b. University of Chinese Academy of Sciences, Beijing, 100049, China;
c. School of Ecological and Environmental Sciences, East China Normal University, Shanghai, 200241, China;
d. Department of Biological Sciences, University of Notre Dame, Notre Dame, IN, 46556, USA;
e. Center of Plant Ecology, Core Botanical Gardens, Chinese Academy of Sciences, Mengla, 666303, Yunnan, China
Predicting species abundance is one of the most fundamental pursuits of ecology (Laughlin et al., 2012) and has inspired tremendous interest and debate (Shipley et al., 2006; Laughlin and Laughlin, 2013). A rich body of literature has arisen to model species abundance patterns among assemblages (McGill et al., 2007). These range from the classic descriptive analyses of species abundance distributions (Preston, 1948) to mechanistic models such as those derived from neutral theory (Caswell, 1976; Hubbell, 1979, 2001) and niche theory (MacArthur, 1957; Tokeshi, 1990). Understanding the mechanisms and processes that generate and shape the differences in species abundance are crucial steps toward a predictive theory for forest dynamics. A trait-based theory of community assembly posits that local abiotic conditions and biotic interactions impose deterministic filters on the membership and relative abundance of species in local communities (Shipley, 2010; Laughlin and Laughlin, 2013). However, local communities exist in a regional metacommunity context. Therefore, the processes that underlie species abundances at the metacommunity scale may also contribute to local community assembly via mass effects on a species pool (Leibold et al., 2004; Ricklefs, 2008). Ecologists have translated this intuitive notion into mathematical models that encode functional traits and metacommunity data to predict species abundance in local communities (Shipley, 2010; Laughlin and Laughlin, 2013). Unfortunately, the importance of traits, metacommunities and stochasticity for species abundance and distributions in different forest types has not been widely investigated.
An intriguing approach has been proposed that leverages statistical mechanics and the principle of maximum entropy (MaxEnt) to predict species abundances in communities based on their traits (Shipley et al., 2006; Shipley, 2010). The Shipley community assembly by trait selection model rests on the principle that abundances are determined by the match between a trait value and the environment. Here, the environment is the abiotic environment, where the abundance-weighted trait mean of a community reflects the optimal trait value for the given abiotic environment (Shipley, 2010). Thus, in a sense, it is a maximum entropy formalization of trait-based abiotic filtering. In their original work, Shipley et al. (2006) demonstrated that the MaxEnt model, also known as community assembly by trait selection (CATS) model, could be utilized to predict the relative abundances of species with surprising success, however, some have claimed that the high predictive ability of the model is largely a statistical artifact when the number of species is close to the number of parameters (i.e., traits) (Marks and Muller-Landau, 2007). Subsequent work has sought to address these concerns and others (e.g., Roxburgh and Mokany, 2007) by using randomizations (Shipley, 2009, 2014). An additional limitation of the original CATS formulation is that it did not consider the metacommunity context. Specifically, the original model assumed the abundance of species at the larger landscape scale was effectively equivalent. However, differences in abundance on the metacommunity scale will influence the probability of arrival in a local community because more abundant species in the metacommunity will tend to produce more propagules. Therefore, metacommunity abundances will contain relevant information and may be used to refine CATS model (Shipley, 2009; Sonnier et al., 2010). However, metacommunity abundances add another constraint to the approach and therefore may be best applied when the number of species in the metacommunity far exceeds the number of constraints used in the inference.
Central to the CATS framework is the idea that individuals are sorted along environmental gradients upon the basis of their functional traits, which determine their fitness and performance in a given environment (McGill et al., 2006; Webb et al., 2010). Therefore, traits are expected to drive differences in the relative abundances of different species across local communities (Warton et al., 2015). However, mechanistic inferences based on this trait-based approach are complicated by the fact that ecologically important sources of trait variation are generated through a combination of interspecific, intraspecific (Jung et al., 2010; Messier et al., 2010), and ontogenetic trait variation (Hérault et al., 2011; Spasojevic et al., 2014; Kariñho-Betancourt et al., 2015). Yet, most studies of trait-based assembly have focused on interspecific trait variation typically measured at a single ontogenetic stage (e.g., Kraft et al., 2008), or used trait data from adults as a crude surrogate for trait values across life stages (e.g., Swenson and Enquist, 2009). Ontogenetic trait variation may reflect community assembly across environmental gradients (Spasojevic et al., 2014). For example, early ontogenetic stages such as seedlings and saplings often live in low-light environments (Zhu et al., 2015). This low-light environment will reduce the probability of colonization for light-demanding species because of their inability to maintain a positive carbon balance in light-limited environments. In contrast, canopy adults in high-light environments may be more strongly limited by nutrient resources, resulting in stronger abiotic filtering of leaf traits across soil-resource gradients (Spasojevic et al., 2014). Thus, trait-based assembly based on a single ontogenetic stage may mask inferences about the relative importance of community assembly processes (Swenson and Enquist, 2009).
The present study aims to estimate the relative importance of trait-based selection, mass effects, and stochasticity on the assembly of tree communities using the CATS modeling approach (S6). Specifically, we used trait and inventory plot (i.e., metacommunity) data to predict the relative abundances of 490 species from two large forest dynamics plots with different terrains (e.g., seasonal tropical and subtropical forests) in Yunnan, China. Thus, the approach seeks to quantify the relative impact of traits and metacommunity context. Additionally, the work focuses on hundreds of species using relatively few traits. Thus, the work avoids trivial predictions arising from a large number of trait constraints relative to the number of species, thereby providing a robust test of CATS approaches for predicting species abundances. Lastly, we use information on differences in trait values across ontogeny within species across the gradient. Ultimately, the work aims to decompose the deviance in local relative abundances as a proportion of the total biologically relevant deviance to quantify the relative contributions of trait-based filtering and the influence of the known relative abundance of all species in a metacommunity. We ask the following specific questions: (ⅰ) does ontogenetic trait variation influence species relative abundance in tree community assembly? (ⅱ) what is the relative importance of metacommunity context, trait-based filtering and stochasticity in the assembly of tree communities? and (ⅲ) does the importance of the metacommunity context, trait-based filtering and stochasticity vary between subtropical and tropical forests?
2. Materials and methods 2.1. Study siteThis study was conducted in two large forest dynamics plots, one from a tropical and the other from a subtropical area in southwestern China. The tropical area is the 20-ha Xishuangbanna Forest Dynamics Plot (FDP) (21°36′N, 101°34′E, Fig. 1). The elevation in this plot varies greatly with the lowest elevation 709.27 m and the highest elevation 869.14 m. Moreover, the plot contains three gullies with a gentle bottom but steep slopes on both sides. The subtropical area is the 20-ha Ailaoshan FDP (24°32′N, 102°01′E, Fig. 1). The elevation of this plot is relatively high, but the terrain is flat, and elevation changes within the plot are small. The species richness of the two FDPs is, respectively, 390 and 101. The Xishuangbanna FDP is characterized as a seasonal tropical rainforest and dominated by large individuals of Parashorea chinensis (Dipterocarpaceae). The Ailaoshan FDP is characterized as an evergreen broad-leaved forest and dominated by Castanopsis wattii (Fagaceae) and Lithocarpus xylocarpus (Fagaceae). All freestanding woody stems with DBH (Diameter at Breast Height, 1.3 m above ground) ≥ 1 cm were measured, mapped, tagged and identified to species. We used the second census data from the Xishuangbanna plot in 2012, and the first census data from the Ailaoshan in 2014 in the present study. Summary information of the two FDPs is given in Table S1. Detailed information of the two plots regarding the species composition and distribution can be found in Cao et al. (2008), Gong et al. (2011) and Huang et al. (2017).
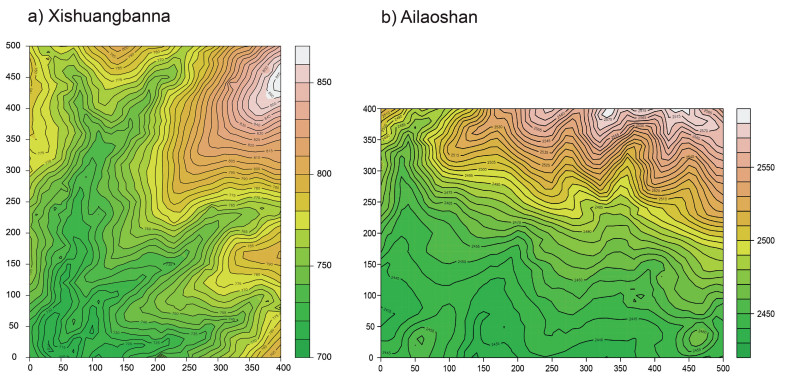
|
| Fig. 1 Xishuangbanna 20-ha forest dynamics plot and Ailaoshan 20-ha forest dynamics plot. |
To make inferences about the importance of ontogenetic trait variation, we divided individuals within species into 4 size classes: saplings (1 cm ≤ dbh < 5 cm), small trees (5 cm ≤ dbh < 10 cm), intermediates (10 cm ≤ dbh < 20 cm) and large trees (dbh ≥20 cm). The number of individuals in the different size classes in each plot is given in Table S2. Classification of ontogenetic stages based on species age would be preferable, but estimating the age of trees across species based upon their diameters is fraught with difficulty and building species-specific models of diameter–age relationships is not feasible for the large number of species studied and particularly for tropical trees that frequently do not have growth rings. Thus, we use the size cut-offs instead of age as an imperfect estimate of ontogenetic stage in this study. Lastly, because we are interested in how traits vary with the environment (e.g., light), and the environment experienced by an individual is likely more related to size than age per se, traits specific to size classes may actually be preferable to traits specific to age. For example, a five-year-old pioneer tree that is 15 cm in diameter and in the canopy cannot be logically compared to a five-year-old, shade-tolerant sapling in the shaded understory.
For each tree size, leaf traits were measured using at least 15 sun-exposed leaves (for shaded plants, leaves from the crowns), sampled from 5 individuals. We measured 9 key functional traits in each ontogenetic stage: leaf thickness, leaf toughness, specific leaf area (SLA), relative leaf chlorophyll content, leaf dry matter content (LDMC), leaf N content, leaf C content, leaf P content, leaf K content. Information on these 9 functional traits is given in Table S3. All measurements of functional traits followed the same methodologies used in Yang et al. (2014) with one exception. Leaf toughness was measured via a punch test, which measures the maximum force to punch out the leaf lamina (originally measured as kilogram-force). In this instance, toughness was determined using the mass needed to penetrate a leaf, employing a penetrometer with a column of 3 mm in diameter (digital force gauges: Imada Co., Ltd, Aichi Japan). Due to differences in sample sizes between ontogenetic stages and species, we focus here on species-level trait means for each ontogenetic stage, calculated as the mean trait value across all the measured individuals of the given ontogenetic stage.
2.3. Environmental variables and spatial scalesFollowing the sampling design described in John et al. (2007), we measured variations in topography (aspect, convexity, mean elevation and slope) at both FDPs. For the Xishuangbanna FDP, we also measured 9 soil nutrients (pH, total N, total P, total K, available N, extractable P, extractable K, total C and bulk density) in the plot. All measurements of topography and soil data followed the same methodologies used in Yang et al. (2014). For the Ailaoshan FDP, we also measured soil moisture and canopy openness. Soil moisture (per volume) was measured with a soil conductivity probe meter (Theta probe MPM-160B, ICT International Proprietary Limited, Armidale, Australia). Canopy openness was measured using a digital camera (Nikon Coolpix 4500, Nikon Corporation, Japan) with a fisheye lens (Nikon FC-E8 Fisheye Converter, Nikon Corporation, Japan) to take hemispherical photographs. Detailed information of soil moisture and canopy openness in the Ailaoshan FDP can be found in Song et al. (2017). Finally, we used Kriging to estimate values of all environmental variables in each 20 × 20 m quadrat, 50 × 50 m quadrats and 100 × 100 m quadrats using the geoR package (Ribeiro and Diggle, 2001) in R.
The process of community assembly was conceptualized by the CATS model, which explains significant variation in species abundances based on their functional traits (Shipley et al., 2006, 2012; Shipley, 2014). In the CATS model, the metacommunity was defined local communities that may exchange propagules and that experience different environmental conditions on a larger spatial scale, and the local communities were defined plants in an area with a sufficiently small spatial scale with no obvious environmental gradients occurring (Shipley, 2010). Based on the terms "metacommunity" and "local community" in the CATS model (Shipley, 2014), we defined each FDP as a "metacommunity", and the 3 spatial scales (20 × 20 m quadrats, 50 × 50 m quadrats and 100 × 100 m) of each FDP as the "local community" (Table S4). The local communities were always nested within the corresponding metacommunity in each analysis. However, not all of the 20 × 20 m quadrats had the requisite number of species (10, traits + 1) for MaxEnt analysis. To maintain a relatively high degree of freedom, we therefore performed our analysis on quadrats in which the number of species was more than 10.
2.4. Statistical analysesTo predict the relative abundances of the local community, we constructed four different CATS models by using two different priors and two different trait constraints based on the CATS model across the scales (Shipley, 2014; Table S4). We used uniform abundance distribution of species as a non-informative prior and the relative abundances for each species in the metacommunity as an informative prior. These two priors represent the two extreme scenarios of neutral or trait-based selection dominant community (Shipley, 2014). The trait constraints were expressed using observed local mean traits versus randomly permuted traits. To distinguish the ontogenetic trait variation effects on species relative abundance in tree community assembly, we calculated both the average value per trait for each species as species mean traits, and the average value per trait for each ontogeny for each species in a quadrat as species ontogenetic mean traits in a local community. Then we used the species mean traits and species ontogenetic mean traits as the actual trait constraints. We performed a randomization test following Shipley (2010) to test the significance of the trait constraints in predicting the species relative abundance in each local community across scales.
To examine the relative importance of the trait-based selection, metacommunity effects and unidentified causes, we decomposed the variation of the relative abundance distribution in each local community into separate components using the Kullback-Leibler index (RKL2) given the four different models (Table 1 and Table S4; Shipley et al., 2012; Shipley, 2014). We used the RKL2 here instead of the classical model R2 because of the exponential regression of CATS model and the R2 is not suitable for measuring the goodness of fit of the exponential regression model (Shipley, 2014). We decomposed the total explained uncertainty into four biological relevant components: (1) pure trait effects; (2) pure metacommunity effects; (3) joint effects; (4) unexplained deviance. The details of the four proportions are shown as follows.
| Plot | RkL2 value | Ontogenetic trait means | Metacommunity trait means | |||||
| 20 × 20 | 50 × 50 | 100 × 100 | 20 × 20 | 50 × 50 | 100 × 100 | |||
| Xishuangbanna | 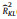 (uniform, null) (uniform, null) |
0.25 | 0.21 | 0.21 | 0.14 | 0.10 | 0.10 | |
| RKL2(uniform, trait) | 0.35 | 0.50 | 0.65 | 0.19 | 0.21 | 0.25 | ||
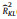 (meta, null) (meta, null) |
0.57 | 0.64 | 0.69 | 0.34 | 0.41 | 0.49 | ||
| RKL2(meta, trait) | 0.65 | 0.77 | 0.85 | 0.40 | 0.47 | 0.52 | ||
| Ailaoshan | 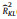 (uniform, null) (uniform, null) |
0.22 | 0.19 | 0.17 | 0.12 | 0.10 | 0.09 | |
| RKL2(uniform, trait) | 0.41 | 0.50 | 0.68 | 0.17 | 0.20 | 0.25 | ||
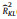 (meta, null) (meta, null) |
0.60 | 0.62 | 0.64 | 0.35 | 0.40 | 0.41 | ||
| RKL2(meta, trait) | 0.66 | 0.75 | 0.82 | 0.44 | 0.48 | 0.54 | ||
| The results show both in ontogenetic-level traits means and plot species traits means. | ||||||||
The contributions due to trait differences among species RKL2(uniform, traits):
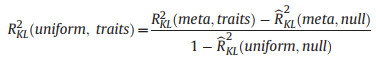
|
(1) |
The contributions due to the metacommunity neutral prior 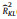
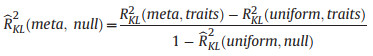
|
(2) |
The contributions due jointly to species trait differences and the metacommunity priorRKL2(meta, traits):
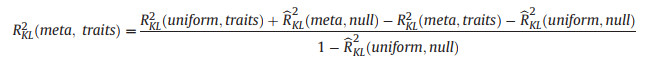
|
(3) |
The unexplained information of residuals 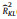
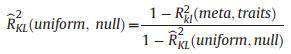
|
(4) |
To describe the pure trait effects variation in the environmental conditions among the different scales, we used a principal component analysis (PCA) of the environmental variables described above. Because our primary focus was to test the influence of ontogenetic trait variation on trait selection environmental gradient rather than to compare its importance on different environmental conditions, in our PCA we focused on the axis that described the most variation among the environmental variables (Table S5). We tested our hypotheses that pure trait effect relationships differ between ontogenetic stage using a simple linear model with PC1 as a continuous independent variable and pure trait-selected values as response variables. To compare trait-environment patterns among the ontogeny stages, we estimated the strength of trait–environment relationships (r2) across the environmental gradient described by PC1.
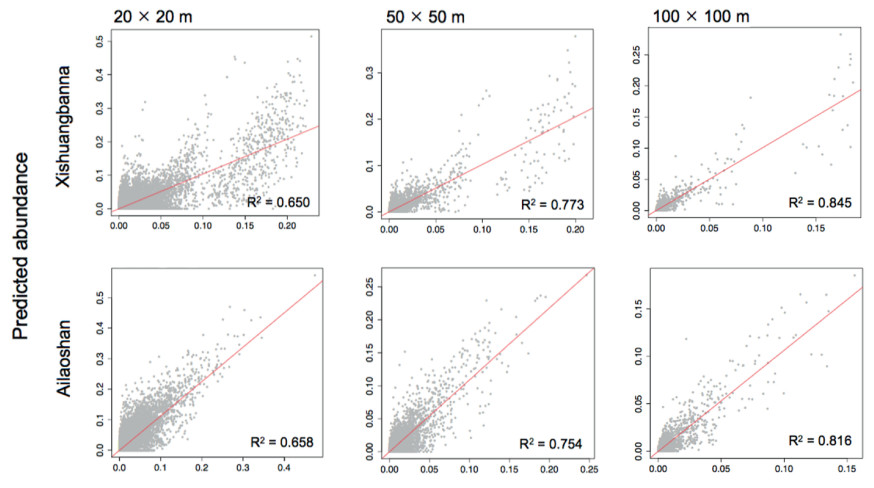
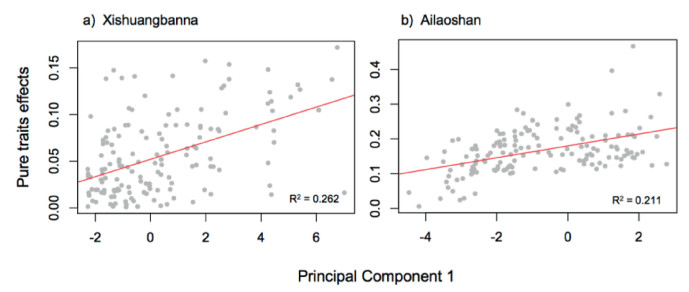
3. Results 3.1. Predicted species relative abundance in local communitiesComparison of observed and predicted relative abundances illustrates that the relative abundances prior and traits constraint was the best model for prediction (Table 1). The relative abundances and traits combined model explained up to 84.5% of the relative abundances at the 100 × 100 m scale, although this percentage decreased to 65% at the 20 × 20 m scale (Fig. 2). However, the pure trait-based model explained 68% of the relative abundances at the 100 × 100 m scale, which decreasing to 35% at the 20 × 20 m scale. The neutral prior model explained 25% of the relative abundance at the three scales of the two plots (Table 1). The predictive ability of the four models was always greater when using ontogenetic traits than species mean traits (Table 1). The trait constraints were significant in 34.5% and 32.1% of the 0.04-ha scale when using the metacommunity prior, percentages of which increased with spatial scale (Fig. 3). Significant trait constraints were positively correlated with the environmental PC1 axis at the 20 × 20 m scale (Fig. 4).
|
| Fig. 2 Predicted relative abundance of the CATS model using observed relative abundances prior and ontogenetic trait means constraint over all quadrats at three scales in each forest dynamics plot.S |
|
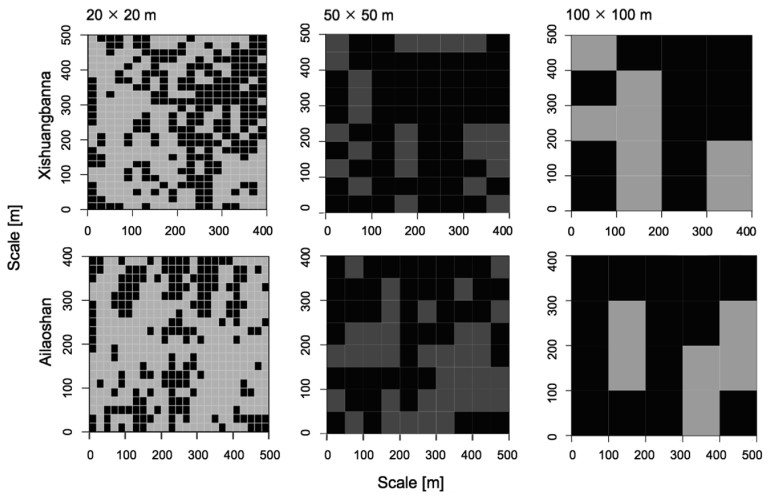
| Fig. 3 Significance of contribution from trait constraints in predicting local species relative abundance in CATS models using observed relative abundances prior and ontogenetic traits means constraint over all quadrats at three scales in each forest dynamics plot. Black quadrats are those in which trait constraints significantly (p < 0.05) improve model fit while grey quadrats are not significant. |
|
| Fig. 4 Ontogenetic trait selection across environmental gradients at 20 × 20 m scale. Regression lines indicate significant linear relationship (p < 0.05) between pure trait effects and the environmental gradient from linear regression.s |
We found a strong scale-dependent effect of processes on the relative species abundance distribution (Table 2). The metacommunity effects explained species abundance up to 39% at the 20 × 20 m scale and this percentage decreased to 16% at the 100 × 100 m scale. The residual explained up to 46% at the 20 × 20 m scale and this percentage decreased to 20% at the 100 × 100 m scale. However, the estimated contribution of pure trait-based selection was very low at three scales in the two plots, which explained 0.08%–0.21% from fine to large scale (Table 2). The estimated joint contribution of trait-based selection and the metacommunity prior increased across scales, being relatively lower at a fine scale but up to the largest components at the large scale.
| Plots | Scale | N | Ontogenetic trait means | Metacommunity trait means | |||||||
| a | b | c | d | a | b | c | d | ||||
| Xishuangbanna | 20 × 20 m | 500 | 0.10 | 0.39 | 0.04 | 0.46 | 0.02 | 0.20 | 0.04 | 0.74 | |
| 50 × 50 m | 500 | 0.17 | 0.35 | 0.19 | 0.29 | 0.06 | 0.29 | 0.05 | 0.59 | ||
| 100 × 100 m | 625 | 0.20 | 0.25 | 0.36 | 0.20 | 0.04 | 0.30 | 0.14 | 0.53 | ||
| Ailaoshan | 20 × 20 m | 80 | 0.08 | 0.33 | 0.16 | 0.44 | 0.04 | 0.25 | 0.01 | 0.70 | |
| 50 × 50 m | 80 | 0.16 | 0.31 | 0.23 | 0.30 | 0.09 | 0.31 | 0.03 | 0.58 | ||
| 100 × 100 m | 100 | 0.21 | 0.16 | 0.41 | 0.22 | 0.13 | 0.32 | 0.04 | 0.51 | ||
| a Pure trait-based selection. b Pure mass effect from the metacommunity prior. c Joint effect of trait-based selection and mass effect from the metacommunity prior. d Residual. Numbers show the average proportion of variation explained across scales in each forest dynamics plot. N is the number of quadrats at each scale of each plot. |
|||||||||||
The predictive ability of the four models between different forest types was always greater when using ontogenetic traits than species mean traits (Table 1). The relative abundances prior and traits constraint explained 84.5% of the species abundance in the tropical forest at the 100 × 100 m scale (Fig. 2). The effect of pure trait-based selection was found to depend on the scale of analyses, with the great contribution for tropical community assembly at fine and medium scales, and subtropical community assembly at a larger scale. The estimated contribution of metacommunity effects, and the joint contribution of trait-based selection and the metacommunity prior were larger for tropical forest than subtropical forest. However, the largest part of the decomposed variation was the residuals in tropical forest across scales (Table 2).
4. DiscussionOur study aimed to predict relative species abundances from metacommunities using the CATS model, which obtains the evenest probability distribution subject to community-weighted mean trait constraints. We used four different models and estimated the relative importance of trait effects, metacommunity effects, joint effects and stochasticity effects across scales in two forest dynamic plots. The present results support the hypothesis that ontogenetic trait variation and metacommunity effects together influence species relative abundance in tree community assembly in two forest types. Our results also quantify the relative importance of metacommunity context, trait-based filtering and stochasticity from the fine to large scales of tree communities that varied between tropical and subtropical forests.
4.1. Ontogenetic trait variation influences relative abundanceOur results show the overall predictive ability of the four models was always greater when using ontogenetic traits instead of species mean traits (Table 1). These results provide evidence that ontogenetic trait variation influences relative abundance in community assembly. Specifically, pure trait selection and the joint contribution of trait-based selection and metacommunity effects generally had a greater predictive ability when ontogenetic trait variation was considered, suggesting that trait selection is an important factor when species co-occur at different stages of ontogeny (Table 2). This result is consistent with the finding that ontogenetic trait variation influences tree community assembly across environmental gradients in the temperate forests in North America (Spasojevic et al., 2014). The relative abundance of a species at a place is the outcome of the species' suitability to the site. This is the central concept of the CATS model (Shipley et al., 2012) and also the hierarchical community assembly rule via trait-based environmental filtering (Keddy, 1992). This concept posits that individuals at a local site have arisen from a regional pool after having passed through an environmental filtering process, expressed as selective forces on functional traits. Thus, we can predict the relative abundance of species from the species pool using the differences in their functional traits; species with traits more suited to the environment at a particular site are expected to increase in abundance, whereas species that are imperfectly adapted to the local environment are expected to decrease (Shipley, 2010). However, mechanistic inferences based on patterns of functional diversity are complicated by the fact that trait variation can emerge through a combination of functional trait variation across ontogenetic stages (Poorter, 2007; Lasky et al., 2015). For example, light limitation strongly affects sapling and small tree leaf traits (Laurans et al., 2012; Dent et al., 2013), while water and nutrient resources affect adult tree traits. Our predictions of species abundances were more accurate than those of previous studies because we used variation in ontogenetic traits, which provides information on how individuals at different stages in life history respond to trait selection via environmental filtering; in contrast, most studies use interspecific trait variation, which is usually measured at a single ontogenetic stage (Table 2). Accurate prediction of the abundance of species is increasingly crucial given the need to understand the species composition and distribution in such rapidly changing world (Laughlin and Laughlin, 2013). Previous applications of the CATS model on herbaceous plant communities have detected strong signals of trait-based community processes (Shipley et al., 2016; Mokany and Roxburgh, 2010). However, this same approach in studies of forest trees has failed to provide strong evidence for trait-based selection in shaping tree communities at either a local or regional scale (Shipley et al., 2012; Xing et al., 2014). Many of these previous applications used metacommunity trait means in tree assemblage, but ignored intraspecific trait variation and ontogenetic trait variation, which are ecologically important sources of trait variation in plant communities. Note that another predictive model, which contains intraspecific trait variation, showed that there are significant positive correlations between observed relative abundances along a temperature gradient (Laughlin et al., 2012). Despite this exception, meager evidence for trait-based community assembly in forests may result from analyzing single ontogenetic stages, which mask inferences about the trait-based selection of community assembly processes (Swenson and Enquist, 2009).
4.2. Relative importance of different processes in the community assemblyThe amount of variation in species composition among local sites, or beta diversity, is a primary tool for connecting the pattern of species assemblages to ecological processes (Legendre et al., 2005; Legendre, 2014), e.g., partitioning beta diversity between environmental variables or/and spatial variables (Legendre and De Cáceres, 2013). However, in a metacommunity context, where local communities are connected to each other through dispersal, spatial differences in community composition can be due to a combination of different processes, including selection by species sorting, dispersal, and stochastic events (Leibold et al., 2004). The local dispersal or demographic effects from a metacommunity is difficult to detect by partitioning the variation with spatial scales. Therefore, we disentangled the degree to which the relative importance of the trait-selected, metacommunity effects (dispersal from metacommunity), joint effects and unexplained effects contribute to the observed abundance of each species in local community using the CATS model.
Our results show little support for pure trait-based selection in shaping tree communities at the fine scale and medium scale in tropical forest or subtropical forest, although this support is stronger at a larger scale. Specifically, we found that at the large scale pure trait-based selection can explain 21% of species abundance in a community, although this contribution decreases to 8% at a fine scale. This pattern is consistent with previous studies (Siefert et al., 2013; Xing et al., 2014; Shipley et al., 2012). As previously discussed, trait variation within species may explain the weak evidence for trait-based selection in the forest. Two additional explanations are possible. First, traits that are strongly selected by the environment may not have been measured and may be largely independent of traits that were measured. For instance, in our study physiological traits related to tree hydraulic and photosynthetic physiological processes that determine species fitness and performance in tree communities are not considered. As a result, the effect of trait-based filtering could be underestimated. Second, the selection pressure for functional traits more often results from strong environmental gradients occurring mostly at large spatial scale, an effect small scales may largely cancel out when averaged (Shipley et al., 2012). This is, thus, may explain the weak relationship between pure trait selection and the environment at small scales in our study plots.
This is also consistent with empirical tests that decomposed phylogenetic and functional beta diversity patterns between local communities and environmental types using spatial information and environmental variables to estimate the relative importance of deterministic and stochastic differences in the Xishuangbanna plot (Yang et al., 2015). Moreover, we found that the joint effect of metacommunity and the ontogenetic trait constraints increased from fine to large scales. However, the proportion of metacommunity effects declined across spatial scales. This may imply that the trait-selection effects have greater weight than the pure trait-selection we observed (Table 2).
The proportion of the residual declined with spatial scale, suggesting that stochastic processes would be weaker across scales. If all important functional traits are included in the work, then this residual component would represent local demographic stochasticity. However, although we used 9 key functional traits in our study, we may have excluded relevant traits that could provide additional ecological axes of the species. Also, there are some processes occurring in the larger landscape that have not been completely captured in the data; thus, the decomposition of this component only provides an upper bound on the importance of demographic stochasticity at different scales.
4.3. Community assembly in different forest typesAlthough the CATS model has been used to examine different vegetation types, studies that compare tropical and subtropical forests across spatial scales are still lacking. We found that the predictive power of the CATS model both for the tropical forest and subtropical forest is good (Fig. 2, Fig. 4). Our results indicate that the CATS model can be used to study community assembly in forests in different climate zones. However, the relative abundance predicted by the CATS model suggests that in subtropical forest the distribution of local abundance may be influenced greatly by trait-based selection, whereas in tropical forest local abundance may be more strongly affected by stochastic forces (Fig. 2). The subtropical forest environment is more stressful than that of the tropical forest. In subtropical forests, resources are scarcer, the climate is harsher and forest dynamics are relatively slow. Under these conditions, trait selection may have a greater influence on the metacommunity. Our results from the decomposed information content of relative abundance support this inference. Specifically, we found the signal for trait selection was stronger in subtropical than tropical forest, indicating that the contribution of the pure trait-selection and its joint effect with metacommunity increased from tropical to subtropical forest. Conversely, tropical forest harbors a wider range of functional traits, reducing the trait selection signal. In addition, the unexplained residual variation, which can be attributed to demographic stochasticity, increased from subtropical to tropical forest likely because subtropical forests have relatively more stable demographic rates than those of tropical forest.
5. ConclusionPredicting species abundance is one of the most fundamental goals of community ecology. We applied a community assembly by trait selection model to predict quadrat-scale species abundances using functional trait variation on ontogenetic stages and metacommunity information for over 490 plant species in a subtropical forest and a lowland tropical forest. Our results support the idea that ontogenetic traits variation influence species relative abundance in tree community assembly. At the same time, both mass effects and trait selection played an important role in tree community assembly, with the former significant at small spatial scales, whereas the latter was stronger at large spatial scales and in subtropical forest. We believe that future community trait-based research will be more effective in inferring the ecological mechanisms at the individual level by incorporating ontogenetic trait variation into prediction frameworks.
、Author contributionsJ.Y., Y.H., and M.C. designed the study; J.Y., D.X., Y.H. and M. A. performed the analysis; J.Y., X.S. and H.W. collected the field data; Y.H., K.S. and N.G.S. wrote the manuscript, and all authors provided comments.
、Declaration of competing interestWe declare that there is no commercial or associated conflict of interest that could influence the review of the manuscript entitled "Ontogenetic trait variation and meta-community effects influence species relative abundances during tree community assembly".
、AcknowledgementsThis research was supported by the National Natural Science Foundation of China (31800353), the Yunnan Fundamental Research Projects (202101AV070005), Yunnan High Level Talents Special Support Plan (YNWR-QNBJ-2018-309), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Y202080), the West Light Foundation of the Chinese Academy of Sciences. We thank Xishuangbanna Station for Tropical Rain Forest Ecosystem Studies and Ailaoshan Station for Subtropical Forest Ecosystem Studies of Xishuangbanna Tropical Botanical Garden for assistance with the fieldwork. NGS was supported by the National Science Foundation United States (NSF DEB-2029997).
、Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.09.002.
Cao, M. et al., 2008. Xishuangbanna Tropical Seasonal Rainforest Dynamics Plot: Tree Distribution Maps, Diameter Tables and Species Documentation. Yunnan Science and Technology Press, Kunming, China
|
Caswell, H., 1976. Community structure-neutral model analysis. Ecol. Monogr., 46: 327-354. DOI:10.2307/1942257 |
Dent, D.H., DeWalt, S.J., Denslow, J.S., 2013. Secondary forests of central Panama increase in similarity to old-growth forest over time in shade tolerance but not species composition. J. Veg. Sci., 24: 530-542. DOI:10.1111/j.1654-1103.2012.01482.x |
Gong, H., Zhang, Y., Lei, Y., et al., 2011. Evergreen broad-leaved forest improves soil water status compared with tea tree plantation in Ailao Mountains, Southwest China. Acta Agric. Scand., 61: 384-388. DOI:10.1080/09064710.2010.494615 |
Huang, H., Chen, Z., Liu, D., et al., 2017. Species composition and community structure of the Yulongxueshan (Jade Dragon Snow Mountains) forest dynamics plot in the cold temperate spruce-fir forest, Southwest China. Biodivers. Sci., 25: 255-264. DOI:10.17520/biods.2016274 |
Hubbell, S.P., 1979. Tree dispersion, abundance and diversity in a dry tropical forest. Science, 203: 1299-1309. DOI:10.1126/science.203.4387.1299 |
Hubbell, S.P., 2001. A Unified Theory of Biodiversity and Biogeography. Princeton University Press, Princeton
|
Hérault, B., Bachelot, B., Poorter, L., et al., 2011. Functional traits shape ontogenetic growth trajectories of rain forest tree species. J. Ecol., 99: 1431-1440. DOI:10.1111/j.1365-2745.2011.01883.x |
John, R., Dalling, J.W., Harms, K.E., et al., 2007. Soil nutrients influence spatial distributions of tropical tree species. Proc. Natl. Acad. Sci. U.S.A., 104: 864-869. DOI:10.1073/pnas.0604666104 |
Jung, V., Violle, C., Mondy, C., et al., 2010. Intraspecific variability and trait-based community assembly. J. Ecol., 98: 1134-1140. DOI:10.1111/j.1365-2745.2010.01687.x |
Kariñho-Betancourt, E., Agrawal, A.A., Halitschke, R., et al., 2015. Phylogenetic correlations among chemical and physical plant defenses change with ontogeny. New Phytol., 206: 796-806. DOI:10.1111/nph.13300 |
Keddy, P., 1992. Assembly and response rules: two goals for predictive community ecology. J. Veg. Sci., 3: 157-164. DOI:10.2307/3235676 |
Kraft, N.J.B., Valencia, R., Ackerly, D.D., 2008. Functional traits and niche-based tree community assembly in an Amazonian forest. Science, 322: 580-582. DOI:10.1126/science.1160662 |
Lasky, J.R., Bachelot, B., Muscarella, R., et al., 2015. Ontogenetic shifts in trait-mediated mechanisms of plant community assembly. Ecology, 96: 2157-2169. DOI:10.1890/14-1809.1 |
Laughlin, D.C., Laughlin, D.E., 2013. Advances in modelling trait-based plant community assembly. Trends Plant Sci., 18: 584-593. |
Laughlin, D.C., Joshi, C., van Bodegom, P.M., et al., 2012. A predictive model of community assembly that incorporates intraspecific trait variation. Ecol. Lett., 15: 1291-1299. DOI:10.1111/j.1461-0248.2012.01852.x |
Laurans, M., Martin, O., Nicolini, E., et al., 2012. Functional traits and their plasticity predict tropical trees regeneration niche even among species with intermediate light requirements. J. Ecol., 100: 1440-1452. DOI:10.1111/j.1365-2745.2012.02007.x |
Legendre, P., De Cáceres, M., 2013. Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecol. Lett., 16: 951-963. DOI:10.1111/ele.12141 |
Legendre, P., 2014. Interpreting the replacement and richness difference components of beta diversity. Global Ecol. Biogeogr., 23: 1324-1334. DOI:10.1111/geb.12207 |
Legendre, P., Borcard, D., Peres-Neto, P.R., 2005. Analyzing beta diversity: partitioning the spatial variation of community composition data. Ecol. Monogr., 75: 435-450. DOI:10.1890/05-0549 |
Leibold, M.A., Holyoak, M., Mouquet, N., et al., 2004. The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett., 7: 601-613. |
MacArthur, R.R., 1957. On the relative abundance of species. Proc. Natl. Acad. Sci. U.S.A., 43: 293-295. DOI:10.1073/pnas.43.3.293 |
Marks, C.O., Muller-Landau, H.C., 2007. Comment on "From plant traits to plant communities: a statistical mechanistic approach to biodiversity. Science, 316: 1425c. |
McGill, B.J., Enquist, B.J., Weiher, E., et al., 2006. Rebuilding community ecology from functional traits. Trends Ecol. Evol., 21: 178-185. |
McGill, B.J., Etienne, R.S., Gray, J.S., et al., 2007. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecol. Lett., 10: 995-1015. DOI:10.1111/j.1461-0248.2007.01094.x |
Messier, J., McGill, B.J., Lechowicz, M.J., 2010. How do traits vary across ecological scales? A case for trait-based ecology. Ecol. Lett., 13: 838-848. DOI:10.1111/j.1461-0248.2010.01476.x |
Mokany, K., Roxburgh, S.H., 2010. The importance of spatial scale for trait-abundance relations. Oikos, 119: 1504-1514. DOI:10.1111/j.1600-0706.2010.18411.x |
Poorter, L., 2007. Are species adapted to their regeneration niche, adult niche, or both?. Am. Nat., 169: 433-442. |
Preston, F.W., 1948. The commonness, and rarity, of species. Ecology, 29: 254-283. DOI:10.2307/1930989 |
, Diggle, P.J., 2001. geoR: a package for geostatistical analysis. R. News, 1: 15-18. |
Ricklefs, R.E., 2008. Disintegration of the ecological community. Am. Nat., 172: 741-750. DOI:10.1086/593002 |
Roxburgh, S.H., Mokany, K., 2007. Comment on "From plant traits to plant communities: a statistical mechanistic approach to biodiversity. Science, 316: 1425b. |
Shipley, B., 2009. Limitations of entropy maximization in ecology: a reply to Haegeman and Loreau. Oikos, 118: 152-159. DOI:10.1111/j.1600-0706.2008.17179.x |
Shipley, B., 2010. From Plant Traits to Vegetation Structure: Chance and Selection in the Assembly of Ecological Communities. Cambridge University Press, Cambridge, UK
|
Shipley, B., 2014. Measuring and interpreting trait-based selection versus meta-community effects during local community assembly. J. Veg. Sci., 25: 55-65. DOI:10.1111/jvs.12077 |
Shipley, B., Paine, C.E.T., Baraloto, C., 2012. Quantifying the importance of local niche-based and stochastic processes to tropical tree community assembly. Ecology, 93: 760-769. DOI:10.1890/11-0944.1 |
Shipley, B., Vile, D., Garnier, E., 2006. From plant traits to plant communities: a statistical mechanistic approach to biodiversity. Science, 314: 812-814. DOI:10.1126/science.1131344 |
Shipley, B., De Bello, F., Cornelissen, J.H., et al., 2016. Reinforcing loose foundation stones in trait-based plant ecology. Oecologia, 180: 923-931. DOI:10.1007/s00442-016-3549-x |
Siefert, A., Ravenscroft, C., Weiser, M.D., et al., 2013. Functional beta-diversity patterns reveal deterministic community assembly processes in eastern North American trees. Global Ecol. Biogeogr., 22: 682-691. DOI:10.1111/geb.12030 |
Song, X., Hogan, J.A., Brown, C., et al., 2017. Snow damage to the canopy facilitates alien weed invasion in a subtropical montane primary forest in southwestern China. For. Ecol. Manag., 391: 275-281. |
Sonnier, G., Shipley, B., Navas, M.L., 2010. Plant traits, species pools and the prediction of relative abundance in plant communities: a maximum entropy approach. J. Veg. Sci., 21: 318-331. DOI:10.1111/j.1654-1103.2009.01145.x |
Spasojevic, M.J., Yablon, E.A., Oberle, B., et al., 2014. Ontogenetic trait variation influences tree community assembly across environmental gradients. Ecosphere, 5: 129. |
Swenson, N.G., Enquist, B.J., 2009. Opposing assembly mechanisms in a Neotropical dry forest: implications for phylogenetic and functional community ecology. Ecology, 90: 2161-2170. DOI:10.1890/08-1025.1 |
Tokeshi, M., 1990. Niche apportionment or random assortment: species abundance patterns revisited. J. Anim. Ecol., 59: 1129-1146. DOI:10.2307/5036 |
Warton, D.I., Shipley, B., Hastie, T., 2015. CATS regression - a model-based approach to studying trait-based community assembly. Methods Ecol. Evol., 6: 389-398. |
Webb, C.O., Slik, J.W.F., Triono, T., 2010. Biodiversity inventory and informatics in southeast asia. Biodivers. Conserv., 19: 955-972. DOI:10.1007/s10531-010-9817-x |
Xing, D., Swenson, N.G., Weiser, M.D., et al., 2014. Determinants of species abundance for eastern North American trees. Global Ecol. Biogeogr., 23: 903-911. DOI:10.1111/geb.12167 |
Yang, J., Swenson, N.G., Zhang, G., et al., 2015. Local-scale partitioning of functional and phylogenetic beta diversity in a tropical tree assemblages. Sci. Rep., 5: 12731. |
Yang, J., Zhang, G., Ci, X., et al., 2014. Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Funct. Ecol., 28: 520-529. DOI:10.1111/1365-2435.12176 |
Zhu, Y., Comita, L.S., Hubbell, S.P., et al., 2015. Conspecific and phylogenetic density-dependent survival differs across life stages in a tropical forest. J. Ecol., 103: 957-966. DOI:10.1111/1365-2745.12414 |



