b. Research and Collections Center, Illinois State Museum, 1011 East Ash Street, Springfield, IL, 62703, USA
Phylogenetic trees serve as tools that facilitate systematic, evolutionary and ecological analyses (Smith and Brown, 2018). For example, phylogenies have been used to address ecological questions (Beaulieu et al., 2012; Cornwell et al., 2014), evolution of climate tolerance (Smith et al., 2009; Edwards and Smith, 2010; Zanne et al., 2014), and diversification (Smith et al., 2011). Since Webb (2000) developed an approach for analyzing phylogenetic structure of biological communities and used the phylogenetic approach to address issues of community assembly two decades ago, the use of phylogenetic trees to investigate patterns of community structure has blossomed (Qian and Jin, 2016; Jin and Qian, 2019).
There are about 357, 000 vascular plant species in the world (Freiberg et al., 2020) but only ~20% of these species have been sequenced, according to gene sequence data with GenBank. Because well resolved phylogenies that include all plant species of a study area are rare, botanists and ecologists often use mega-tree approaches to generate plant phylogenetic trees for their studies.
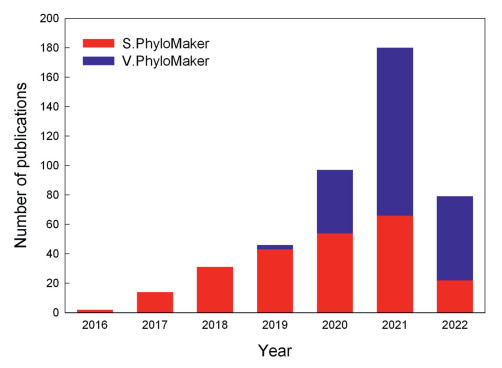
Zanne et al. (2014) published a phylogeny that includes 31, 749 plant species. Qian and Jin (2016) developed an R package, S.PhyloMaker, to use an updated version of Zanne et al.'s phylogeny as a backbone to generate phylogenetic trees for seed plants. S.PhyloMaker has been used in 232 published studies according to Thompson Reuters ISI Web of Science (access on April 21, 2022) (Fig. 1).
|
| Fig. 1 The number of the publications that used either S.PhyloMaker (Qian and Jin, 2016) or V.PhyloMaker (Jin and Qian, 2019) in each year, based on Thompson Reuters ISI Web of Science (access on April 21, 2022). The total number of the publications using these two packages is 449. |
Smith and Brown (2018) published a phylogeny for seed plants (i.e. GBOTB.tre), which includes 79, 881 taxa at and below the species rank. Jin and Qian (2019) updated and expanded this phylogeny by standardizing botanical nomenclature, removing duplicate species, including pteridophyte species from Zanne et al.'s (2014) phylogeny, and including those families that are missing from the two phylogenies. The resulting phylogeny (i.e. GBOTB.extended.tre) includes 74, 531 species of vascular plants in 10, 587 genera (Jin and Qian, 2019). Jin and Qian (2019) developed an R package, V.PhyloMaker, to use GBOTB.extended.tre as a backbone to generate phylogenetic trees for vascular plants. V.PhyloMaker has been used in 217 published studies according to Thompson Reuters ISI Web of Science (access on April 21, 2022) (Fig. 1).
Taken together, S.PhyloMaker and V.PhyloMaker have been used in a total number of 449 studies, according to Thompson Reuters ISI Web of Science (access on April 21, 2022) (Fig. 1). The number of the publications using the packages increases exponentially from 2016 to 2021 (Fig. 1), and these packages were used to generate plant phylogenies in 180 published studies in 2021 alone (Fig. 1), suggesting that the two packages have been considered as useful tools to generate plant phylogenetic trees.
There is a drawback in S.PhyloMaker and V.PhyloMaker. Plant names in the phylogenies implemented in both S.PhyloMaker and V.PhyloMaker were standardized according to The Plant List (TPL; http://www.theplantlist.org), which has been static since 2013. The botanical nomenclature in TPL is outdated and 23% of plant names in TPL are unsolved. Considering that several global plant databases have been recently developed, it is time to update V.PhyloMaker by improving its phylogenetic backbone so that it reflects the up-to-date botanical nomenclature. Here, we present a new version of V.PhyloMaker, which is called V.PhyloMaker2. In this new version, in addition to retaining the phylogeny based on the botanical nomenclature of TPL (we retain this phylogeny because TPL remains to be widely used as a botanical nomenclature standardization source in the current literature; e.g. Kinlock et al., 2022), we include two new phylogenetic backbones, one based on the botanical nomenclature of the Leipzig catalogue of vascular plants (LCVP) database (Freiberg et al., 2020), and the other based on the botanical nomenclature of the World Plants (WP) database (https://www.worldplants.de).
2. Package descriptionThis package includes two major components: the R package 'V.PhyloMaker2', which included a set of functions and a set of data files. Details about the package were described in the document entitled "Introduction to the 'V.PhyloMaker2' package", which was provided along with the package (see below for website address), and the document entitled "Descriptions of scenarios, functions and data files.doc", which is one of the supplementary files of this paper. We focus on several key components of the package below.
2.1. The mega-treesThree mega-trees were generated, each of which is in the Newick format.
2.1.1. GBOTB.extended.TPL.treThis mega-tree, which was based on the TPL nomenclature standardization system, is the same as GBOTB.extended.tre in the previous version of V.PhyloMaker reported in Jin and Qian (2019), except for the following changes. (1) One genus (with a single species, Agdestis clematidea) in the family Phytolaccaceae and one genus (with two species, Dactylaena microphylla, D. pauciflora) in the family Cleomaceae were separated from the other genera of their respective families in GBOTB.extended.tre, causing these two families being non-monophyletic. These two genera were removed. (2) Eleven species in the ten genera (i.e. Acanthosyris, Cervantesia, Comandra, Geocaulon, Jodina, Okoubaka, Pilgerina, Pyrularia, Scleropyrum, and Staufferia) were placed in the family Santalaceae in GBOTB.extended.tre, but they do not belong to the clade with all other species of the family in GBOTB.extended.tre. These ten genera belong to two families, Comandraceae (Comandra and Geocaulon) and Cervantesiaceae (the other eight genera), which were placed in the family Santalaceae in APG IV (Angiosperm Phylogeny Group, 2016) but were recognized as two separate families in some botanical literature (e.g. Flora of North America Editorial Committee, 2016). We retained these ten genera in GBOTB.extended.TPL.tre but we placed them in Comandraceae and Cervantesiaceae. The family Tiganophytaceae, with a single genus and a single species (Tiganophyton karasense), was established by Swanepoel et al. (2020) with the following phylogenetic relationships with its most closely related families: (Koeberliniaceae(Tiganophytaceae(Bataceae, Salvadoraceae))). We included the family Tiganophytaceae in GBOTB.extended.TPL.tre based on the phylogenetic relationships reported in Swanepoel et al. (2020). We estimated the divergence time of Tiganophytaceae (about 41.1 million years ago) based on data reported in Smith and Brown (2018) and Swanepoel et al. (2020). As a result, GBOTB.extended.TPL.tre included 74, 529 species of vascular plants in 10, 597 genera and 482 families.
2.1.2. GBOTB.extended.LCVP.treThis mega-tree, which was based on the LCVP nomenclature standardization system, was generated as follows. (1) We extracted the clade of pteridophytes from Zanne et al.'s (2014) phylogeny and attached it to GBOTB from Smith and Brown (2018). The divergence time between pteridophytes and seed plants was set according to Zanne et al.'s (2014) phylogeny, as in GBOTB.extended.tre (Jin and Qian, 2019). (2) We used the LCVP database (Freiberg et al., 2020) to standardize spellings and nomenclature for the plant names in the phylogeny, and removed duplicate plant names. (3) For those families that were present in GBOTB.extended.TPL.tre but were absent from the phylogeny derived from Zanne et al. (2014) and Smith and Brown (2018), we added them to the phylogeny, based on their locations in GBOTB.extended.TPL.tre. (4) As in GBOTB.extended.TPL.tre, we removed the genera Agdestis and Dactylaena from the phylogeny. Consequently, GBOTB.extended.LCVP.tre included 73, 420 species of vascular plants in 10, 134 genera and 482 families.
2.1.3. GBOTB.extended.WP.treThis mega-tree was generated by following the same procedure used to generate GBOTB.extended.LCVP.tre, except that the WP database (https://www.worldplants.de) was used to standardize spellings and nomenclature for the plant names in the phylogeny. GBOTB.extended.WP.tre included 72, 570 species of vascular plants in 10, 581 genera and 482 families.
A hybrid sign (×, x, or X) in front of a genus name may cause a problem when using V.PhyloMaker to generate a phylogeny. Consequently, hybrid sign indicating a hybrid genus was removed from each of the three mega-trees. Each hybrid species in each of the three mega-trees was indicated with "X" (e.g. Abelia_X_grandiflora).
2.2. The V.PhyloMaker2 packageThe V.PhyloMaker2 package contains three sets of functions and data files, and each set of functions and data files work with one of the three above-described mega-trees. The user chooses one mega-tree from the three based on the user's preference. The main function of the package is phylo.maker. All other functions and data files were shown in Table 1. Note that all functions are the same for the previous version and this new version of the software and work for the three mega-trees (Table 1). Descriptions for each type of functions and data files are available in table 1 of Jin and Qian (2019) and in the "Descriptions of scenarios, functions and data files.doc" file in Supplementary data with the present article. When the user loads V.PhyloMaker2 in R by the command library ("V.PhyloMaker2"), all functions shown in Table 1 are automatically loaded. The four types of data files are the mega-trees, the data frames with tip information (i.e. tips.info) that contains the family and genus assignments of every tip species in each mega-tree, and the two data frames (e.g. nodes.info.1.TPL and nodes.info.2.TPL generated by build.nodes.1 and build.nodes.2, respectively, based on the mega-tree GBOTB.extended.TPL.tre) that contain the genus- and family-level node and age information of each mega-tree. The user determines whether the function build.nodes.1 or build.nodes.2 is used to generate a phylogeny.
| V.PhyloMaker | V.PhyloMaker2 | |||
| TPL | TPL | LCVP | WP | |
| Function | ||||
| at.node | at.node | at.node | at.node | |
| bind.relative | bind.relative | bind.relative | bind.relative | |
| build.nodes.1 | build.nodes.1 | build.nodes.1 | build.nodes.1 | |
| build.nodes.2 | build.nodes.2 | build.nodes.2 | build.nodes.2 | |
| ext.node | ext.node | ext.node | ext.node | |
| int.node | int.node | int.node | int.node | |
| phylo.maker | phylo.maker | phylo.maker | phylo.maker | |
| Data | ||||
| GBOTB.extended.tre | GBOTB.extended.TPL.tre | GBOTB.extended.LCVP.tre | GBOTB.extended.WP.tre | |
| nodes.info.1.csv | nodes.info.1.TPL.csv | nodes.info.1.LCVP.csv | nodes.info.1.WP.csv | |
| nodes.info.2.csv | nodes.info.2.TPL.csv | nodes.info.2.LCVP.csv | nodes.info.2.WP.csv | |
| tips.info.csv | tips.info.TPL.csv | tips.info.LCVP.csv | tips.info.WP.csv | |
The phylo.maker function makes phylogenetic hypotheses under three scenarios (i.e. S1, S2 and S3), which are the same three scenarios as in S.PhyloMaker (Qian and Jin, 2016) and V.PhyloMaker (Jin and Qian, 2019), where details about the three scenarios are available (also see the "Descriptions of scenarios, functions and data files.doc" file in Supplementary data with the present article). The user determines which scenario is used to generate a phylogeny. Of the three scenarios, scenario 3 (i.e. S3) has been most commonly used.
The V.PhyloMaker2 package was written in RStudio (RStudio Team, 2015), is for use in the R language (R Core Team, 2016), and requires a standard installation of R and the 'ape' package (Paradis et al., 2004). When the user loads V.PhyloMaker2, the 'ape' package will be automatically loaded. The V.PhyloMaker2 package has been tested by different users on different computers with different versions of R (e.g. v.3.5.0, v.3.5.3, v.4.0.3, v.4.0.5, v.4.1.1, v.4.1.2, v.4.1.3, v.4.2.0), and all the tests were completed with success. If the user of V.PhyloMaker2 encounters a problem when installing the package, the problem may be resolved when the user installs any of the above-mentioned versions or a newer version of R.
V.PhyloMaker2 is an open-source package (published under GPL-2). The R package of V.PhyloMaker2, together with documentation, is available from GitHub (https://github.com/jinyizju/V.PhyloMaker2). The package can be installed in R using the install_github function in the 'devtools' package (Wickham et al., 2018), as follows, devtools: : install_github("jinyizju/V.PhyloMaker2").
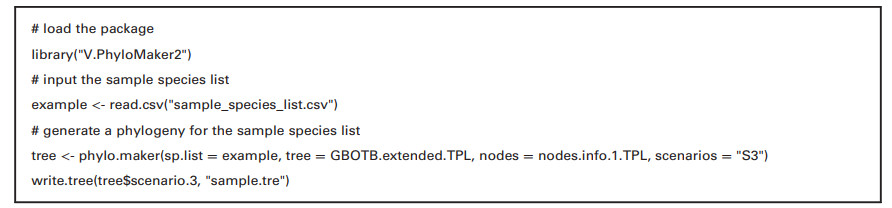
We included a sample species list (i.e. 'sample_species_list.csv') in Supplementary data, which the user of V.PhyloMaker2 may use as a sample species list to test this package. We provided an R script in Box 1 to run V.PhyloMaker2 on the sample species list. The same R script is also available in the file 'R script to run V.PhyloMaker2.txt' in Supplementary data.
We tested V.PhyloMaker2 on a species list extracted from LCVP (including 365, 198 accepted species) on an HP EliteBook Laptop with the following features: Windows 7 Enterprise; Intel® Core (TM) i5-2540 CPU @ 2.60 GHz; 4 GB RAM; 64-bit Operating System). After running about 42 h, V.PhyloMaker2 successfully generated a phylogeny for the species.
2.3. Preparation of species list for V.PhyloMaker2A species list to be used by V.PhyloMaker2 should be in the format of comma-separated values (csv) with five columns as shown in the file 'sample_species_list.csv' in Supplementary data. The first three columns have to be filled and the last two columns are optional. Because the spelling and nomenclature of species names in each of the three mega-trees were standardized according to its plant database (i.e. TPL, LCVP or WP), to maximize matching in species names between the user's species list and a mega-tree chosen by the user, we suggest the user standardize spelling and nomenclature of species according to the respective plant database. All names in the user's species list that are considered as synonyms in the plant database should be replaced with their accepted names in the plant database. Because taxa with terminal branches in each mega-tree are species-level taxa (i.e. binomials), infraspecific taxa (e.g. subspecies, variety, and forma) in the user's species list should be combined with their parental species. Duplicate names should be removed. A hybrid sign in front of a genus name should be removed, and a hybrid species should be indicated with "X" (e.g. Abelia_X_grandiflora) to maximize the matching of species names in the user's species list and species names in the mega-tree chosen by the user.
Box 1
R script for using V.PhyloMaker2 to generate a phylogenetic tree for the species in 'sample_species_list.csv' based on GBOTB.extended.TPL.tre, nodes.info.1.TPL, and scenario 3 (i.e. S3). When "S3" in the script is changed to "S2" or "S1", and "tree$scenario.3" is changed to "tree$scenario.2" or "tree$scenario.1", the script generates a phylogeny based on scenario S2 or S1, respectively, using GBOTB.extended.TPL.tre and nodes.info.1.TPL. When "TPL" in the script is changed to "LCVP" or "WP", the script generates a phylogeny based on GBOTB.extended.LCVP.tre or GBOTB.extended.WP.tre, respectively. When nodes.info.1 is changed to nodes.info.2, the script generates a phylogeny based on the nodes.info.2 data file.
|
The file 'family_list_for_V.PhyloMaker2.csv' in Supplementary data included a complete list of families of the world's flora of vascular plants. All the families in this family list were included in each of the three mega-trees with V.PhyloMaker2. Each species in the user's species list must be assigned with one of the families shown in the family list. The relationships between genera and families may be obtained from WP (https://www.worldplants.de) for pteridophytes and gymnosperms, and from Angiosperm Phylogeny Website (http://www.mobot.org/MOBOT/research/APweb/) for angiosperms. Species in the user's species list that do not have a family name in column 'family' will not be included in a phylogeny generated by V.PhyloMaker2. For a species that is present in both the mega-tree chosen by the user and in the user's species list, V.PhyloMaker2 will ignore the family name of the species in the user's species list.
Some species or genera that are absent from the mega-trees are sister to or closely related with some species or genera in the mega-trees. When the information of their closely related species or genera is given in the user's species list (columns 'species.relative' and 'genus.relative'), these species or genera will be attached to their closely related species or genera as sister species or genera in a phylogeny generated by V.PhyloMaker2 (see Fig. 1 of Jin and Qian (2019) for details).
3. DiscussionV.PhyloMaker2 is an updated and enlarged version of V.PhyloMaker published in Jin and Qian (2019). The major difference between the two versions is that V.PhyloMaker2 offers the user with more options to generate phylogenetic trees based on different botanical nomenclature systems. V.PhyloMaker2 is implemented with three botanical nomenclature systems (i.e. TPL, LCVP, and WP). Future versions of V.PhyloMaker may include more botanical nomenclature systems.
Because V.PhyloMaker2 is largely based on the version of V.PhyloMaker published in Jin and Qian (2019), we recommend the user of V.PhyloMaker2 reads the article by Jin and Qian (2019). When the user publishes the study based on a phylogenetic tree generated by V.PhyloMaker2, we suggest the user cites both the present article and the article by Jin and Qian (2019) in the user's publication. We also suggest the user cites the original articles of the mega-trees (i.e. Zanne et al., 2014; Smith and Brown, 2018) and the botanical nomenclature sources used in the mega-trees (see above for details).
The mega-trees GBOTB.extended.TPL.tre, GBOTB.extended.LCVP.tre and GBOTB.extended.WP.tre included 70%, 75% and 74% of all genera of vascular plants in TPL, LCVP and WP global databases, respectively. Our empirical studies show that GBOTB.extended.TPL.tre commonly includes 85%–95% of all genera in a regional or local species list (e.g. 87% for the seed plant flora of China, Qian et al., 2019; 94% for the Arctic flora, Qian et al., 2022). A recent study (Qian and Jin, 2021) shows that the results of a study using a species-level phylogeny resolved at the genus level with species being attached to their respective genera as polytomies is nearly identical to the results of a study based on a phylogeny fully resolved at the species level. Thus, phylogenetic trees generated by V.PhyloMaker2 based on each of the three mega-trees are expected to be robust for studies on community phylogenetics, and may be used in other phylogeny-based studies.
Author contributionH.Q. developed the idea of the software and worked on species lists and nomenclature in the species lists; Y.J. produced the package and wrote all R codes; H.Q. and Y.J. wrote the paper.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgementsWe are grateful to anonymous reviewers for their constructive comments on the manuscript and V.PhyloMaker2. This work was supported by the Natural Science and Technology Foundation of Guizhou Province [[2020]1Z013] (to Y.J.); and the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou Province [U1812401] (to Y.J.).
Angiosperm Phylogeny Group, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 181: 1-20. |
Beaulieu, J.M., Ree, R.H., Cavender-Bares, J., et al., 2012. Synthesizing phylogenetic knowledge for ecological research. Ecology, 93: S4-S13. DOI:10.1890/11-0638.1 |
Cornwell, W.K., Westoby, M., Falster, D.S., et al., 2014. Functional distinctiveness of major plant lineages. J. Ecol., 102: 345-356. DOI:10.1111/1365-2745.12208 |
Edwards, E.J., Smith, S.A., 2010. Phylogenetic analyses reveal the shady history of C4 grasses. Proc. Natl. Acad. Sci. U.S.A., 107: 2532-2537. DOI:10.1073/pnas.0909672107 |
Flora of North America Editorial Committee, 2016. Flora of North America North of Mexico. vol. vol. 12. Oxford, New York: Oxford University Press.
|
Freiberg, M., Winter, M., Gentile, A., et al., 2020. LCVP, the Leipzig catalogue of vascular plants, a new taxonomic reference list for all known vascular plants. Sci. Data, 7: 416. DOI:10.1038/s41597-020-00702-z |
Jin, Y., Qian, H., 2019. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography, 42: 1353-1359. DOI:10.1111/ecog.04434 |
Kinlock, N.L., Dehnen-Schmutz, K., Essl, F., et al., 2022. Introduction history mediates naturalization and invasiveness of cultivated plants. Glob. Ecol. Biogeogr.. DOI:10.1111/geb.13486 |
Paradis, E., Claude, J., Strimmer, K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20: 289-290. DOI:10.1093/bioinformatics/btg412 |
Qian, H., Jin, Y., 2016. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J. Plant Ecol., 9: 233-239. DOI:10.1093/jpe/rtv047 |
Qian, H., Jin, Y., 2021. Are phylogenies resolved at the genus level appropriate for studies on phylogenetic structure of species assemblages?. Plant Divers., 43: 255-263. DOI:10.1016/j.pld.2020.11.005 |
Qian, H., Deng, T., Ricklefs, R.E., 2022. Evolutionary assembly of the Arctic flora. Glob. Ecol. Biogeogr., 31: 396-404. DOI:10.1111/geb.13434 |
Qian, H., Deng, T., Jin, Y., et al., 2019. Phylogenetic dispersion and diversity in regional assemblages of seed plants in China. Proc. Natl. Acad. Sci. U.S.A., 116: 23192-23201. DOI:10.1073/pnas.1822153116 |
R Core Team, 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
|
RStudio Team, 2015. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, USA. URL http://www.rstudio.com/.
|
Smith, S.A., Brown, J.W., 2018. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot., 105: 302-314. DOI:10.1002/ajb2.1019 |
Smith, S.A., Beaulieu, J.M., Donoghue, M.J., 2009. Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol. Biol., 9: 37. DOI:10.1186/1471-2148-9-37 |
Smith, S.A., Beaulieu, J.M., Stamatakis, A., et al., 2011. Understanding angiosperm diversification using small and large phylogenetic trees. Am. J. Bot., 98: 404-414. DOI:10.3732/ajb.1000481 |
Swanepoel, W., Chase, M.W., Christenhusz, M.J.M., et al., 2020. From the frying pan: an unusual dwarf shrub from Namibia turns out to be a new brassicalean family. Phytotaxa, 439: 171-185. DOI:10.11646/phytotaxa.439.3.1 |
Webb, C.O., 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat., 156: 145-155. |
Wickham, H., Hester, J., Chang, W., 2018. devtools: tools to Make Developing R Packages Easier. R package version 1.13.6. https://CRAN.R-project.org/package=devtools.
|
Zanne, A.E., Tank, D.C., Cornwell, W.K., et al., 2014. Three keys to the radiation of angiosperms into freezing environments. Nature, 506: 89-92. DOI:10.1038/nature12872 |



