b. Royal Botanic Garden Edinburgh, 20A Inverleith Row, Edinburgh EH3 5LR, UK;
c. College of Science, Tibet University, Lhasa, Tibet 850000, China;
d. Department of Geography, Philipps-Universität Marburg, Deutschhausstraße 10, Marburg 35032, Hessen, Germany
Alpine habitats are characterised by challenging environmental conditions: sharp gradients, limited nutrient resources, strong natural fragmentation, and a high frequency of disturbance (Körner, 2003). As growth is limited by harsh climatic conditions, establishment from seeds is greatly restricted (Urbanska and Schütz, 1986). A number of alpine plants are therefore capable of reproducing both sexually and asexually (Bengtsson and Ceplitis, 2000). Sexual reproduction can lead to mutations that may facilitate the ability to adapt to new environments. Sexually reproducing populations may also differentiate and form new species over time. Sexual reproduction is generally characterized by the investment of resources into reproductive organs rather than vegetative growth (Bazzaz et al., 1987; Rusterholz et al., 2009; Yang and Kim, 2016). In contrast, asexual reproduction allocates more resources for the maintenance of life functions, such as cellular respiration, carbon dioxide assimilation, uptake of water and minerals, and is more commonly found in extreme habitats (Beatty et al., 2008; Heinken and Weber 2013; Stein et al., 2013; Weppler et al., 2006; Wilk et al., 2009; Yang and Kim, 2016). However, the very limited dispersal distances of clones in addition to a lack of new genetic variation in populations undergoing asexual reproduction often results in low genetic diversity and adaptive potential (Douhovnikoff and Dodd, 2015). Accordingly, the genetic diversity of plants growing in alpine regions have been affected by changes in reproductive strategies in these harsh regions (Ellegren and Galtier, 2016).
The relationship between elevational gradients and genetic diversity can result in four different patterns (Shen et al., 2014): (1) genetic diversity is highest in sub-populations at mid-elevation area, due to mild environmental conditions and local adaption (Oyama et al., 1993; Taira et al., 1997); (2) genetic diversity increases along elevational gradients, which is common in highland plants, if the sub-population's fittest sites have similar conditions to high-elevation habitats (Jump et al., 2006); (3) genetic diversity decreases along elevational gradients, due to severe environment conditions at high elevations impede growth and expanding (Mathiasen and Premoli, 2013); (4) genetic diversity is not significantly associated with elevational gradients, which may be due to limited study scale or strong gene flow among (sub-)populations at different elevations (Korshikov and Mudrik, 2006).
Many natural populations of junipers in high-elevation habitats consist of erect or creeping shrubs, such as Juniperus squamata Buch.-Ham. ex D. Don populations of the Hengduan Mountains. As junipers can reproduce both asexually and sexually, it can be difficult to ascertain the structure of a population. Due to clonal reproduction, two spatially separated individuals might in fact be the same genetic individual. Juniper seed cones are mainly dispersed by birds. The seed coat thins after passing through the digestive tract of the bird, thus increasing the success of germination (Traveset and Verdú, 2002; Jordano, 1993; Chambers et al., 1999; García, 2001). Although J. squamata produce one seed per cone (birds usually feed on multiple seed cones each time), it is likely that some patches which appear to be one individual are actually mosaics of different genetic individuals that originated from the excretion of multiple seeds in the same place. Thus, there are three possible structures of juniper patches, where a 'patch' is defined as an area covered by a dense assembly of juniper shrubs (see Fig. 1a): (1) a patch consists of one genetic individual, (2) a patch is a mosaic of multiple genetic individuals, and (3) physically separated patches consist of the same genetic individual (clonal). Excavating roots is the most accurate way to establish the patch structure. Due to the laborious process and the ensuing damage to the plants and the habitat, however, rarely have researchers adopted this approach (Miwa et al., 2001). Indirect methods using molecular markers are therefore needed for reliable assessment of patch structure.

|
| Fig. 1 Photos of Juniperus squamata. (a) Patches are clear from the photo: two shrubs with clear borders are treated as two separate patches. The aim of this study was to determine the genetic structure of these patches. (b) Photo of Juniperus squamata branches, left: female branch with seed cones; right: male branch with pollen cones. |
Here we present the first comprehensive, fine-scale case study of juniper shrubs in alpine habitats of the Hengduan Mountains. The aim of the study was to answer the following questions:
1. Does Juniperus squamata patch structure (single, mosaic or clonal) change over elevation differences of about 500 m?
2. What is the pattern of genetic diversity along this elevational gradient?
3. Which elevation has the fittest populations?
2. Materials and methods 2.1. Study location and sampling strategyJuniperus squamata (Fig. 1) is an evergreen coniferous shrub or small tree belonging to the Cupressaceae family (Adams, 2014). The species is an endemic conifer of Asia, which grows at elevations ranging from 1340 to 4850 m a.s.l. in forests, thickets, and along roadsides (Farjon, 2005; Adams, 2014). It is characterized by its dimorphism of either growing as creeping scrub at higher elevations or as a tree (5–10 m high and up to 1 m diameter at breast height) at lower elevations. J. squamata can be monoecious or dioecious. The seed cones are ovoid or subglobose, which are green before ripening, then turn black or dark blue when ripe (Farjon, 2005).
The study area is located on the Hengduan Mountains, which is characterised by large elevational gradients and steep terrain. The main vegetation type is alpine shrub-meadow. In this region, J. squamata grows exclusively on south-facing slopes (Zhang et al., 2009) as creeping shrubs in roundish patches of 1.5–10 m in diameter. The sampling sites were located in Litang County, Sichuan and Zuogong County, Tibet (Xizang), 240 km apart at elevations above 4000 m (Fig. 2a). Both sites have a continental climate that is semi-humid in Litang and semi-arid in Zuogong (Zheng et al., 2013). Mean annual precipitation at Litang and Zuogong is 765 mm and 456 mm, respectively. The average annual temperature in Litang is 4.6 ℃ and in Zuogong it is 5.7 ℃ (Data: China Meteorological Data Service Centre, climate details in Fig. 2c). In both regions, frosts occur regularly from October to April, and number of growing degree days (number of days at which mean daily maximum air temperature > 5 ℃) are very similar: 126 days in Litang and 127 days in Zuogong (data: CHELSA v.2.1, Karger et al., 2017).
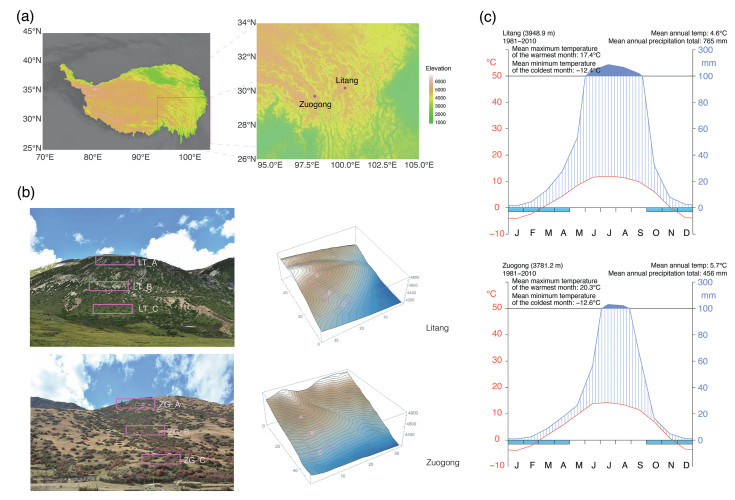
|
| Fig. 2 Topography and climate of the sampling sites. (a) Sampling localities of the studied populations of Juniperus squamata (Elevation data source: Terrain Tiles on AWS, Larrick et al., 2020). Left: elevation map of Hengduan Mountains and the Qinghai-Tibet Plateau (Zhang et al., 2014); right: elevation map of the sampling sites and surrounding area. (b) Photos and terrains of sampling sites, with boxes indicating plot localities. LT_A, high-elevation Litang plot; LT_B, mid-elevation Litang plot; LT_C, low-elevation Litang plot; ZG_A, high-elevation Zuogong plot; ZG_B, mid-elevation Zuogong plot; ZG_C, low-elevation Zuogong plot. (c) Climate diagrams of the nearest weather station from sampling localities (Source: China Meteorological Data Service Centre). Blue bars under the horizontal axis represent months with freezing temperatures. |
Three plots, each 3000 m2 (100 m 30 m) in size, with elevational gradients of 150 m between them, were set up at each of the two locations (Fig. 2b) to cover the whole population from the foot of the hill to the shrub line. We surveyed all patches of J. squamata in the six plots, recording size and location of each patch. To characterize patch structure, a total of 730 leaf samples of 332 patches were collected in silica gel from the six plots. The sampling strategy depended on the size of the patches: for patches with a radius of 5 m or more, five leaf samples were collected; for those with a radius of 2.5–5 m, four samples were taken; and for the patches with radius smaller than 2.5 m, one sample was taken (Fig. 3b, Table 1) as the physical connection between rooting stem and branches could be easily checked. To control the cost of the study, we ranked the surveyed patches in each of the six plots by size, taking five leaf samples from the top 1% of patches, four, three and two samples from the 3%, 5% and 7% of patches respectively, and one sample from each of the remaining patches for genotyping. Based on the criterion, a total of 420 leaf samples were used for subsequent genetic analysis and the remaining 310 were stored in the herbarium. We also collected all seed cones along ca. 20 cm of 3–5 seed cone bearing branches from female and monoecious individuals to evaluate populations fitness (sampling criteria in Fig. 1b).
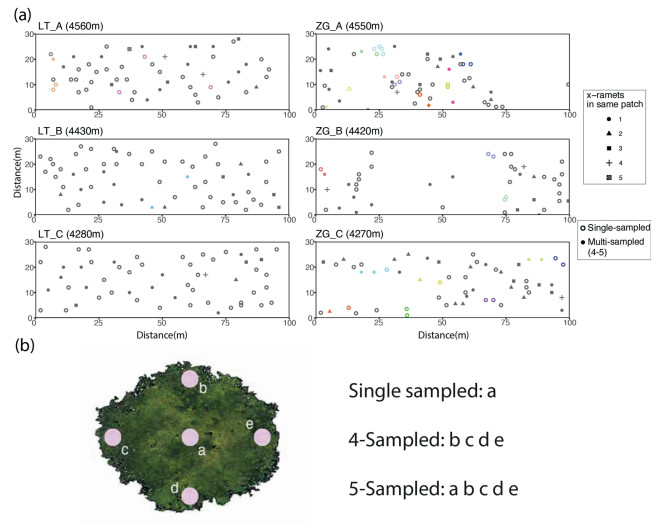
|
| Fig. 3 Spatial distribution of Juniperus squamata patches sampled at three different elevations in Litang (LT) and Zuogong (ZG). (a) Each circle represents one patch with clone patches appearing in the same colour and ramets in different shapes. Number of genetic samples per patch are indicated by solid and hollow shapes. (b) Schematic diagram of sampling strategy. |
| Site | Plot | Elevation (m) | Latitude (°N) | Longitude (°E) | Patch | |||
| ≥5 m | 2.5 m~5 m | ≤2.5 m | Total | |||||
| Litang | LT_A | 4560 | 30°11′21.08″ | 99°58′22.10″ | 11 | 33 | 13 | 57 |
| LT_B | 4430 | 30°11′12.95″ | 99°58′21.73″ | 18 | 29 | 12 | 59 | |
| LT_C | 4280 | 30°11′08.12″ | 99°58′15.64″ | 4 | 34 | 16 | 55a | |
| Zuogong | ZG_A | 4550 | 29°42′29.55″ | 97°55′35.56″ | 22 | 24 | 14 | 60 |
| ZG_B | 4420 | 29°42′28.06″ | 97°55′45.92″ | 20 | 21 | 2 | 43 | |
| ZG_C | 4270 | 29°42′27.98″ | 97°56′0.47″ | 14 | 36 | 8 | 58 | |
| a One patch in LT_C did not have the usual round or elliptical shape; thus, no diameter was recorded. | ||||||||
To identify the number of individuals in one shrub patch, we used microsatellite genotypes as DNA markers. DNA was extracted from leaf tissue using a modified Cetyltrimethylammonium Bromide protocol (CTAB; Doyle and Doyle, 1987), followed by the amplification and sequencing of 18 microsatellite markers (for details of DNA extraction and amplification, see Ju et al., 2020). Microsatellites were scored using GeneMarker v.2.4.0 (Soft Genetics, State College, PA, USA) and automatically binned by the software FlexiBin (Amos et al., 2007). CERVUS 3.0 (Kalinowski et al., 2007) was used to check for null alleles: when the frequency (null) > 0.4, the allele was treated as null allele and excluded from subsequent analyses. Hardy–Weinberg equilibrium (HWE) tests were performed using the function 'hw.test ()' from the pegas package (v.1.0; Paradis, 2010) in R 4.0.5 (R Core Team, 2021). Different patches with the same genotype are regarded as clones, whereas patches with two or more genotypes are classified as mosaic. We plotted the clonal/mosaic status together with the GPS coordinates of the patches in Mathematica 12 to examine the relationship between patches in space. We first put the standardized allele frequencies — obtained by 'scaleGen' function, from adegenet R package (v.2.1.3; Jombart, 2008) — into principle components space using the function 'dudi.pca ()' implemented in ade4 R package (v.1.7; Dray and Dufour, 2007), then we applied Moran's I test (Moran, 1950) on the first two PCs and a Mantel test (Mantel, 1967) on the whole data set using spdep package (v.1.1; Bivand et al., 2013) in R. Moran's I is a correlation coefficient that measures the overall spatial autocorrelation of a data set, indicating how similar one sample is to the others surrounding it. The spatial analysis and subsequent analysis of genetic diversity was performed after clone correction, which means ramets are excluded.
2.3. Evaluation of fitnessTo evaluate fitness at the population level, we used seed cone weight and the genetic diversity of plots. As cone and seed weight are correlated, both traits are often used as proxies for fitness Matziris, 1998, Sevik and Topacoglu, 2015, Zas and Sampedro, 2015. We also used the dry weight of seed cones as a proxy for investment in sexual reproduction. This was measured as the dry weight of all cones from the same patch divided by the number of cone-bearing branches. Population genetic diversity described by DNA markers (i.e. AFLP) has been previously found to be significantly correlated with population fitness. In particular, when genetic diversity is low, the fitness of the population tends to be low as well (Westemeier et al., 1998; Maehr et al., 2006). Although a few studies have found weak or no relationships between genetic diversity and population fitness, these studies were based on interspecific rather than intraspecific comparisons (Reed and Frankham, 2003; Teixeira and Huber, 2021), or were on invasive species that have experienced the founder effect (Kettenring and Mock, 2012; Selechnik et al., 2019). For intra-specific comparisons of indigenous species (here, J. squamata) at fine scale, genetic diversity can still reflect fitness levels of (sub-)populations (Markert et al., 2010). We therefore used genetic diversity as a proxy to evaluate population fitness. The software GenAlex 6.5 (Peakall and Smouse, 2006) was used to estimate genetic diversity, including the number of alleles (Na), the effective number of alleles (Ne), observed (Ho) and expected heterozygosity (He), and inbreeding coefficient (FIS). A t-test was performed to establish whether differences between Ho and He were significant.
3. Results 3.1. Microsatellite markersFour out of the 18 SSR loci had a high frequency of null alleles and were therefore excluded from further analyses. Based on 14 SSR loci, a total of 97 alleles were detected (Na) in 420 samples of J. squamata. The number of alleles per SSR locus ranged from 4 to 10, with a mean of 6.93. The number of effective alleles (Ae) ranged from 1.32 to 4.78 with a mean value of 2.47. The observed heterozygosity (Ho) ranged from Ho = 0.25 to 0.99 with a mean Ho = 0.57, and the expected heterozygosity (He) ranged from H = 0.19 to 0.79 with a mean H = 0.55 (Table S1). The HWE test indicated that loci HEX-43 (p = 0.996), HEX-68 (p = 0.999) and HEX-77 (p = 0.157) differed significantly from Hardy–Weinberg expectations (see details in Appendix, Table S2).
3.2. Patch structureIn total, 390 out of the 420 samples collected from 332 patches, had unique genotypes. Mosaic patches (i.e., patches consisting of multiple genetic individuals) were found in all six plots, whereas clonal patches were found in five plots (no clones in low-elevation Litang plot, LT_C). The number of different genotypes per mosaic patch ranged from 2 to 5, with patches consisting of three genotypes were most common. The fewest mosaic patches were found in the mid–elevation plot in both sites (Fig. 3a). Clonal growth was more common in the highest plots but only accounted for 5.3% in high-elevation Litang plot (LT_A) and 17.8% in high-elevation Zuogong plot (ZG_A) (Table 2). Single patches were recorded in all of the three size categories, and were more common in Litang than in Zuogong; mid–elevation plots had the highest ratio of single patches when compared to high-elevation and low-elevation plots (Tables 2 and S5).
| Site | Plot | N | G | Genets (ramets) | Gm | Gs | Nc | Nm | Ns |
| Litang | LT_A | 76 | 72 | 2 (6) | 27 | 43 | 0.028 | 0.375 | 0.583 |
| LT_B | 68 | 67 | 1 (2) | 15 | 51 | 0.015 | 0.224 | 0.761 | |
| LT_C | 65 | 65 | 0 | 15 | 50 | 0.000 | 0.231 | 0.754 | |
| Zuogong | ZG_A | 73 | 60 | 10 (23) | 22 | 28 | 0.167 | 0.367 | 0.450 |
| ZG_B | 56 | 53 | 3 (6) | 19 | 31 | 0.057 | 0.358 | 0.566 | |
| ZG_C | 82 | 73 | 8 (17) | 44 | 25 | 0.110 | 0.603 | 0.342 | |
| N: Number of tested samples; G: Number of genotypes; Gm: Number of genotypes found in mosaic patches; Gs: Number of genotypes found in single patches; Nc: Average clonal size (Nc = Genets/G); Nm: Average mosaic size (Nm = Gm/G); Ns: Average single size (Ns = Gs/G). | |||||||||
The two most distant clonal patches were 15 m apart (found in the high-elevation Litang plot, LT_A). Because the roots of two patches can grow toward each other, performed Moran's I Test at 30 m. The results of Moran's I test showed that PC1 of high-elevation Litang plot (LT_A) had a significantly lower Moran's I value than I0 (null value, p = 0.007), whereas patches in other plots showed no significant difference between Moran's I and the corresponding I0 for the first two PCs. Only the high-elevation Litang plot (LT_A) had a significant Mantel test score (p = 0.032). Both tests therefore indicated that the high-elevation Litang plot (LT_A) had a positive spatial autocorrelation, suggesting that patches were genetically more similar than expected. In contrast, patches in other plots were randomly distributed (see details in Fig. S1; Tables S3 and S4).
3.3. Population fitnessThe dry weight of cones were significantly higher in both mid–elevation plots than in high-elevation plots (p < 0.05). The dry weight of cones was significantly higher in Litang than in Zuogong (Fig. 4; p < 0.001), which is consistent with the results of the clonal analysis (Table 2), suggesting that plant invest more resources in clonal reproduction in the Zuogong patches than in the Litang patches.

|
| Fig. 4 Dry weight of cones sampled in Litang (a) and Zuogong (b). Note the difference in y-axis scale in the two figures. Lower and upper whiskers show 95% confidence intervals. *p < 0.05; **p < 0.01 after Wilcoxon signed-rank test. |
The comparison of genotypic richness between plots was based on the number of genotypes that would be expected at the same sample size (N = 56, ZG_B). In both sites, shrubs at higher elevations had the lowest genotypic richness compared to shrubs at lower ones (see details in Fig. S2). There was no significant difference between Ho and He (Fig. S3). All genetic diversity parameters (Na, Ne, Ho, He and FIS) showed higher values in the Litang population than in the Zuogong population. Mid–elevation plots had the highest genetic diversity (He, Ho, Ne) in both sites compared to the lower and higher elevation plots (Table 3). Both the dry weight of cones and genetic diversity indicated that the mid-elevation populations were the fittest at both sites.
| Site | Plot | Na | Ne | Ho | He | FIS |
| Litang | LT_A | 6.280 (5.571, 6.786) | 2.69 | 0.57 | 0.536 | −0.064 |
| LT_B | 5.903 (5.429, 6.357) | 2.75 | 0.63 | 0.556 | −0.127 | |
| LT_C | 6.099 (5.643, 6.500) | 2.72 | 0.58 | 0.548 | −0.080 | |
| Mean | 6.094 | 2.72 | 0.60 | 0.547 | −0.090 | |
| Zuogong | ZG_A | 5.255 (4.929, 5.571) | 2.44 | 0.54 | 0.523 | −0.027 |
| ZG_B | 5.391 (5.071, 5.643) | 2.61 | 0.57 | 0.559 | −0.020 | |
| ZG_C | 5.118 (4.857, 5.357) | 2.50 | 0.53 | 0.538 | 0.080 | |
| Mean | 5.255 | 2.52 | 0.55 | 0.540 | 0.011 | |
| Na: Allelic richness (Number of alleles) after sample size standardization, confidence intervals at 95% confidence level are shown in parentheses; Ne: Effective number of alleles; Ho: Observed heterozygosity; He: Expected heterozygosity; FIS: Inbreeding coefficient. | ||||||
Our survey of J. squamata patches along elevational gradients suggests that individuals invest more in sexual reproduction at mid–elevation plots than at higher-elevation patches. This is consistent with results from other studies that showed asexual reproduction is favoured when plants face harsh or challenging environments (Peck et al., 1998; Houle and Duchesne, 1999; García and Zamora, 2003). However, the response of J. squamata to a higher elevation environment seems moderate compared with congeneric species facing other extreme environments. For example, in response to drought stress, Juniperus sabina populations almost exclusively reproduce clonally in semi-arid southern Mongolia, where precipitation is around 130 mm (Wesche et al., 2005); in contrast, in our study, no more than 20% of clonal patches were recorded in the highest plots.
Our results indicate that even a difference in elevation of 150 m can lead to a noticeable change in reproduction strategies of J. squamata populations. Populations from mid elevations in both Litang and Zuogong produced the most and heaviest seed cones and displayed higher genetic diversity than lower or higher elevation populations. Higher elevations often coincide with the shrubline, which is also the case in our two highest plots. Harsher environmental conditions may lead to the selection of a few well adapted genotypes and the fixation of a few specific traits might decrease genetic diversity and increase spatial autocorrelation, both of which have been observed in our high elevation Litang plot (LT_A). This indicates that when faced with a changing climate, plants close to the shrubline may be affected more severely. In contrast, although lower elevation populations might experience more benign environmental conditions than high-elevation and mid-elevation populations, they are closer to human settlements and could therefore be more affected by human activities. We speculate that this is the reason why our lower-elevation populations have lower values for genetic diversity and dry cone weight compared to the mid-elevation populations.
The frequency of mosaic patches also changed along an elevational gradient. In landscape ecology, "mosaics" are often used to describe the spatial and temporal structure of patches and corridors (Naveh and Lieberman, 2013). Our study, however, demonstrates that a similar mosaic of genetic structures exists even at a much finer scale within patches: Mosaic patches (~2.5–10 m in diameter) that consist of two or more different genotypes were found in all plots. The occurrence of mosaic patches is probably linked to the excretion of multiple seeds by birds in the same place. Patches smaller than 2.5 m in diameter were only sampled once, and therefore can only contain a single genotype by definition. Although the visual inspection of these small patches did not suggest multiple individuals, it is possible that some branches that were not physically connected to the main stem might have been overlooked. Thus, the frequency of mosaic patches is likely to be a minimum estimate and mosaic patches may be more common than indicated in the results.
Microhabitat heterogeneity is one of the main ecological factors affecting the distribution patterns of plant individuals, patches, traits and genes at a fine scale. Most related studies have been conducted in tropical rainforest ecological zones (i.e. Baraloto and Couteron, 2010; Torroba-Balmori et al., 2017), where the complex ecological structure and high species abundance tend to create heterogeneous habitats. Few studies have found weak impacts of microhabitat heterogeneity on mountain-inhabited plants (i.e., Paccard et al., 2013). In the present study we focused on alpine habitats, where the vegetation type is homogeneous, the terrain is wide open and the soil type is basically sandy; furthermore, the studied slopes were all south-facing. We therefore conclude that the effect of microhabitat heterogeneity is minimal.
There were fewer clonal and mosaic patches in both mid–elevation plots from Litang and Zuogong than in lower- or higher–elevation plots (Table 2). These results are consistent with the mid-domain effect, although the mid-domain effect hypothesis is applied to the prediction of species abundance rather than genetic diversity (Colwell and Lees 2000). A more likely reason for this phenomenon is the effect of higher levels of human disturbance at lower elevations.
4.2. Human disturbanceA major difference between the two study sites, Litang and Zuogong, is that the distance between Litang and the nearest human settlement is 11 km, whereas the nearest settlement to Zuogong is only one hundred metres away. It is therefore not surprising that the intensity of human activity differs considerably between the two sites. We found that fewer seed cones were produced in the Zugong patches than in the Litang patches (Fig. 4), and that the genetic diversity was higher in the Litang populations than in populations at the same elevation in Zuogong. It is unlikely that this is due to more intense grazing in Zuogong, as a closely related species of Juniperus is poisonous to livestock (Wesche et al., 2005). A more likely explanation is habitat fragmentation due to human activities, such as wood harvesting.
Habitat fragmentation can lead to reduced seed production as well as reduced genetic diversity of populations (Fahrig, 2017). This is consistent with our results, as populations have lower seed cone weight and genetic diversity in Zuogong than in Litang (Fig. 4, Table 3). There were also more clonal patches in Zuogong than in Litang (Table 2), which could be an indication of human disturbance as plants might respond similar to populations at higher elevations when faced with challenging environments by investing more in clonal rather than sexual reproduction.
Patches in Zuogong were distributed more unevenly than in Litang (Fig. 3a), which could have been caused by the differences in human activity as well as climatic differences between the two sites. Vegetation in semi-arid areas is usually fragmented and irregular, which was described by the Klausmeier model (Klausmeier, 1999). As rainwater flows downhill, the water-holding capacity of the vegetation allows more of it to be left on the vegetated surface, while the bare ground is drier so that new vegetation cannot sprout there. This effect becomes more pronounced as precipitation decreases, creating more uneven strips and patches of vegetation. Considering the difference in precipitation between Litang (765 mm) and Zuogong (456 mm) (Fig. 2c), the difference in spatial structure is reasonable.
5. ConclusionsOur results suggest that the genetic structure of Juniperus squamata patches can vary over an elevational gradient of only 150 m, with mid-elevation populations showing higher genetic diversity, fewer clonal patches as well as higher investment in sexual reproduction compared to lower- and higher-elevation populations. This pattern might partly be due to the elevational gradient and human disturbance. Our results can be used to guide vegetation conservation and management in alpine regions of the Hengduan Mountains and the Qinghai-Tibet Plateau.
、Author contributionsK.M., T.J. and Z.H. designed research; T.J., Z.H., J.L. and W.T. performed research; T.J. designed primer; Z.H. and T.J. analysed data; Z.H., T.J. and K.M. wrote the paper; M.R., S.T. and G.M. revised the paper.
、Declaration of competing interestThe authors declare no competing financial interest.
、AcknowledgementsWe thank to Wentao Wang and Le Ren, Sichuan University, for their generous help during field research. This study was funded by the National Natural Science Foundation of China (grant number: U20A2080, 31622015), Sichuan University (Fundamental Research Funds for the Central Universities, SCU2021D006, SCU2020D003).
、Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.12.002.
Adams, R.P., 2014. Junipers of the world: The genus Juniperus. Trafford Publishing, British Columbia
|
Amos, W., Hoffman, J., Frodsham, A., et al., 2007. Automated binning of microsatellite alleles: problems and solutions. Mol. Ecol. Notes, 7: 10-14. |
Baraloto, C., Couteron, P., 2010. Fine-scale microhabitat heterogeneity in a French Guianan forest. Biotropica, 42: 420-428. DOI:10.1111/j.1744-7429.2009.00620.x |
Bazzaz, F.A., Chiariello, N.R., Coley, P.D., et al., 1987. Allocating resources to reproduction and defense. Bioscience, 37: 58-67. DOI:10.2307/1310178 |
Beatty, G.E., McEvoy, P.M., Sweeney, O., et al., 2008. Range-edge effects promote clonal growth in peripheral populations of the one-sided wintergreen Orthilia secunda. Divers. Distrib., 14: 546-555. DOI:10.1111/j.1472-4642.2008.00469.x |
Bengtsson, B.O., Ceplitis, A., 2000. The balance between sexual and asexual reproduction in plants living in variable environments. J. Evol. Biol., 13: 415-422. |
Bivand, R.S., Pebesma, E.J., Gomez-Rubio, V., et al., 2013. Applied Spatial Data Analysis with R (Vol. 2). Springer, Berlin
|
Chambers, J.C., Vander Wall, S.B., Schupp, E.W., 1999. Seed and seedling ecology of pinon and juniper species in the pygmy woodlands of western North America. Bot. Rev., 65: 1-38. |
Colwell, R.K., Lees, D.C., 2000. The mid-domain effect: geometric constraints on the geography of species richness. Trends Ecol. Evol., 15: 70-76. |
Douhovnikoff, V., Dodd, R.S., 2015. Epigenetics: a potential mechanism for clonal plant success. Plant Ecol., 216: 227-233. DOI:10.1007/s11258-014-0430-z |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Dray, S., Dufour, A.B., 2007. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Software, 22: 1-20. |
Ellegren, H., Galtier, N., 2016. Determinants of genetic diversity. Nat. Rev. Genet., 17: 422-433. DOI:10.1038/nrg.2016.58 |
Fahrig, L., 2017. Ecological responses to habitat fragmentation per se. Annu. Rev. Ecol. Evol. Syst., 48: 1-23. DOI:10.1146/annurev-ecolsys-110316-022612 |
Farjon, A., 2005. Monograph of Cupressaceae and Sciadopitys. Royal Botanic Gardens, Kew
|
García, D., 2001. Effects of seed dispersal on Juniperus communis recruitment on a Mediterranean mountain. J. Veg. Sci., 12: 839-848. DOI:10.2307/3236872 |
García, D., Zamora, R., 2003. Persistence, multiple demographic strategies and conservation in long-lived Mediterranean plants. J. Veg. Sci., 14: 921-926. |
Heinken, T., Weber, E., 2013. Consequences of habitat fragmentation for plant species: do we know enough? Perspect. Plant Ecol. Evol. Syst., 15: 205-216. |
Houle, G., Duchesne, M., 1999. The spatial pattern of a Juniperus communis var. depressa population on a continental dune in subarctic Québec, Canada. Can. J. For. Res., 29: 446-450. |
Jombart, T., 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24: 1403-1405. DOI:10.1093/bioinformatics/btn129 |
Jordano, P., 1993. Geographical ecology and variation of plant-seed disperser interactions: southern Spanish junipers and frugivorous thrushes. Vegetatio, 107: 85-104. |
Ju, T., Farhat, P., Tao, W., et al., 2020. Development and characterization of EST-SSR markers for Juniperus squamata (Cupressaceae), an ecologically important conifer in Asian mountains. Silvae Genet., 69: 116-122. DOI:10.2478/sg-2020-0016 |
Jump, A.S., Hunt, J.M., Martínez–Izquierdo, J.A., et al., 2006. Natural selection and climate change: temperature-linked spatial and temporal trends in gene frequency in Fagus sylvatica. Mol. Ecol., 15: 3469-3480. |
Kalinowski, S.T., Taper, M.L., Marshall, T.C., 2007. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol., 16: 1099-1106. |
Karger, D.N., Conrad, O., Böhner, J., et al., 2017. Climatologies at high resolution for the earth's land surface areas. Sci. Data, 4: 1-20. DOI:10.1080/1369118X.2017.1418016 |
Kettenring, K.M., Mock, K.E., 2012. Genetic diversity, reproductive mode, and dispersal differ between the cryptic invader, Phragmites australis, and its native conspecific. Biol. Invasions, 14: 2489-2504. DOI:10.1007/s10530-012-0246-5 |
Klausmeier, C.A., 1999. Regular and irregular patterns in semiarid vegetation. Science, 284: 1826-1828. |
Körner, C., 2003. The alpine life zone. Alpine plant life. Springer, berlin
|
Korshikov, I., Mudrik, E., 2006. Elevation-dependent genetic variation of plants and seed embryos in the Crimea mountain population of Pinus pallasiana D. Don. Russ. J. Ecol., 37: 79-83. |
Larrick, G., Tian, Y., Rogers, U., et al., 2020. Interactive visualization of 3d terrain data stored in the cloud. 2020 11th IEEE Annual Ubiquitous Computing, Electronics & Mobile Communication Conference (UEMCON), 0063-0070
|
Maehr, D., Crowley, P., Cox, J., et al., 2006. 2006. Of cats and haruspices∗: genetic intervention in the Florida panther. response to Pimm et al. Anim. Conserv., 9: 127-132. DOI:10.1111/j.1469-1795.2005.00019.x |
Mantel, N., 1967. The detection of disease clustering and a generalized regression approach. Cancer Res., 27: 209-220. |
Markert, J.A., Champlin, D.M., Gutjahr-Gobell, R., et al., 2010. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol., 10: 1-13. |
Mathiasen, P., Premoli, A.C., 2013. Fine-scale genetic structure of Nothofagus pumilio (lenga) at contrasting elevations of the altitudinal gradient. Genetica, 141: 95-105. DOI:10.1007/s10709-013-9709-6 |
Matziris, D., 1998. Genetic variation in cone and seed characteristics in a clonal seed orchard of Aleppo pine grown in Greece. Silvae Genet., 47: 37-41. |
Miwa, M., Tanaka, R., Yamanoshita, T., et al., 2001. Analysis of clonal structure of Melaleuca cajuputi (Myrtaceae) at a barren sandy site in Thailand using microsatellite polymorphism. Trees (Berl.), 15: 242-248. |
Moran, P.A., 1950. Notes on continuous stochastic phenomena. Biometrika, 37: 17-23. |
Naveh, Z., Lieberman, A.S., 2013. Landscape ecology: Theory and application. Springer Science & Business Media, Berlin
|
Oyama, K., Ito, M., Yahara, T., et al., 1993. Low genetic differentiation among populations of Arabis serrata (Brassicaceae) along an altitudinal gradient. J. Plant Res., 106: 143-148. |
Paccard, A., Vance, M., Willi, Y., 2013. Weak impact of fine-scale landscape heterogeneity on evolutionary potential in Arabidopsis lyrata. J. Evol. Biol., 26: 2331-2340. DOI:10.1111/jeb.12220 |
Paradis, E., 2010. Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics, 26: 419-420. DOI:10.1093/bioinformatics/btp696 |
Peakall, R., Smouse, P.E., 2006. Genalex 6: genetic analysis in excel. population genetic software for teaching and research. Mol. Ecol. Notes, 6: 288-295. DOI:10.1111/j.1471-8286.2005.01155.x |
Peck, J.R., Yearsley, J.M., Waxman, D., 1998. Explaining the geographic distributions of sexual and asexual populations. Nature, 391: 889-892. |
R Core Team. 2021. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria
|
Reed, D.H., Frankham, R., 2003. Correlation between fitness and genetic diversity. Conserv. Biol., 17: 230-237. |
Rusterholz, H.P., Kissling, M., Baur, B., 2009. Disturbances by human trampling alter the performance, sexual reproduction and genetic diversity in a clonal woodland herb. Perspect. Plant Ecol. Evol. Systemat., 11: 17-29. |
Selechnik, D., Richardson, M.F., Shine, R., et al., 2019. Increased adaptive variation despite reduced overall genetic diversity in a rapidly adapting invader. Front. Genet., 10: 1221. |
Sevik, H., Topacoglu, O., 2015. Variation and inheritance pattern in cone and seed characteristics of Scots pine (Pinus sylvestris L.) for evaluation of genetic diversity. J. Environ. Biol., 36: 1125. |
Shen, D., Bo, W., Xu, F., et al., 2014. Genetic diversity and population structure of the Tibetan poplar (Populus szechuanica var. tibetica) along an altitude gradient. BMC Genet., 15: 1-10. DOI:10.1080/00207179.2014.986762 |
Stein, B.A., Staudt, A., Cross, M.S., et al., 2013. Preparing for and managing change: climate adaptation for biodiversity and ecosystems. Front. Ecol. Environ., 11: 502-510. DOI:10.1890/120277 |
Taira, H., Tsumura, Y., Tomaru, Y., et al., 1997. Regeneration system and genetic diversity of cryptomeria japonica growing at different altitudes. Can. J. For. Res., 27: 447-452. |
Teixeira, J.C., Huber, C.D., 2021. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. U.S.A., 118: e2015096118. |
Torroba-Balmori, P., Budde, K.B., Heer, K., et al., 2017. Altitudinal gradients, biogeographic history and microhabitat adaptation affect fine-scale spatial genetic structure in African and Neotropical populations of an ancient tropical tree species. PLoS One, 12: e0182515. DOI:10.1371/journal.pone.0182515 |
Traveset, A., Verdu, M., 2002. A meta-analysis of the effect of gut treatment on seed germination, in: Silva, W.R., Galetti, M. (Eds.), Seed dispersal and frugivory: Ecology, evolution, and conservation. CAB International, New York, pp. 339-350
|
Urbanska, K.M., Schütz, M., 1986. Reproduction by seed in alpine plants and revegetation research above timberline. Bot. Helv., 96: 43-60. |
Weppler, T., Stoll, P., Stöcklin, J., 2006. The relative importance of sexual and clonal reproduction for population growth in the long-lived alpine plant Geum reptans. J. Ecol., 94: 869-879. DOI:10.1111/j.1365-2745.2006.01134.x |
Wesche, K., Ronnenberg, K., Hensen, I., 2005. Lack of sexual reproduction within mountain steppe populations of the clonal shrub Juniperus sabina L. in semi-arid southern Mongolia. J. Arid Environ., 63: 390-405. |
Westemeier, R.L., Brawn, J.D., Simpson, S.A., et al., 1998. Tracking the long-term decline and recovery of an isolated population. Science, 282: 1695-1698. |
Wilk, J.A., Kramer, A.T., Ashley, M.V., 2009. High variation in clonal vs. sexual reproduction in populations of the wild strawberry, Fragaria virginiana (Rosaceae). Ann. Bot., 104: 1413-1419. DOI:10.1093/aob/mcp239 |
Yang, Y.Y., Kim, J.G., 2016. The optimal balance between sexual and asexual reproduction in variable environments: a systematic review. J. Ecol. Environ., 40: 1-18. |
Zhang, Q., Zhang, Y., Peng, S., et al., 2009. Spatial structure of alpine trees in Mountain Baima Xueshan on the southeast Tibetan plateau. Silva Fenn., 43: 197-208. |
Zas, R., Sampedro, L., 2015. Heritability of seed weight in Maritime pine, a relevant trait in the transmission of environmental maternal effects. Heredity, 114: 116-124. DOI:10.1038/hdy.2014.76 |
Zhang, Y., Li, B., Zheng, D., 2014. Datasets of the boundary and area of the Tibetan Plateau. Acta Geograph. Sin., 69: 164-168. |
Zheng, J., Bian, J., Ge, Q., et al., 2013. The climate regionalization in China for 1981-2010. Sci. Bull., 58: 3088-3099. |



