the mature fruits, sown in pots (30 cm × 30 cm) containing sand and perlite (3:1) in a greenhouse located at the experiment station of Hubei University, China under room temperature conditions. For the seed stratification: 1000 peeled matured D. involucrata seeds were spread on 5 cm wet sand (85% moisture content) in the experimental field. The buried D. involucrata seeds were sampled at different stratification times: 0M, 6M, 12M, 18M, 24M and 30M (M = months). Following that, the moist sand stratified seeds from at time point were sown in the pots, and the germination rate was evaluated. For this, the seed samples from each of the stratification stages were sown evenly in a glass culture box filled with moist perlite and covered with 3 cm thick moist (moisture content of 85%) after sowing. The cultivated seeds were kept at a constant temperature of 25 ℃ with alternating light conditions at 12 h light/12 h dark daily. Germination rate was computed based on the 30 seeds sown for each treatment at 30 days with 4 replications. Germination test was done when the radicle broke through 1 mm of the seed coat. Finally, the stratified seeds were pulverized into a fine powder in liquid nitrogen. The powdered samples were kept in the −80 ℃ ultra-low deep-freezer for subsequent metabolome analyses.
2.2. Sampling for biochemical studyThe skin of the mature D. involucrata seeds were removed, washed and packed. A portion of mature D. involucrata seeds (5 g of the embryo and cotyledons) that have not been sand stratified (0M as control-CK) were transferred into separate packs for storage. The seeds were kept under a shed for wet sand stratification, and transferred between the time intervals (0M, 6M, 12M, 18M, 24M and 30M). Five grams (5 g) of intact embryos were harvested. The cotyledons were put in liquid nitrogen for 24 h, and then stored in an ultra-low temperature refrigerator at −80 ℃ for subsequent metabolomics determination.
2.3. Metabolites extractionFifty milligrams (50 mg) of samples were weighed into an EP tube. After the addition of 1000 μL of extract solvent (acetonitrile: methanol: water, 2:2:1, containing internal standard 1 μg/mL), the samples were vortexed for 30 s, homogenized at 45 Hz for 4 min, and sonicated for 5 min in ice-water bath. The homogenate and sonicate cycles were repeated three times, followed by incubation at −20 ℃ for 1 h and centrifuged at 12, 000 rpm and 4 ℃ for 15 min. The resulting supernatants were transferred to liquid chromatography mass spectrometry (LC-MS) vials and stored at −80 ℃ until the ultra-performance liquid chromatography-tandem mass spectrometer-quercetin equivalent (UPLC-QE) Orbitrap/MS analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatants from all of the samples.
2.4. Liquid chromatography with tandem mass spectrometry analysisLiquid chromatography with tandem mass spectrometry (LC-MS/MS) analyses were performed using an UHPLC system (1290, Agilent Technologies) with a UHPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm) coupled to Q Exactive (Orbitrap MS, Thermo Fisher). The mobile phase A was 0.1% formic acid in water for positive, and 5 mmol/L ammonium acetate in water for negative, and the mobile phase B was acetonitrile. The elution gradient was set as follows: 0 min, 1% B; 1 min, 1% B; 8 min, 99% B; 10 min, 99% B; 10.1 min, 1% B; 12 min, 1% B. The flow rate was 0.5 mL/min. The injection volume was 2 μL. The QE mass spectrometer was used to acquire tandem mass spectrometry (MS/MS) spectra on an information-dependent basis during the LC/MS experiment. In this mode, the acquisition software (Xcalibur 4.0.27, Thermo Fisher) continuously evaluated the full scan survey MS data as it collected and triggered the acquisition of MS/MS spectra according to preselected criteria. ESI source conditions were set as the following: sheath gas flow rate as 45 Arb, Aux gas flow rate as 15Arb, capillary temperature 320 ℃, full ms resolution as 70, 000, MS/MS resolution as 17, 500, collision energy as 20/40/60 eV in NCE model, Spray Voltage as 3.8 kV (positive) or −3.1 kV (negative), respectively.
2.5. Metabolite functional classificationIn order to fully mine the biological information contained in the quantitative data, we used the Gene Ontology (GO) (Young et al., 2010), Cluster of Orthologous Groups of proteins (COG) (Roman et al., 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) databases to analyze the pathways and functionally annotate the metabolites detected in D. involucrata seeds.
2.6. Data processing and annotationMS raw data (.raw) files were converted to the mzML format using ProteoWizard, and processed by the R package XCMS (version 3.2). The pre-processing results generated a data matrix that consisted of the retention time (RT), mass-to-charge ratio (m/z) values, and peak intensity. OSI-SMMS (version 1.0, Dalian Chem Data Solution Information Technology Co. Ltd, China) was used for peak annotation after XCMS data processing with in-house MS/MS database. We performed a widely targeted metabolome profiling to identify changes in metabolites and associated pathways in the stratified seeds at different germination stages using the ultra-high-performance liquid chromatography-Q Exactive Orbitrap-Mass spectrometry (UHPLC-QE-MS).
2.7. Statistical analysesQuality control (QC) analysis was conducted to confirm the reliability of the data prior to the overall analyses. Data matrices with the intensity of the metabolite features from the stratified seeds were uploaded to the Analyst software (v. 1.6.1; AB Sciex, Canada) for statistical analyses. The partial least squares discriminant analysis (PLS-DA) was performed to maximize the metabolome differences between stratified sample pairs. The relative importance of each metabolite to the PLS-DA model was tested using the variable importance in projection (VIP) as a parameter. Metabolites with VIP ≥1 and fold change ≥2 or fold change ≤0.5 were considered as differential metabolites for group discrimination (Kanehisa and Goto, 2000). PCA and hierarchical clustering heatmap were performed using R software (v.3.3.2; www.r-project.org). Consequently, a metabolic pathway was constructed according to KEGG (http://www.genome.jp/kegg/) (Kanehisa and Goto, 2000); and pathway analysis was performed using MetaboAnalyst (http://www.metaboanalyst.ca/) based on the change in metabolite concentration compared with the corresponding controls. Analysis of variance (ANOVA) was performed to assess the variations in the sample extracts content using the GenStat Statistical Software (v. 12; VSN International, UK).
3. Results 3.1. Germination rate of stratified and unstratified seeds in Davidia involucrataErratic germination is one of the major limitations in D. involucrata as a result of the double dormancy which delays the plant growth and development (Debeaujon et al., 2000). The fruit and seed have a thick cover (Fig. 1A and B) which may impede germination. Likewise, the cotyledon is enclosed in an envelope-like cover (Fig. 1C) which may contribute to the unpredictable germination pattern. The hypocotyl which represents the embryonic axis below the point of attachment of the cotyledon(s), connecting the epicotyl and radicle, being the stem–root transition zone (Fig. 1D) may also delay root formation and development. Our previous work (yet to be published) revealed that D. involucrata seeds have physical dormancy + deep physiological dormancy based on a recent description by Jaganathan (2020).
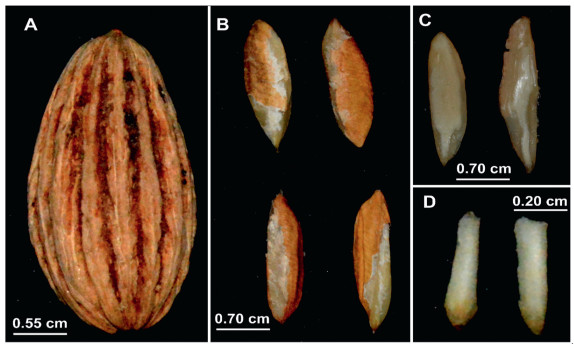
|
| Fig. 1 Organs of Davidia involucrata. (A). Fruit. (B). Seed. (C). Cotyledon. (D). Hypocotyl. |
In the current study, the D. involucrata seeds were moist sand stratified for 6, 12, 18, 24 and 30 months and their germination rates were compared with unstratified seeds (CK). Germination rate (mean ± standard error in %) was highest in 30M (83.00 ± 0.91%) followed by 24M (75.00 ± 1.08%), 18M (64.00 ± 1.08%) and 12M (14.75 ± 0.85%), while CK and 6M recorded no germination (Fig. 2). This suggests that moist sand seed stratification enhances seed germination in D. involucrata. However, the molecular mechanism underlying such enhancement largely remains unknown.
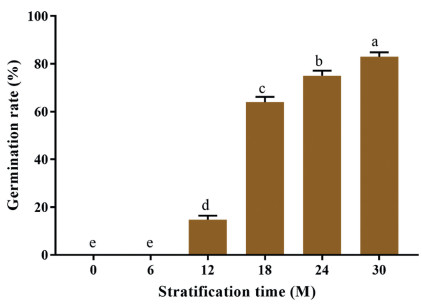
|
| Fig. 2 Germination rate (mean ± standard error in %) of Davidia involucrata seeds subjected to different durations of stratification. Bars with different alphabets indicate significant difference at 5% probability with Post-hoc mean separation by Duncan Multiple Range Test. The standard errors were computed from 4 replicated values. |
In order to understand the molecular basis of erratic germination in D. involucrata, the present study adopted a widely targeted metabolome profiling approach to identify changes in metabolites and associated pathways in germinating seeds obtained from different moist sand stratification experiments (6M, 12M, 18M, 24M and 30M) and CK with five biological repeats. These were profiled via ultra-high-performance liquid chromatography-Q Exactive Orbitrap-Mass spectrometry (UHPLC-QE-MS).
The total ion chromatograms from the germinating seeds (Fig. S1A) indicate that the number of metabolite peaks and the intensities varied among the six samples (CK, 6M, 12M, 18M, 24M and 30M) of germinating seeds of D. involucrata. In all, we detected 10, 008 and 6952 metabolites in the germinated seeds at the positive and negative ion nodes, respectively. However, further comparative analyses between metabolites in positive or negative ion nodes revealed that nearly all metabolites detected in the negative ion node were in the positive ion node. Hence, further downstream analyses were focused on the positive ion node only.
Principal component analysis (PCA) using ion intensities of the detected metabolites was conducted to assess the extent of variability among the six time points in the germinating seeds. Biological replicates were grouped together indicating a good sampling and metabolite quantification in the present study (Fig. 3). The PCA nearly followed the same trend as observed in the germination rate (Fig. 1), thus the stratified seeds for 6 months were identical against those of the unstratified seeds (CK). Conversely, the samples at 12M were more distinct relative to either of the other five stages.
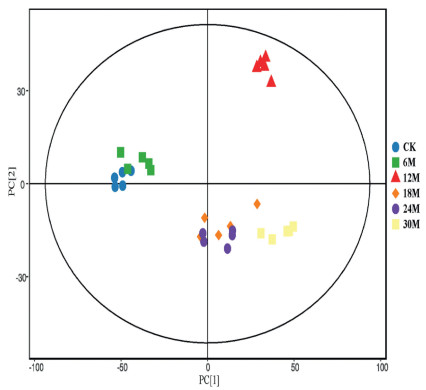
|
| Fig. 3 Principal component analysis based on ion intensities of metabolites detected in germinating seeds Davidia involucrata obtained from different durations of stratification. CK represented unstratified seeds. 6M, 12M, 18M, 24M and 30M denote seed stratified at 6, 12, 18, 24 and 30 months, respectively. |
The 10, 008 metabolites detected in the germinated seeds at the six stages were subjected to differential accumulation analysis in pairwise comparisons (with every 6 months interval) using OPLS-DA at a threshold of log fold change (logFC) ≥ 1 and variable importance in projection (VIP) ≥ 1. The goodness of prediction model of the samples from the six time points of germinating seeds accounted for more than 81.00% of the total variation in the metabolites (Table 1).
| Pairwise group | R2X | R2Y | Q2 |
| 6M-CK | 0.44 | 1.00 | 0.90 |
| 12M–6M | 0.47 | 1.00 | 0.93 |
| 18M–12M | 0.48 | 1.00 | 0.94 |
| 24M–18M | 0.34 | 1.00 | 0.91 |
| 30M–24M | 0.42 | 0.99 | 0.81 |
| R2X and R2Y represent the cumulative degree of interpretation of the model to the X and Y variables, respectively. Q2 denotes goodness of prediction. | |||
In all, 463, 526, 590, 448 and 365 differentially accumulated metabolites (DAMs) were detected in 6M−CK, 12M–6M, 24M–18M and 30M−24M, respectively, in the germinating seeds obtained from different duration of moist sand stratification (Fig. 4A; Tables S1A–E). Globally, 12M seems to be an important stage for the germination process in the moist sand stratified seeds because the highest DAMs were obtained from 6M to 12M and 12M–18M.
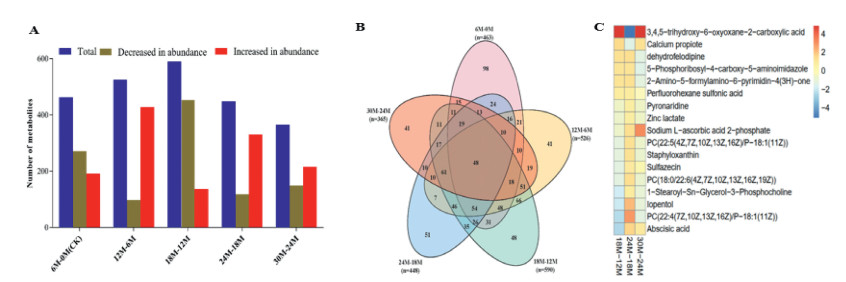
|
| Fig. 4 Number of differentially accumulated metabolites (DAMs) detected in pairwise comparisons of different duration of moist sand seed stratification of Davidia involucrata. (A). Total number of DAMs and their abundance levels. The partial least squares discriminant analysis of log fold change ≥1 and variable importance in projection ≥1 were used to screen for DAMs. (B). Venn diagram of DAMs among the pairwise comparisons. (C). Seventeen common differentially accumulated metabolites detected in 18M–12M, 24M–18M and 30M–24M. Log fold change of between the three pairwise groups 12M, 18M, 24M and 30M denote seed stratified at 12, 18, 24 and 30 months, respectively. CK represents control (unstratified seeds), and 6M, 12M, 18M, 24M and 30M denote seed stratified at 6, 12, 18, 24 and 30 months, respectively. |
We further identified conserved DAMs among the 5 pairwise groups from the germinating seeds (Fig. 4B). A total of 48 DAMs were conserved among all the 5 pairwise groups (Fig. 4B; Table S2), 7–66 DAMs were found concurrently between 2 and 4 pairwise groups, while 98, 41, 48, 51 and 41 DAMs were unique in 6M−CK, 12M–6M, 18M–12M, 24M−18M and 30M−24M, respectively, in the seed (Fig. 4B; Table S1).
In the comparisons of CK to 6M, 12M, 18M, 24M and 30M, a range of 273–386 DAMs accumulated higher in CK compared to either of the 5 stratified seeds. In contrast, we identified a range of 183–444 DAMs higher accumulated in either of the 5 stratified seeds than the CK (Tables S3A–E). These results suggest that stratification largely induces more metabolites relative to the CK.
Seventeen DAMs were exclusively detected from 12M to 30M (Fig. 4B and C). Among these, sodium l-ascorbic acid 2-phosphate showed a decreasing accumulation with over seed stratification period from 12M to 30M. This compound has been reported to inhibits germination at higher concentration but promotes germination at lower concentration (Chen et al., 2021; Li et al., 2021).
3.4. Abscisic acid and its related metabolitesAbscisic acid (ABA) is known to play an important role in regulating germination in plants by the induction and the maintenance of seed dormancy (Li et al., 2021b) ABA accumulated higher in 12M than either 18M or 24M, but contrary was observed in 30M. This trend suggests that ABA contributes markedly in the regulation of germination of seed dormancy in Davidia involucrata. Relative to the CK, ABA accumulated 0.18 and 0.55 times lower in 18M and 24M, respectively (Tables S3A–E). There is one well known metabolite (l-glutamine) which is involved in the regulation of germination by suppressing ABA activity (Qiu et al., 2020). This metabolite was more abundant with prolonging duration of seed stratification (Tables S2–S3). For instance, l-glutamine accumulated 1.23, 10.13, 1.41, 6.56 and 2.66 higher in 6M, 12M, 18M, 24M and 30M, respectively, compared with the CK (Tables S3A–E).
In addition to the above phytohormone (ABA), auxin is reported to control seed dormancy through simulation of ABA signaling (Liu et al., 2013). One auxin metabolite (indole-3-acetic acid, IAA) only accumulated in seeds from CK and 6M (Table S1). This gives an indication that IAA may have contributed to the suppressed germination observed in CK and 6M (Fig. 1). Another metabolite known to interfere with ABA is Sinapic acid (Bi et al., 2017). This compound was highly accumulated in seeds from 12M treatment and above, suggesting that Sinapic acid is negatively correlated with seed dormancy in D. involucrata. Overall, the above results suggest the important role of ABA in regulating dormancy and germination in D. involucrata.
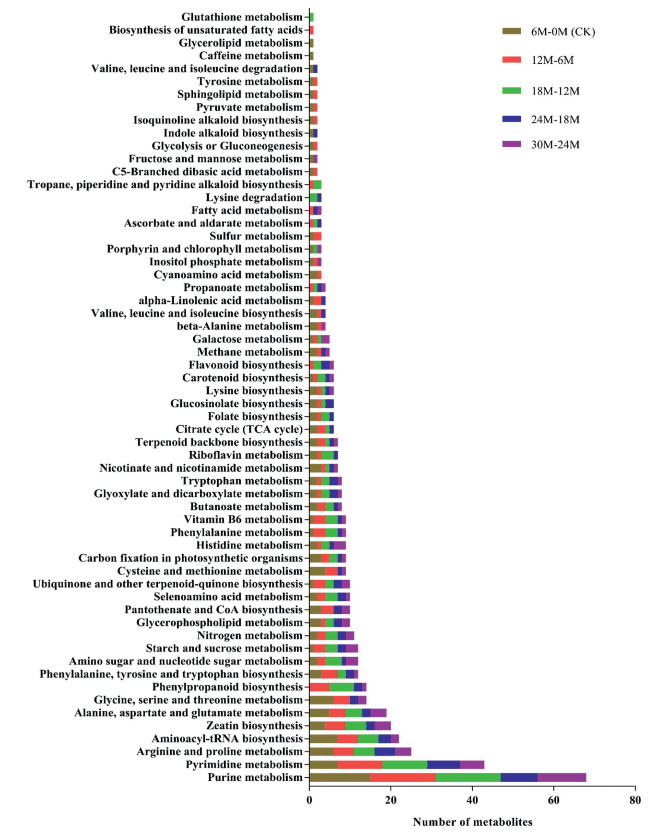
3.5. Functional analyses of differentially accumulated metabolites in Davidia involucrata seeds at different germination stagesTo identify major metabolic pathways regulating germination of D. involucrata seeds, we further analyzed the DAMs in the germinated seeds for their pathway enrichment with the help of Kyoto Encyclopedia of Genes and Genomes (KEGG) compound database (Kanehisa and Goto, 2000). In all, 60 metabolic pathways were identified among the 5 pairwise groups in germinating seeds (Fig. 5). Three prominent pathways: purine metabolism, pyrimidine metabolism and, arginine and proline metabolism were common in germinating stratified and unstratified seeds (Fig. S2). However, glutathione metabolism was only enriched in 18M–12M with l-Glutamic acid the most represented compound (Fig. 5). l-Glutamic acid may be the basis for the initiation of germination in 12M stratified seeds compared with either CK or 6M seeds (Fig. 1). We further delved into the three prominent pathways to assess the DAM and to gain potential mechanistic insights into dormancy regulation in unstratified (CK) and stratified seeds D. involucrata seeds.
|
| Fig. 5 Number of metabolites enriched in metabolic pathways among the 5 pairwise groups in the germinating seeds of Davidia involucrata. CK represents control (unstratified seeds), and 6M, 12M, 18M, 24M and 30M denote seed stratified at 6, 12, 18, 24 and 30 months, respectively. The chocolate, red, green, blue and purple colors represent 6M−CK, 12M–6M, 18M–12M, 24M–18M and 30M–24M, respectively. |
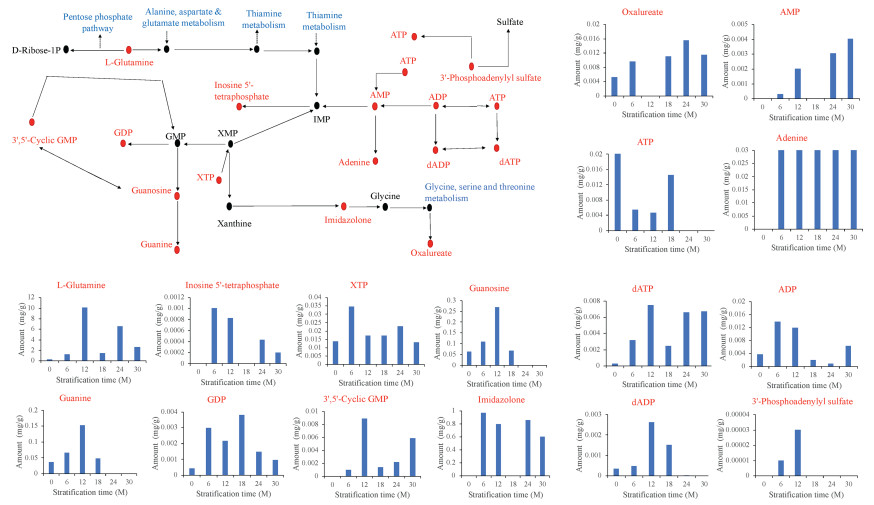
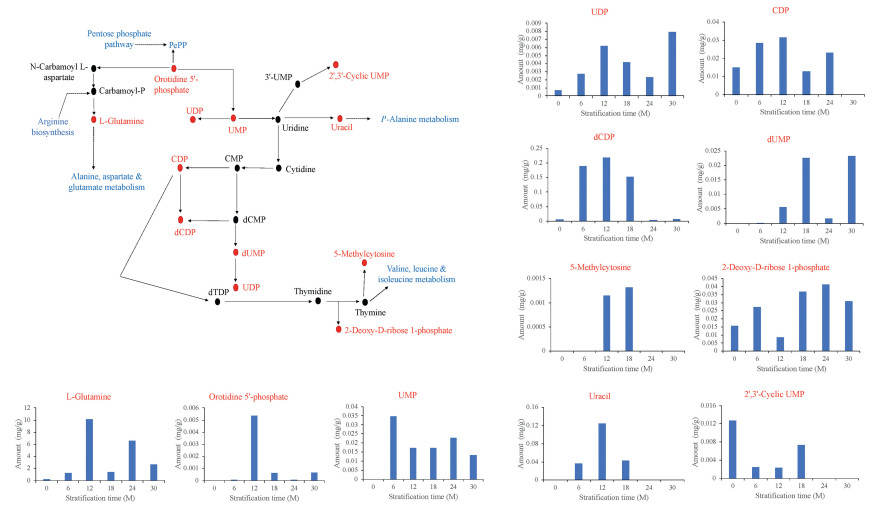
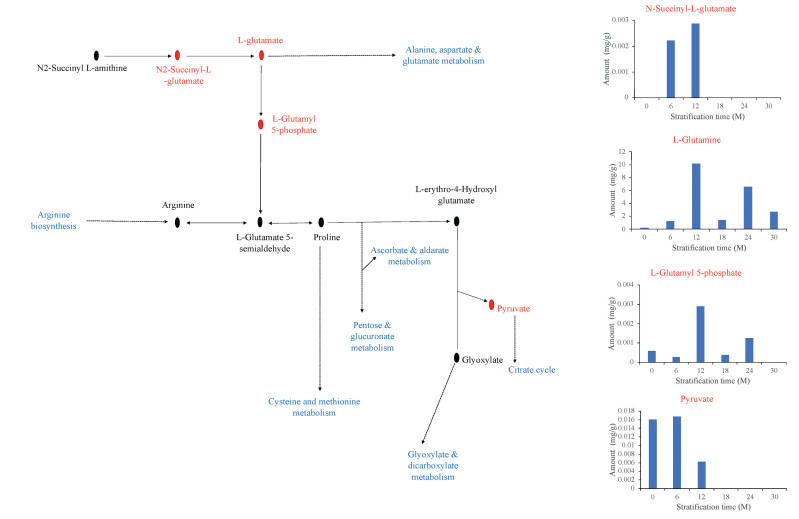
Largely, DAMs enriched in purine metabolism (Fig. 6), pyrimidine metabolism (Fig. 7) and, arginine and proline metabolism (Fig. 8) increased in accumulation over the seed stratification periods. These give an indication that moist sand seed stratification at relatively longer periods induced germination as a result of increased metabolites accumulation to suppress seed dormancy.
|
| Fig. 6 Metabolic changes in a component of purine metabolism in the germinating seeds of Davidia involucrata obtained from different duration of stratification. The key metabolites differentially accumulated in this study are presented in red font and their accumulations are shown in the bar graph. The biosynthetic pathways in the component of purine metabolism presented are shown in blue font. CK represents control (unstratified seeds), and 6M, 12M, 18M, 24M and 30M denote seed stratified at 6, 12, 18, 24 and 30 months, respectively. Key metabolites detected in this study include: Adenosine diphosphate (ADP), deoxynucleotides ADP (dADP), Adenosine triphosphate (ATP), Deoxynucleotides ATP (dATP), Adenosine monophosphate (AMP), Guanosine diphosphate (GDP), Guanosine 5′-monophosphate (GMP), Guanosine triphosphate (GTP) and Xanthine triphosphate (XTP), while the other abbreviation IMP stands for Inosine monophosphate. |
|
| Fig. 7 Metabolic changes in a component of pyrimidine purine metabolism in the germinating seeds of Davidia involucrata obtained from different duration of stratifications. The key metabolites differentially accumulated in this study are presented in red font and their accumulations are shown in the bar graphs. The biosynthetic pathways in the component of pyrimidine metabolism presented are shown in blue font. CK represents control (unstratified seeds), and 6M, 12M, 18M, 24M and 30M denote seed stratified at 6, 12, 18, 24 and 30 months, respectively. Key metabolites detected in this study include: Uridine 5′-monophosphate (UMP), Choline diphosphate (CDP), Deoxyuridine CDP (dCDP), Uridine diphosphate (UDP), Deoxyuridine UDP (dUDP), Uridine-5′-triphosphate (UTP), Deoxyuridine and monophosphate (dUMP). |
|
| Fig. 8 Metabolic changes in a component of arginine and proline metabolism in the germinating seeds of Davidia involucrata obtained from different duration of stratifications. The key metabolites differentially accumulated in this study are presented in red font and their accumulations are shown in the bar graphs. The biosynthetic pathways in the component of arginine and proline metabolism presented are shown in blue font. CK represents control (unstratified seeds), and 6M, 12M, 18M, 24M and 30M sand seed stratified at 6, 12, 18, 24 and 30 months, respectively. |
Uniform germination and growth of Davidia involucrata is currently impossible to achieve due to the double dormancy (Zhang et al., 1995; Mochida and Shinozaki, 2011). With the recent advancement in omics coupled with drastic reduction in the profiling cost and easy to use bioinformatics tools, metabolome profiling has been state-of-the-art technique in understanding metabolites in biological cells, tissues, organs, or organisms, which are the end products of cellular processes (Finch-Savage and Leubner-Metzger, 2006; Mochida and Shinozaki, 2011). The present study employed a widely targeted metabolome profiling to identify and quantify key metabolites as well as their pathways that modulate germination in D. involucrata.
As evidenced in Fig. 1, seed of the Davidia involucrata possesses thick layers which may impede water imbibition as the first step for germination (Finch-Savage and Leubner-Metzger, 2006). The germination process begins with the consumption of water by the seed (imbibition) and ends when a part of the embryonic axis (usually the radicle) extends and crosses the seed coat (emergence) (Weitbrecht et al., 2011; Chen et al., 2021). Dormancy was broken with 12M moist sand stratification, however the germination rate of D. involucrata seeds were enhanced with increased duration of moist sand stratification treatment (Fig. 2). This is not surprising as it has earlier been reported that moist stratification can be used to break seed dormancy, thereby enhancing germination of dormant seeds (Hance and Bevington, 1992; Song and Zhu, 2016). The initiation of germination and the highest number of metabolites detected in 12M indicate that 12M moist sand seed stratification may be ideal for breaking D. involucrata seeds dormancy.
The role of phytohormones in seed dormancy and germination is well documented (Kucera et al., 2005; Liu et al., 2013). ABA is a major phytohormone known to induce and maintain seed dormancy and inhibit germination (Ali et al., 2021; Zhao et al., 2021). This hormone was found differentially accumulated in 18M–12M, 24M–18M and 30M–24M stratified germinating seeds but contrary was observed in 6M−CK and 12M–6M (Tables S1A–E). This suggests that dormancy in D. involucrata is a primary dormancy, which according to Hilhorst (1995) and Kucera et al. (2005a) is induced by ABA during seed maturation on the mother plant. ABA is important in the early phase of seed germination (Cornelius et al., 2011). ABA inhibits endosperm rupture and seedling growth; however, its contents decline sharply during stratification (Weitbrecht et al., 2011). Though, this was inconsistent in our current study.
We further screened the identified metabolites for compounds that are antagonistic to ABA. One of such compounds is l-Glutamine and its derivatives which increased in germinating seeds from 0M to 24M (Tables S1A–E; Table S2). Overexpression of an Arabidopsis gene linked to glutamate receptor homolog3.5 (At2g32390: AtGLR3.5) which largely expressed in germinating seeds led to increase in cytosolic Ca2+ concentration that counteracts the influence of ABA to enhance germination (Kong et al., 2015). On the other hand, repression of AtGLR3.5 impairs Ca2+ concentration and significantly delays germination, enhancing ABA sensitivity in seeds thereby delaying germination. Hence, the metabolite, l-Glutamine and its derivatives could be targeted to overcome seed dormancy in D. involucrata (Kong et al., 2015). Seed stratified at 12M is suggested as the ideal period to break the dormancy. This was evidenced from the comparison of stratified seeds (6M, 12M, 18M, 24M and 30M) and CK, the highest accumulation of l-glutamine (10.13) in 12M compared to the other stratified seeds and CK (Supplementary Table S3 A-E).
ABA signaling pathway in regulating seed dormancy requires auxin action (and vice versa) in interdependent manner (Liu et al., 2013). Auxin controls seed dormancy by simulating ABA signaling through induction of AUXIN RESPONSE FACTORS 10 and 16-mediated AB13 activation in Arabidopsis (Liu et al., 2013). We detected IAA only in 0M–6M stratified germinating seeds. This may explain the break in dormancy and initiation of germination observed in the 12M germinating seeds of D. involucrata (Fig. 2). Exogenous application of auxin enhanced the inhibition of seed germination by ABA in Arabidopsis (Brady et al., 2003; Liu et al., 2007) and delayed seed germination in wheat (Brady et al., 2003; Liu et al., 2007).
Sinapic acid and its esters is reported to have diverse functions in different stages of seed germination and plant development (Bi et al., 2017; Kong et al., 2015; Strack, 1981). This compound accumulated in germinating seeds from 6M to 12M (Table S1B). It has been demonstrated that Sinapic acid affects ABA catabolism by reducing ABA levels and increasing levels of ABA-glucose ester (Bi et al., 2017). This was demonstrated by using mutants deficient in the synthesis of sinapate esters which was more sensitive to ABA than the wild type. In addition, Arabidopsis mutants deficient in either ABA deficient 2 (ABA2) or ABA insensitive 3 (ABI3) showed increased expression of the sinapoyglucose: choline sinapoyltransferase and sinapoylcholine esterase genes with Sinapic acid treatment thereby affecting accumulation of sinapoylcholine and free choline during germination (Bi et al., 2017). Earlier studies have showed that a number of phenolic compounds such as gallic acid, salicylic acid (SA), cinnamic acid, p-coumaric acid, ferulic acid and coumarin can antagonize the effects of ABA by reversing ABA-induced abscission, hypocotyl growth and seed germination (Apte and Laloraya, 1982; El-Araby et al., 2006). Gallic acid, SA, cinnamic acid, p-coumaric acid, ferulic acid and coumarin were found to accumulate higher in 12M germinating seeds and hypocotyl than either of the stages (0M, 6M, 18M, 24M or 30M) (Tables S1A–E).
Phenylpropanoid metabolism produces a diverse group of compounds that are derived from the carbon skeleton of phenylalanine which are involved in plant defense, structural support and survival. For example, flavonoid biosynthesis in which hydroxycinnamic acids are activated by CoA-esters in the Sinapic ester metabolism, 1-O-acyl-glucose esters serve as the energy-rich metabolites during seed maturation and germination (Laloraya et al., 1986; Mock and Strack, 1993). Another propelling case is where Arabidopsis mutants impaired flavonoid metabolism (i.e., transparent testa mutant) exhibited a reduction of both seed longevity and dormancy (Debeaujon et al., 2000, 2003) leading to a better germination performance under optimal conditions.
In order to provide useful information for metabolic pathway engineering to overcome dormancy (Ma et al., 2017) in Davidia involucrata, we subjected the DAMs to KEGG pathway enrichment analysis. In all, we detected 60 pathways modulating germination of D. involucrata seeds (Fig. 7), of which nearly all have been reported to play roles in maintaining primary dormancy in Pinus koraiensis seeds (Song and Zhu, 2019) and herbaceous peony (Paeonia lactiflora Pall.) (Kucera et al., 2005b). Among these, the three prominent pathways were purine metabolism (Fig. 7, Fig. 8), pyrimidine metabolism (Fig. 5, Fig. 7) and, arginine and proline metabolism (Figs. 7 and 9) which are associated to l-Glutamine. It is well known that rehydration after imbibition activates respiration and amino synthesis as early metabolic events (Weitbrecht et al., 2011). Purine and pyrimidine metabolism produce nucleotides which are important for life because they are coenzymes for redox reaction (Zheng et al., 2005). Purine and pyrimidine nucleotides are major energy carriers, subunits nucleic acids and precursors for the synthesis of nucleotide cofactors (Moffatt and Ashihara, 2002). In addition to l-Glutamine, one metabolite worth elaborating is uracil which was in highest abundance in 12M in the pyrimidine metabolic pathway could further explain the break in dormancy (Fig. 7). Uracil or thymine degradation is initiated by dihydrouracil dehydrogenase (designated as PYD1). When PYD1 linked gene in Arabidopsis, At3g17810 was knocked out, there was a delayed germination and seedling development as a result of alteration in the accumulation of sucrose due to decreased cytosolic invertase activity (Cornelius et al., 2011).
One metabolite involved in arginine and proline metabolism deserving further attention is Pyruvate (Fig. 8; Tables S1A–E). This metabolite has been implicated in seed dormancy regulation (Gianinetti et al., 2018). In the current study, the CK and 6M seeds which did not germinate had the highest pyruvate accumulation suggesting that this metabolite may be key contributor to seed dormancy in D. involucrata. Therefore, l-Glutamine, uracil and other beneficial metabolites could be used as candidate biomarkers for dormancy release in D. involucrata (Guillamón et al., 2020).
5. ConclusionInduced metabolites in stratified seeds of Davidia involucrata were profiled using a widely targeted metabolomics technique. A large number of metabolites were detected and characterized. Our metabolomic characterization of the moist stratified seeds revealed the presence of distinct metabolites that were induced by the stratification treatments, which could be used to study germination-induction and biomarker development for D. involucrata. The findings provide useful foundation for breeding to overcome erratic germination in D. involucrata.
Author contributionsZhijun Deng and Qiang Xiao conceived and designed the study; Dan Jiang, and Jian Hong collected the samples, and performed the experiments. Shiming Deng analyzed the data and drafted the manuscript. Shijia Luo provided financial support, supervised the study and revised the first drafts of the manuscript. All authors have read and approved the final version of this manuscript.
FundingThis work was funded by the National Natural Sciences Foundation of China (31460203, 31860073) and Research Center for Germplasm Engineering of Characteristic Plant Resources in Enshi Prefecture (2019–2021). Hubei Key Laboratory of Biological Resources Protection and Utilization (PT012204).
Availability of data and materialsThe datasets used and/or analyzed during the current study are available in the manuscript and its supplementary files.
Declaration of competing interestThe authors declare no conflict of interest.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.12.001.
Ali, F., Qanmber, G., Li, F., et al., 2021. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res.. DOI:10.1016/j.jare.2021.03.011 |
Ali Reza Einali, H.R.S., 2007. Alleviation of dormancy in walnut kernels. Tree Physiol., 27: 519-525. DOI:10.1093/treephys/27.4.519 |
Apte, P.v., Laloraya, M.M., 1982. Inhibitory action of phenolic compounds on abscisic acid-induced abscission. J. Exp. Bot., 33: 826-830. DOI:10.1093/jxb/33.4.826 |
Arc, E., Chibani, K., Grappin, P., et al., 2012. Cold stratification and exogenous nitrates entail similar functional proteome adjustments during Arabidopsis seed dormancy release. J. Proteome Res., 11: 5418-5432. DOI:10.1021/pr3006815 |
Bi, B., Tang, J., Han, S., et al., 2017. Sinapic acid or its derivatives interfere with abscisic acid homeostasis during Arabidopsis thaliana seed germination. BMC Plant Biol., 17: 10.1186/s12870-017-1048-9. DOI:10.1186/s12870-017-1048-9 |
Brady, S.M., Sarkar, S.F., Bonetta, D., et al., 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J., 34: 67-75. DOI:10.1046/j.1365-313X.2003.01707.x |
Cadman, C.S.C., Toorop, P.E., Hilhorst, H.W.M., et al., 2006. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J., 46: 805-822. DOI:10.1111/j.1365-313X.2006.02738.x |
Chen, Z., Cao, X., Niu, J., 2021. Effects of exogenous ascorbic acid on seed germination and seedling salt-tolerance of alfalfa. PLoS One, 16: e0250926. DOI:10.1371/JOURNAL.PONE.0250926 |
Cornelius, S., Witz, S., Rolletschek, H., et al., 2011. Pyrimidine degradation influences germination seedling growth and production of Arabidopsis seeds. J. Exp. Bot., 62: 5623-5632. DOI:10.1093/jxb/err251 |
Das, A., Kim, D.W., Khadka, P., et al., 2017. Unraveling key metabolomic alterations in wheat embryos derived from freshly harvested and water-imbibed seeds of two wheat cultivars with contrasting dormancy status. Front. Plant Sci., 8: 1203. DOI:10.3389/fpls.2017.01203 |
Debeaujon, I., Léon-Kloosterziel, K.M., Koornneef, M., 2000. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol., 122: 403-413. DOI:10.1104/pp.122.2.403 |
Debeaujon, I., Nesi, N., Perez, P., et al., 2003. Proanthocyanidin-accumulating cells in arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell, 15: 2514-2531. DOI:10.1105/tpc.014043 |
Downie, B., Bewley, J.D., 2000. Soluble sugar content of white spruce (Picea glauca) seeds during and after germination. Physiol. Plantarum, 110: 1-12. DOI:10.1034/j.1399-3054.2000.110101.x |
El-Araby, M.M., Moustafa, S.M.A., Ismail, A.I., et al., 2006. Hormone and phenol levels during germination and osmopriming of tomato seeds, and associated variations in protein patterns and anatomical seed features. Acta Agron. Hung., 54: 441-457. DOI:10.1556/AAgr.54.2006.4.7 |
Finch-Savage, W.E., Leubner-Metzger, G., 2006. Seed dormancy and the control of germination. New Phytol., 171: 501-523. DOI:10.1111/j.1469-8137.2006.01787.x |
Gang, W., Shan-heng, H., Hong-chang, W., et al., 2004. Living characteristics of rare and endangered species—Davidia involucrata. J. For. Res., 15: 39-44. DOI:10.1007/bf02858008 |
Gao, F., Jordan, M.C., Ayele, B.T., 2012. Transcriptional programs regulating seed dormancy and its release by after-ripening in common wheat (Triticum aestivum L. ). Plant Biotechnol. J., 10: 465-476. DOI:10.1111/j.1467-7652.2012.00682.x |
Gianinetti, A., Finocchiaro, F., Bagnaresi, P., et al., 2018. Seed dormancy involves a transcriptional program that supports early plastid functionality during imbibition. Plants, 7: 35. DOI:10.3390/plants7020035 |
Hance, B.A., Bevington, J.M., 1992. Changes in protein synthesis during stratification and dormancy release in embryos of sugar maple (Acer saccharum). Physiol. Plantarum, 86: 365-371. DOI:10.1111/j.1399-3054.1992.tb01332.x |
Guillamón, J.G., Prudencio, Á. S., Yuste, J.E., et al., 2020. Ascorbic acid and prunasin, two candidate biomarkers for endodormancy release in almond flower buds identified by a nontargeted metabolomic study. Hortic. Res., 7: 1-13. DOI:10.1038/s41438-020-00427-5 |
Han, C., Yang, P., 2015. Studies on the molecular mechanisms of seed germination. Proteomics, 15: 1671-1679. DOI:10.1002/pmic.201400375 |
He, Z.C., Li, J.Q., Wang, H.C., 2004. Karyomorphology of Davidia involucrata and Camptotheca acuminata, with special reference to their systematic positions. Bot. J. Linn. Soc., 144: 193-198. DOI:10.1111/j.1095-8339.2003.00241.x |
Hilhorst, H.W.M., 1995. A critical update on seed dormancy. Ⅰ. Primary dormancy. Seed Sci. Res., 5: 61-73. DOI:10.1017/S0960258500002634 |
Jaganathan, G.K., 2020. Defining correct dormancy class matters : morphological and morphophysiological dormancy in Arecaceae. Ann. For. Sci., 77: 100. DOI:10.1007/s13595-020-01010-7 |
Jiang, Z., Xu, G., Jing, Y., et al., 2016. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun., 7: 1-10. DOI:10.1038/ncomms12377 |
Kanehisa, M., Goto, S., 2000. KEGG: Kyoto Encyclopedia of genes and Genomes. Nucleic Acids Res., 28: 27-30. DOI:10.1093/nar/28.1.27 |
Kong, D., Ju, C., Parihar, A., et al., 2015. Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol., 167: 1630-1642. DOI:10.1104/pp.114.251298 |
Kucera, B., Cohn, M.A., Leubner-Metzger, G., 2005. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res., 15: 281-307. DOI:10.1079/ssr2005218 |
Li, A., Li, S., Wu, X., et al., 2016. Effect of light intensity on leaf photosynthetic characteristics and accumulation of flavonoids in Lithocarpus litseifolius (Hance) Chun. (Fagaceae). Open J. For., 6: 445-459. DOI:10.4236/ojf.2016.65034 |
Li, Y.X., Chen, L., Juan, L., et al., 2002. Suppression subtractive hybridization cloning of cDNAs of differentially expressed genes in dovetree (Davidia involucrata) bracts. Plant Mol. Biol. Rep., 20: 231-238. DOI:10.1007/BF02782458 |
Li, M., Dong, X., Peng, J., et al., 2016. De novo transcriptome sequencing and gene expression analysis reveal potential mechanisms of seed abortion in dove tree (Davidia involucrata Baill.). BMC Plant Biol., 16: 82. DOI:10.1186/s12870-016-0772-x |
Li, B., Zhang, P., Wang, F., et al., 2021. Integrated analysis of the transcriptome and metabolome revealed candidate genes involved in ga3-induced dormancy release in Leymus chinensis seeds. Int. J. Mol. Sci., 22: 10.3390/ijms22084161. DOI:10.3390/ijms22084161 |
Li, B., Zhang, P., Wang, F., et al., 2021. Transcriptome and metabolome analyses revealing the potential mechanism of seed germination in Polygonatum cyrtonema. Sci. Rep., 11. DOI:10.1038/s41598-021-91598-1 |
Liu, P.P., Montgomery, T.A., Fahlgren, N., et al., 2007. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J., 52: 133-146. DOI:10.1111/j.1365-313X.2007.03218.x |
Liu, S., Yang, L., Li, Jialong, et al., 2021. FHY3 interacts with phytochrome B and regulates seed dormancy and germination. Plant Physiol.. DOI:10.1093/plphys/kiab147 |
Laloraya, M.M., Nozzolillo, C., Purohit, S., et al., 1986. Reversal of abscisic acid-induced stomatal closure by trans-cinnamic and p-coumaric acid. Plant Physiol., 81: 253-258. DOI:10.1104/pp.81.1.253 |
Liu, X., Zhang, H., Zhao, Y., et al., 2013. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A., 110: 15485-15490. DOI:10.1073/pnas.1304651110 |
Ma, Z., Bykova, N. v., Igamberdiev, A.U., 2017. Cell signaling mechanisms and metabolic regulation of germination and dormancy in barley seeds. Crop J.. DOI:10.1016/j.cj.2017.08.007 |
Mares, D.J., Mrva, K., Cheong, J., et al., 2021. Dormancy and dormancy release in white-grained wheat (Triticum aestivum L.). Planta, 253: 10.1007/s00425-020-03518-8. DOI:10.1007/s00425-020-03518-8 |
Mesihovic, A., Iannacone, R., Firon, N., Fragkostefanakis, S., 2016. Heat stress regimes for the investigation of pollen thermotolerance in crop plants. Plant Reprod.. DOI:10.1007/s00497-016-0281-y |
Mochida, K., Shinozaki, K., 2011. Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol.. DOI:10.1093/pcp/pcr153 |
Mock, H.P., Strack, D., 1993. Energetics of the uridine 5′-diphosphoglucose: hydroxycinnamic acid acyl-glucosyltransferase reaction. Phytochemistry, 32: 575-579. DOI:10.1016/S0031-9422(00)95139-2 |
Moffatt, B.A., Ashihara, H., 2002. Purine and pyrimidine nucleotide synthesis and metabolism. The Arabidopsis Book/American Society of Plant Biologists, 1: e0018. DOI:10.1199/TAB.0018 |
Noland, T.L., Brad Murphy, J., 1986. Protein synthesis and aminopeptidase activity in dormant sugar pine seeds during stratification and warm incubation. J. Plant Physiol., 124: 1-10. DOI:10.1016/S0176-1617(86)80172-9 |
Park, C.J., Seo, Y.S., 2015. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J.. DOI:10.5423/PPJ.RW.08.2015.0150 |
Qi, G., Li, J.T., Ruan, Q.P., et al., 2009. An optimised, small-scale preparation of high-quality RNA from dry seeds of Davidia involucrata. Phytochem. Anal., 20: 139-142. DOI:10.1002/pca.1108 |
Qiu, X.M., Sun, Y.Y., Ye, X.Y., et al., 2020. Signaling role of glutamate in plants. Front. Plant Sci., 10: 1743. DOI:10.3389/fpls.2019.01743 |
Roman, T.L., Galperin, M.Y., Natale, D.A., et al., 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28: 33-36. DOI:10.1093/nar/28.1.33 |
Song, Y., Zhu, J., 2019. The roles of metabolic pathways in maintaining primary dormancy of Pinus koraiensis seeds. BMC Plant Biol., 19. DOI:10.1186/s12870-019-2167-2 |
Shimozu, Y., Kimura, Y., Esumi, A., et al., 2017. Ellagitannins of Davidia involucrata. Ⅰ. Structure of davicratinic acid a and effects of davidia tannins on drug-resistant bacteria and human oral squamous cell carcinomas. Molecules, 22. DOI:10.3390/molecules22030470 |
Song, Y., Zhu, J.J., 2016. How does moist cold stratification under field conditions affect the dormancy release of Korean pine seed (Pinus koraiensis)?. Seed Sci. Technol., 44: 27-42. DOI:10.15258/SST.2016.44.1.06 |
Strack, D., 1981. Sinapine as a supply of choline for the biosynthesis of phosphatidylcholine in Raphanus sativus seedlings. Z. Naturforsch. C, 36: 215-221. DOI:10.1515/znc-1981-3-407 |
Su, Z.X., Zhang, S.L., 1999. The reproductive phenology and the influencing factors of Davidia involucrata population. J. Coll. Sci. Teach., 20: 313-318. |
Sun, J.F., Gong, Y.B., Renner, S.S., et al., 2008. Multifunctional bracts in the dove tree Davidia involucrata (Nyssaceae: Cornales): rain protection and pollinator attraction. Am. Nat., 171: 119-124. DOI:10.1086/523953 |
Szczotka, Z., Pawłowski, T., Krawiarz, K., 2003. Proteins and polyamines during dormancy breaking of European beech (Fagus sylvatica L.) seeds. Acta Physiol. Plant., 25: 423-435. DOI:10.1007/s11738-003-0025-0 |
Tang, C.Q., Dong, Y.F., Herrando-Moraira, S., et al., 2017. Potential effects of climate change on geographic distribution of the Tertiary relict tree species Davidia involucrata in China. Sci. Rep., 7: 10.1038/srep43822. DOI:10.1038/srep43822 |
Wahid, A., Gelani, S., Ashraf, M., et al., 2007. Heat tolerance in plants: an overview. Environ. Exp. Bot., 62: 199-223. DOI:10.1016/j.envexpbot.2007.05.011 |
Weitbrecht, K., Mü Ller, K., Leubner-Metzger, G., 2011. First off the mark: early seed germination. J. Exp. Bot., 62: 3289-3309. DOI:10.1093/jxb/err030 |
Yang, Y.G., Lv, W.T., Li, M.J., et al., 2013. Maize membrane-bound transcription factor zmbzip17 is a key regulator in the cross-talk of er quality control and aba signaling. Plant Cell Physiol., 54: 2020-2033. DOI:10.1093/pcp/pct142 |
Young, M.D., Wakefield, M.J., Smyth, G.K., et al., 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol., 11: 10.1186/gb-2010-11-2-r14. DOI:10.1186/gb-2010-11-2-r14 |
Zhang, J., L, J., Z, B., et al., 1995. Research on the protection of Davidia involucrata populations, a rare and endangered plant endemic to China. J. Beijing For. Univ., 17: 25-30. |
Zhao, J., Li, W., Sun, S., et al., 2021. The rice small auxin-up rna gene ossaur33 regulates seed vigor via sugar pathway during early seed germination. Int. J. Mol. Sci., 22: 1-17. DOI:10.3390/ijms22041562 |
Zheng, X., Hayashibe, E., Ashihara, H., 2005. Changes in trigonelline (N-methylnicotinic acid) content and nicotinic acid metabolism during germination of mungbean (Phaseolus aureus) seeds. J. Exp. Bot., 56: 1615-1623. DOI:10.1093/JXB/ERI156 |



