b. Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China, Ministry of Education, Southwest Forestry University, China
Coffee (Coffea arabica L.) is one of the most valued and exportable commodities in the world; a major source of revenue and employment in tropical countries (International Coffee Organization Trade Statistics, 2021). Coffee originated in Africa and belongs to the Rubiaceae family. The genus, Coffea, comprises of more than 124 species (Davis et al., 2011). C. arabica and C. canephora Pierre ex Froehner are the most commercially important cultivars and account for 70% and 30% of global production, respectively (International Coffee Organization Trade Statistics, 2021). Coffee is predominantly diploid, except for C. arabica, which is tetraploid due to a recent whole genome duplication between C. canephora (Robusta) and C. eugenioides in the Central Ethiopian plateaus (Geleta et al., 2012). There are over 40 C. arabica mutants reported so far, but only a few have traits of commercial interest. The most desirable traits comprise bean color, bean size, special flavor, male sterility or low caffeine content (Sant'Ana et al., 2018).
Coffee quality is a very complex trait influenced by numerous factors, such as environment and genotype (Cheng et al., 2016). Color variations in coffee have an impact on the aesthetics, quality, and commercial value of the fruit (Ságio et al., 2013). C. arabica's purple and yellow color variants account significantly for its demand and use in coffee beverages. Coffee is also high in bioactive compounds and metabolites that are beneficial to human health, such as those that nourish the kidneys, boost immunity, and fight cancer (Neto et al., 2011; Mishra and Slater, 2012; Denoeud et al., 2014; Knevitt, 2016). To support increased coffee production and genetic improvement, a better understanding of the molecular basis of fruit composition is required (Fenilli et al., 2007; Reis et al., 2009; Neto et al., 2011). Several biochemical studies have been conducted on the genetic and environmental factors that influence the accumulation of key fruit metabolites and compounds (Cheng et al., 2016; Sant'Ana et al., 2018). Changes in gene expression during coffee fruits ripening and coloration, including transcripts that regulate bean filling, and in response to stress, can be used to investigate genetic control of these processes (Sant'Ana et al., 2018; Mekbib et al., 2020). Earlier studies applied RT-PCR or microarrays to study coffee leaves or seedlings (Cheng et al., 2016; Sant'Ana et al., 2018). More recently, different tissues of Arabica coffee including flowers, leaves and fruit pericarp have been subjected to transcriptome analysis (Mishra and Slater, 2012; Knevitt, 2016; Mofatto et al., 2016). A genome-wide study recently reported identification of key genes regulating the lipid and diterpene contents of Arabica coffee fruits (Sant'Ana et al., 2018). However, analysis of the genetics of other essential components of the fruits contributing to coffee quality was not included in these studies (Mofatto et al., 2016; Sant'Ana et al., 2018; Tran et al., 2018). Additionally, gaps remain in knowledge of gene interactions and how expression varies at the global scale at specific ripening stages and finally determines the quality and regeneration capability of coffee fruits.
Plant volatiles, strigolactones, and phytohormones such as abscisic acids are produced when carotenoids are degraded (Tanaka et al., 2008; Wang et al., 2010; Yahia et al., 2017). Furthermore, carotenoids are widely used in the food and pharmaceutical industries; for example, lutein and similarly structured carotenoids can protect retinal cells in the eye from oxidative stress, and several studies have suggested that supplementing with lutein can help maintain eye health and reduce the risk of various chronic eye diseases (Inbaraj et al., 2008, Wang et al., 2010).
Genes that code for carotenoid biosynthesis and degradation enzymes are expressed in specific patterns in different organs and at different stages of development. Various transcription factors finely regulate their expression patterns, but only a few studies have investigated these regulatory pathways thoroughly (Riechmann et al., 2000; Dubos et al., 2010; Licausi et al., 2013). Welsch et al. (2007) showed that the PSY binding factor (AP2/ERF), RAP2.2, binds to the Arabidopsis thaliana-APETALA2 (AP2) and PDS promoters. The knockdown mutant was shown to have a decreased PSY and PDS expression and lowers its carotenoid content by 30% (Welsch et al., 2007; Zhang et al., 2020). There has also been evidence that a Phytochrome-Interacting Factor (PIF) binds to and inhibits PSY promoter expression (Li et al., 2010). Similarly, the RIN (MADS box transcription factor), which interacts with the promoter of PSY and participates in carotenoid accumulation in tomatoes (Solanum lycopersicum) fruits, has been identified (Ye et al., 2015). These transcription factors generally regulate the accumulation of carotenoids in plants. Zhang et al. (2020) revealed recently in a transcriptome study that carotenoid degradation genes are key determinants of carotenoid content of marigold flowers. Flavonoids, which are phenolic compounds determine flower, fruit, and seed color in a variety of plant species (Falcone Ferreyra et al., 2012; Rashid et al., 2019). Anthocyanins are water-soluble, synthesized in the cytosol, stored in the vacuole, and encode for the blue, purple, red, or white colors of flowers and fruits (Ben-Meir et al., 2002; Dai et al., 2009; Yan and Kerr, 2013). They are also key members of the phenylpropanoid biosynthetic pathway (Malien-Aubert et al., 2001; Dai et al., 2009). These compounds have three aromatic rings, which can be replaced with acyl, hydroxyl, methyl, or sugar depending on the plant species (Ben-Meir et al., 2002). Anthocyanin biosynthesis is regulated by structural and regulatory genes involved in the formation of enzymes and the regulation of specific enzyme expression (Zhuang et al., 2019). The formation of spots in petunia flowers is caused by differential expression of anthocyanin structural genes (Koseki et al., 2005). Important genes participating in the anthocyanin biosynthetic pathway encode 3-hydroxylase flavanone, 3′-hydroxylase flavonoid, 4-reductase dihydroflavonol synthase chalcone, isomerase chalcone and 4-hydroxylase cinnamate, 4-coumaroyl CoA ligase, S-transference glutathione, S-dioxygenase glutathione, ammonia-lyase phenylalanine, and GL-UDP-flavonoid transmission (Holton and Cornish, 1995; Malien-Aubert et al., 2001; Tanaka et al., 2008; Kumar and Yadav, 2013; Yan and Kerr, 2013).
Transcriptome profiling allows for the identification and quantification of transcripts, differentially expressed genes and has been shown to be effective in detecting molecular differences in various tissues (Ye et al., 2015; Yuyama et al., 2016; Cheng et al., 2017, 2018). RNA-Seq has been used to investigate the effects of altitude on coffee gene expression. RNA-Seq was used to investigate the effects of temperature on coffee sub-genomes and regulation of coffee gene expression (Cheng et al., 2018). On an Illumina HiSeq 2000, researchers compared de novo transcriptome assemblies between Coffea arabica and C. eugenioides (Yuyama et al., 2016). Using Illumina HiSeq 2000 technology, a transcriptome analysis of Arabica coffee leaves, flowers, and perisperm development revealed a similar pattern of gene transcription and diterpenes synthesis (Ivamoto et al., 2017).
In plants, transcription factors (TFs) regulate gene expression in a variety of biological processes (Riechmann et al., 2000; Licausi et al., 2013). In plants, the regulatory mechanisms of several genes involved in the anthocyanin biosynthetic pathway have been well studied (Tanaka et al., 2008; Wang et al., 2010; Ságio et al., 2013). Antirrhinum, for example, has been found to have flower-specific MYB protein for activation of genes involved in phenylpropanoid biosynthesis (Allan et al., 2008; Lloyd et al., 2017; Jian et al., 2019). PAP1/MYB75, PAP2/MYB90, MYB113, and MYB114 regulate the biosynthesis of anthocyanins in Arabidopsis (Takos et al., 2006; Machemer, 2011; Xu et al., 2018). TTG1 (WD40), GL3/EGL3/TT8 (bHLH), and PAP1/PAP2/MYB113/MYB114 (MYB) have also been identified as components of a WBM complex that regulates anthocyanin biosynthesis (Varaud et al., 2011; Hao et al., 2012; Ikeda, 2013). In Arabidopsis and Petunia, the role of WRKY TFs in the co-regulation of anthocyanin via the MBW complex has been documented (Verweij et al., 2016; Lloyd et al., 2017). In addition, members of the bZIP TF family are potential regulators of the anthocyanin pathway in apples (Espley et al., 2007).
Despite extensive knowledge of the molecular mechanisms of color formation in various plant species, the role of carotenoids and anthocyanins in Arabica coffee fruit color formation remains unknown. In this study, we used transcriptome sequencing to identify key candidate genes for the carotenoid and anthocyanin biosynthetic pathways that underpin color formation of Coffea arabica fruits.
2. Materials and methods 2.1. Plant materials and growth conditionsThe healthy and quality seeds of the different varieties of Coffea arabica were obtained from the Coffee Research Center, Institute of Tropical and Subtropical Cash Crops, Yunnan Academy of Agricultural Sciences, China. The seeds were planted at the Scientific Research Station of the Institute of Tropical and Subtropical Cash Crops, Lujiang Town, Baoshan City (24°58′17.93″N; 98°52′43.28″E), China. The area belongs to the low-latitude quasi-tropical monsoon rainforest with a dry and hot valley transition type of climate with little rainfall, large evaporation from the ground, dry and wet seasons and at 750 m above sea level. The dry season in the area generally starts from November to May of the following year, with sufficient sunlight, and large temperature difference between day and night. The annual average temperature and rainfall is 21.5 ℃ and 755.3 mm, respectively.
The four fruit-colored coffee varieties (orange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF)) were planted in four rows, with 1 m × 2 m spacing × row spacing and each fruit-colored coffee plant had more than 15 plants. The marginal plants at both ends of each row were removed during sampling. All relevant agronomic practices were performed throughout the study period. Five plants were randomly selected, fully mature and uniformly colored fresh coffee fruits were picked from the first branch in the middle of the plant. Five fresh fruits were picked for each sample. The harvested fruits were labeled and put it in a liquid nitrogen for transfer and storage in −80 ℃ ultra-low temperature refrigerator.
2.2. RNA extraction and cDNA library preparationOrange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF)) of Coffea arabica varieties were harvested from the upper branches. Total isolated RNAs from 12 samples (triplicate for each sample) was processed individually in accordance with Cheng et al. (2018) and Furtado et al. (2015). Total RNA was evaluated with an Agilent RNA 6000 nano kit and chips through a Bioanalyzer 2100 (Agilent Technologies, California, USA). A standard 18× Truseq total RNA library prepared by using an additional Ribo-Zero kit was then conducted. Samples were sequenced on Illumina HiSeq 4000 platform (2–150 bp paired-end reading).
2.3. Reads mining and RNA-Seq analysesCLC Genomic Workbench 10.0.1 (CLC Bio, Denmark) was used to process raw reads as follows: (1) Indexes and adapters were trimmed. (2) Reads that did not match the PHRED score (0.01) or length (40 bp) were discarded. (3) The processed reads were subjected to RNA-Seq analysis (read similarity 0.9, length similarity 0.8). The expression parameter was Transcripts Per Kilobase Million (TPM) and the reference was a recently published long-read coffee bean transcriptome (Denoeud et al., 2014; Tran et al., 2018). This study did not consider outlier expression values, which are classified by coefficient of variation and standard deviation.
2.4. Analysis of transcript levels by qRT-PCRThe qRT-PCR protocol was adapted from Rodriguez-Villalo et al. (2009) and run on a LightCycler 480 instrument with Fast Start Universal SYBR Green Master Mix (Roche, USA). The Actin 7 gene was utilized to normalize for transcript levels. The primer sequences are presented in Table S1.
2.5. Carotenoid quantificationTotal carotenoids were extracted and measured spectrophotometrically according to a previously established procedure (Fraser et al., 2008). Concentration in a given sample was estimated relative to the fresh weight. Data collected were subjected to analysis of variance in GraphPad Prism statistical software (v.8, GraphPad Software, San Diego California USA, www.graphpad.com) at P ≤ 0.05.
2.6. Statistical analysesTranscripts Per Kilobase Million (TPM) (> 1) was used to filter transcripts expressed in each fruit sample. Functional annotations were performed and the BLAST2GO was used to search for GO terms and KEGG pathway distribution (Kanehisa and Goto, 2000; Kanehisa et al., 2016). An online tool, InteractiVenn (Heberle et al., 2015) was used to create the Venn diagrams. TPM (> 500) was used to filter out highly expressed transcripts. For significant fruit coloration, the differential gene expression tool for RNA-seq (CLC) was used. The false discovery rate (FDR) p-value correction (0.01) and maximum group means (TPM 10) were used to filter differentially expressed genes (DEGs). Mercator (Lohse et al., 2015) and Mapman (v.3.6.0) (Usadel et al., 2009) were used to create the key storage component association with DEGs. Mapman-annotated storage DEGs and candidate genes from the targeted analysis were used to build the co-expression network. The WGCNA's built-in Web MEV package in R (cutoff 0.9) (Langfelder and Horvath, 2008) and Cytoscape 3.5 (Lopes et al., 2010) were used to log 2(x) transform gene expression of the transcripts prior to analysis. Before log2 transformation, transcripts with an expression value of 0 were assigned the value 0.0001.
3. Results 3.1. Fruit samples, RNA sequencing, quality control and functional annotationFruits of four Coffea arabica varieties (orange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF)) (Fig. 1) were subjected to transcriptome profiling to identify genes, key biosynthesis pathways and transcription factors responsible for fruit coloration. In total, 12 libraries (4 varieties of different colors × 3 biological repeats) were sequenced, yielding average total reads of 55, 706, 891 in RF and 52, 840, 623 in YF (Table 1). From these, 95.07%, 96.61%, 96.56% and 96.32% were clean reads from ORF, PF, RF and YF, respectively (Table 1). In addition, the error rates, Q30% and GC contents ranged from 0.02% to 0.03%, 94.21% to 94.70% and 43.70% to 44.08%, respectively (Table 1), indicating that the transcriptome results were valid for further downstream analyses.

|
| Fig. 1 Fully matured and ripe fruit samples of four varieties of Coffea arabica used for transcriptome profiling. (A) Purple colored fruits. (B) Red colored fruits. (C) Orange colored fruits. (D) Yellow colored fruits. |
| Samplea | Raw Reads | Clean Readsb | Clean Base (Gb) | Error Rate (%) | Q30 (%) | GC Content (%) |
| ORF1 | 55, 129, 804 | 53, 308, 704 (96.70) | 8.00 | 0.02 | 94.66 | 43.85 |
| ORF2 | 55, 161, 990 | 51, 742, 058 (93.80) | 7.76 | 0.02 | 94.66 | 43.82 |
| ORF3 | 53, 420, 648 | 50, 583, 620 (94.69) | 7.59 | 0.02 | 94.64 | 43.99 |
| Average ORF | 54, 570, 814 | 50, 583, 620 (95.07) | 7.78 | 0.02 | 94.65 | 43.89 |
| PF1 | 55, 278, 072 | 53, 420, 692 (96.64) | 8.01 | 0.02 | 94.51 | 43.93 |
| PF2 | 54, 939, 944 | 53, 042, 492 (96.55) | 7.96 | 0.02 | 94.70 | 43.89 |
| PF3 | 52, 826, 986 | 51, 052, 692 (96.64) | 7.66 | 0.02 | 94.59 | 43.93 |
| Average PF | 54, 348, 334 | 52, 505, 292 (96.61) | 7.88 | 0.02 | 94.60 | 43.92 |
| RF1 | 56, 274, 238 | 54, 411, 070 (96.69) | 8.16 | 0.02 | 94.63 | 43.82 |
| RF2 | 54, 029, 490 | 52, 264, 794 (96.73) | 7.84 | 0.02 | 94.29 | 43.70 |
| RF3 | 56, 816, 946 | 54, 696, 622 (96.27) | 8.20 | 0.02 | 94.65 | 43.77 |
| Average RF | 55, 706, 891 | 53, 790, 829 (96.56) | 8.07 | 0.02 | 94.52 | 43.76 |
| YF1 | 55, 316, 476 | 53, 071, 382 (95.94) | 7.96 | 0.02 | 94.55 | 43.98 |
| YF2 | 52, 473, 072 | 50, 491, 614 (96.22) | 7.57 | 0.02 | 94.39 | 44.08 |
| YF3 | 50, 732, 320 | 49, 129, 920 (96.84) | 7.37 | 0.03 | 94.21 | 44.03 |
| Average YF | 52, 840, 623 | 50, 897, 639 (96.32) | 7.63 | 0.02 | 94.38 | 44.03 |
| a Orange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF). b Those in parenthesis represent proportion of clean reads to total reads expressed in percentage. All analyses were done triplicate. |
||||||
From the above, 39, 938 genes were identified from the 12 libraries. The fragments per kilobase of exon per million fragments mapped (FPKM) distribution and principal components analysis (PCA) (Fig. 2A, B) revealed different transcriptomes in the four Coffea arabica varieties. More specifically, the PCA showed that transcriptional regulation of ORF, PF, RF and YF are different with a 45.51% variation accounted for by the first two principal component axes (Fig. 2B). From the total genes expressed, 29, 081 genes were annotated in at least one of the four public databases (Kyoto Encyclopedia of Genes and Genomes (KEGG), KEGG orthologous group (KOG), Swiss protein (Swissprot) and Gene Ontology (GO)) (Fig. 3). These coupled with high correlation among biological repeats of the same sample (Fig. S1) and thus further validating our transcriptome results for downstream analyses.

|
| Fig. 2 (A) Fragments per kilobase of exon per million fragments mapped (FPKM) distribution among the four varieties of Coffea arabica in triplicate. (B) Principal components analysis of four fruit samples in triplicate based on FPKM. Orange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF). |

|
| Fig. 3 Total expressed genes and their functional annotation among the four varieties (orange, purple, red and yellow colored fruits) of Coffea arabica in four public databases. Kyoto Encyclopedia of Genes and Genomes (KEGG), KEGG orthologous group (KOG), Swiss protein (Swissprot) and Gene Ontology (GO). |
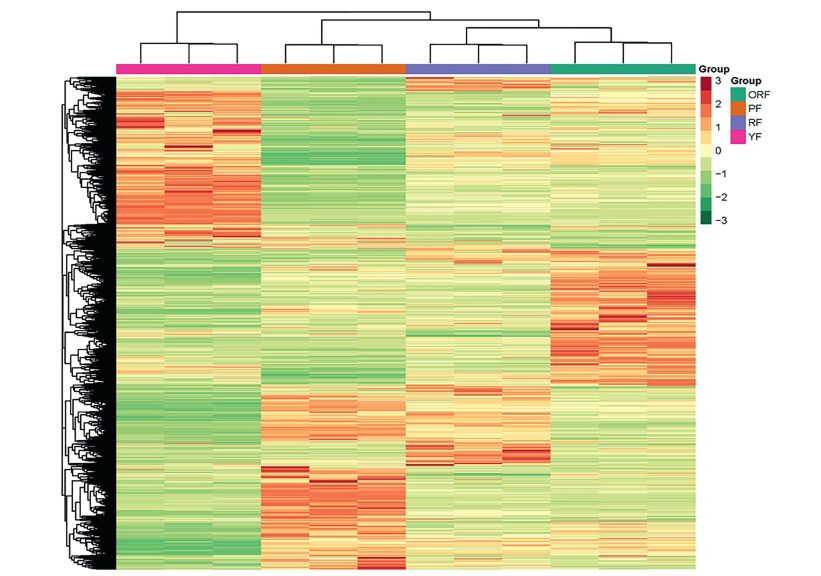
We adopted the stringent criteria of log 2 fold change (log2FC) ≥ 1 and the FDR with adjusted P-value < 0.05 to screen for DEGs in pairwise group comparisons. A total of 9877 genes showed differential expressions among the six pairwise groups (Fig. 4). We used the hierarchical clustering method for the DEGs with their FPKMs to heatmap the four varieties. Two major clusters were obtained (Fig. 5). The Cluster 1 consisted of only YFs whereas Cluster 2 comprised two sub-clusters: PFs as one sub-cluster and, RFs and ORFs formed another. This together with the results of K-means clustering (Fig. S2) indicates that YF transcriptome is quite different from that of PF, RF and YF samples. RF and ORF relatively have a similar transcriptional regulation compared to PF. Therefore, subsequent comparisons were made with YF as control relative to the other three fruit samples (PF, RF and ORF), thus YF_vs_PF, YF_vs_RF and YF_vs_ORF. A total of 2, 745, 781 and 1224 DEGs were detected in YF_vs_PF, YF_vs_RF and YF_vs_ORF, respectively (Fig. S3). By comparing the three list of DEGs, 1732 DEGs were mutually detected among the three pairwise groups (Fig. S3). These indicate that some genes were highly conserved among the fruits of the four varieties.

|
| Fig. 4 Differentially expressed genes among orange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF) of Coffea arabica. Total is the summation of down- and up-regulated genes. |
|
| Fig. 5 Hierarchical clustering heatmap based on fragments per kilobase of exon per million fragments mapped of differentially expressed genes (DEGs) of four Coffea arabica varieties (orange colored fruits (ORF); purple colored fruits (PF); red colored fruits (RF) and yellow colored fruits (YF)). |
To validate the results of RNA-seq, we selected 12 genes involved in the carotenoid and anthocyanin biosynthesis pathways to perform a qRT-PCR analysis. The results from this experiment were consistent with the RNA-seq results which confirms the reliability and consistency of our sequencing results (Fig. S4).
In order to identify the enriched pathways involved in fruit coloration in Coffea arabica, we screened the pathways in each pairwise group based on the P-value ≤ 0.05. A total of 43 pathways were enriched among the three pairwise groups (Table S2). These pathways include ABC transporters; carotenoid biosynthesis; cutin, suberine and wax biosynthesis; diterpenoid biosynthesis; nitrogen metabolism; sesquiterpenoid and triterpenoid biosynthesis; phenylpropanoid/flavonoid/flavone and flavonol biosynthesis and several other metabolic pathways (Table S2). These results indicate that fruit coloration in C. arabica may be correlated with fruit quality traits such as flavor, aroma, and sugar level.
3.3. Targeted analyses of carotenoid biosynthesis associated with differentially expressed genes among yellow colored, purple, red and orange colored fruitsIt is widely reported in literature that pigmentation in plants are determined by four classes of compounds namely anthocyanins, chlorophylls, carotenoids and betalains (Langfelder and Horvath, 2008; Zhao et al., 2017). We illustrated the regulation of carotenoid biosynthesis pathway among the four varieties with YF_vs_PF, YF_vs_RF and YF_vs_ORF (Fig. 6). Thirty-two genes that encode enzymes in the carotenoid biosynthetic pathway were detected in the three pairwise group comparisons. These comprised one phytoene synthase (PSY), one lycopene epsilon-cyclase (LYCE), four beta-ring hydroxylase (CHXB/E or LUT5), two beta-carotene isomerase (BCIS), four carotenoid cleavage dioxygenases 7 and 8 (CCD7 and CCD8), three zeaxanthin epoxidase (ZEP), one violaxanthin de-epoxidase (VDE), two abscisic-aldehyde oxidase (AAO3), six xanthoxin dehydrogenase (ABA2), one 9-cis-epoxycarotenoid dioxygenase (NCED), five (+)-abscisic acid 8′-hydroxylase (CYP7070A) and two abscisate beta-glucosyltransferase (AOG) (Fig. 6). The PSY (gene-LOC113688784) together with β-CHY (gene-LOC113730013), CCD7 (gene-LOC113728842), NCED (gene-LOC113689681) and ABA2 (gene-LOC113729473) were in higher abundance in YF than either PF, RF or ORF (Fig. 6). The higher abundance of these genes may have accounted for more enzymatic activity in the carotenoid biosynthesis and yellow color formation in YF.

|
| Fig. 6 A: Schematic diagram of carotenoid biosynthetic pathway and known enzymes (red font) and B: their corresponding differentially expressed genes in the present study in Coffea arabica (the heatmap gives log2fold change between the yellow (YF) colored fruits relative to purple (PF), red (RF) and orange (ORF) colored fruits. Expression with positive values in the legend of the heatmap at the right side of the figure means lower expression in YF while negative values means higher expression relative to either PF, RF or ORF (See heatmap key in B). GGPP: geranyl–geranyl diphosphate; PSY: phytoene synthase; LYCE: lycopene epsilon-cyclase [EC: 5.5.1.18]; CHXB/E: beta-ring hydroxylase [EC: 1.14.15.24] (or LUT5); BCIS: beta-carotene isomerase, CCD7 and CCD8: carotenoid cleavage dioxygenases 7 and 8, respectively [EC: 1.13.11.68–69]; ZEP: zeaxanthin epoxidase [EC: 1.14.15.21]; VDE: violaxanthin de-epoxidase [EC: 1.23.5.1]; BCIS: beta-carotene isomerase [EC: 5.2.1.14]; AAO3: abscisic-aldehyde oxidase [EC: 1.2.3.14]; ABA2: xanthoxin dehydrogenase [EC: 1.1.1.288]; NCED: 9-cis-epoxycarotenoid dioxygenase [EC: 1.13.11.51]; CYP7070A: (+)-abscisic acid 8′-hydroxylase [EC: 1.14.14.137]; and AOG: abscisate beta-glucosyltransferase [EC: 2.4.1.263]. |
β-Carotene is converted into isomers of which, 9-cis-β-carotene is reported to be the most abundant product of β-Carotene (Guo et al., 2008). Two BCIS (gene-LOC113704159 and gene-LOC113704210) showed higher abundance in either PF, RF or ORF than in YF. Similarly, 1 LYCE (gene-LOC113696970), 2 CHXE (gene-LOC113710427 and gene-LOC113710429), 3 ZEP (gene-LOC113699006, gene-LOC113701789 and gene-LOC113707477), 1 VDE (gene-LOC113734378), 2 AAO3 (gene-LOC113704288 and gene-LOC113725547) and 4 CYP707A (gene-LOC113710025, gene-LOC113729282, gene-LOC113722535 and gene-LOC113731421) had higher transcript abundance in either PF, RF and ORF relative to YF in order of RF > ORF > PF. These imply that these genes play key role in red coloration in coffee fruits.
3.4. Targeted analyses of phenylpropanoid/flavonoid/anthocyanin biosynthesis among the three pairwise groupsIn addition to carotenoid biosynthesis, flavonoids are reported to be major molecules involved in plant pigmentation particularly anthocyanins (Lai et al., 2014). Phenylpropanoids/flavonoids are precursors of anthocyanin biosynthesis, therefore we explored key DEGs involved in phenylpropanoid/flavonoid biosynthesis. A total of 82 genes which encode for 18 enzymes involved in phenylpropanoid-flavonoid biosynthesis in various steps prior to anthocyanin biosynthesis were identified (Fig. S5A–B). Most of these genes were lowly expressed in YF compared to either PF, RF or ORF. For instance, among the 4-phenylalanine ammonia-lyase (PAL) genes (gene-LOC113707625, gene-LOC113720652, gene-LOC113727571 and gene-LOC113725801), only gene-LOC113727571 had higher expression in YF than in PF while the remaining 3 genes had higher expression in either PF, RF or ORF than YF (Fig. S5A). These genes may be responsible for color break from purple to yellow, orange and purple. This is not surprising as the conversion of phenylalanine to anthocyanins requires a series of reactions catalyzed by enzymes initiated by ammonia and the action of PAL plays significant catabolic role in producing nitrogen and carbon (Khoo et al., 2017; Cui et al., 2021). Also, two genes (gene-LOC113706397 and gene-LOC113704221) associated with bi-functional dihydroflavonol 4-reductase (DFR) which has been reported to reduce dihydroflavonol to leucoanthocyanidins in both anthocyanin biosynthesis and proanthocyanidins accumulation were mostly expressed higher in either PF, RF or ORF than YF.
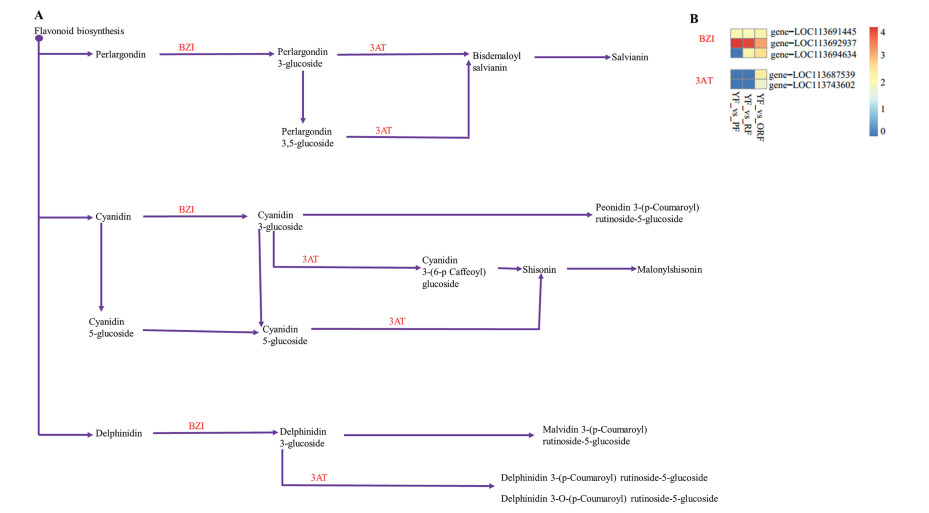
Anthocyanins are responsible for the red, purple, orange and blue colors in fruits and vegetables (Khoo et al., 2017), therefore we mined the expressed anthocyanins-related genes. In all, we detected five genes linked to enzymes, 3 anthocyanidin 3-O-glucosyltransferase (BZI) and 2 anthocyanidin 3-O-glucoside 6″-O-acyltransferase (3AT)) (Fig. 7). The BZI genes (gene-LOC113691445, gene-LOC113692937 and gene-LOC113694634) and 3AT genes (gene-LOC113687539 and gene-LOC113743602) expressed nearly two times higher in either PF, RF or ORF than YF. Cumulatively, these genes expressions followed PF > ORF > RF > YF. These suggest anthocyanin biosynthesis in YF may have been limited during fruit development as compared to either PF, RF or ORF.
|
| Fig. 7 Schema of last steps of the anthocyanin biosynthesis pathway (A) and their corresponding differentially expressed genes in the present study (B) presented in heatmap based on the log2fold change of fragments per kilobase of exon per million fragments mapped of differentially expressed genes involved in anthocyanin biosynthesis between yellow colored fruit against either purple, red or orange color fruit (YF, PF, RF and ORF, respectively) of Coffea arabica. BZI: anthocyanidin 3-O-glucosyltransferase [EC: 2.4.1.115] and 3AT: anthocyanidin 3-O-glucoside 6″-O-acyltransferase [EC: 2.3.1.215]. |
Transcription factors (TFs) regulate gene expression (Ullah et al., 2019; Mitsis et al., 2020), thus they are involved in the regulation of the structural genes of the pathways involved in carotenoid and anthocyanin accumulation in several fruits and vegetables (Stanley and Yuan, 2019; Cui et al., 2021). Majority of the TFs detected in this study were bHLH, MYB, AP2/ERF, NAC, MADS, AUX/IAA and WRKY with varied levels of regulation (Fig. 8). The regulatory mechanisms underlying transcriptional control of anthocyanin biosynthesis have widely been elucidated in various fruits and are known to involve MYB, bHLH and WD40 transcriptional complexes (Lloyd et al., 2017). However, for carotenoid biosynthesis, reports on the major transcriptional regulations are largely inconsistent (Ruiz-Sola and Rodríguez-Concepción, 2012; Lu et al., 2018; Stanley and Yuan, 2019; Zhou et al., 2019). In all, 56 and 43 DEGs encode MYB and bHLH TFs, respectively (Fig. 8A–G). Most of the MYB and bHLH regulated genes expressed higher in either PF, RF or ORF than YF (Fig. 8A, B). This implies that MYB and bHLH TFs may regulate carotenoid biosynthesis positively in yellow colored fruit with same trend. A contrary trend was observed for anthocyanins in either PF, RF or ORF. For example, 4 MYB gene (gene-LOC113728720, gene-LOC113716527, gene-LOC113716576 and gene-LOC113738429) and 4 bHLH gene (gene-LOC113721953, gene-LOC113733739, gene-LOC11374297 and gene-LOC113696134) expressed at least two times higher in either PF, RF or ORF than YF. However, 1 bHLH gene (gene-LOC113734588) was completely not detected in RF or YF; whereas 1 MYB gene (gene-LOC113703529) was completely absent in YF. The genes detected with MYB or bHLH TF for the fruit coloration in the four fruits of C. arabica could further be screened to select candidate genes for functional validation to unravel molecular mechanisms underlying color formation in each of the four varieties.

|
| Fig. 8 Major transcription factors involved in the regulation of differentially expressed genes in Coffea arabica among the three pairwise groups (yellow colored fruit (YF) relative to either purple (PF), red (RF) or orange (ORF) color fruit). Heatmap based on log2fold change values between YF_vs_PF, YF_vs_RF and YF_vs_ORF. (A) MYB. (B) bHLH. (C) AP2/ERF. (D) AUX/IAA. (E) MADS. (F) NAC. (G) WRKY. |
Again, MYB, bHLH TFs, AP2/ERF, NAC, MADS, AUX/IAA, and WRKY TFs encode 47, 12, 21, 26 and 31 DEGs (Fig. 8C–G). Majority of these TFs expressed higher in either PF, RF or ORF than in YF (Fig. 8C–G), indicating that these TFs may positively regulate structural genes involved in color formation in Coffea arabica. These results suggest that other TFs such as AP2/ERF, NAC, MADS, AUX/IAA and WRKY equally warrant further research attention.
3.6. Detection and quantification of carotenoid components among the four varieties of Coffea arabicaWe further profiled quantitatively the various compositions of carotenoid among the four varieties based on the higher number of DEGs involved in carotenoid biosynthesis. We performed analysis of variance (ANOVA) and the means were separated by Duncan Multiple Range Test at 0.05 probability level. In all, 33 constituents of carotenoids were detected among the four varieties with different colors evaluated in this study (Table 2). The predominant constituents include α-carotene, β-carotene, β-cryptoxanthin, lutein, lutein dilaurate, lutein dipalmitate, lutein dimyristate, lutein laurate, lutein palmitate, lutein myristate, zeaxanthin-laurate-palmitate, zeaxanthin dimyristate, and zeaxanthin palmitate (Table 2). Among these, lutein dimyristate was the most abundant in ORF (37.00 ± 0.72 μg/g) and RF (35.63 ± 1.04 μg/g). Lutein myristate was significant in RF (34.20 ± 0.91 μg/g) followed by YF (31.70 ± 0.69 μg/g), ORF (29.37 ± 0.94 μg/g) and PF (21.13 ± 0.35 μg/g) whereas lutein was found at least 1.2 times higher in YF than either in PF, ORF or RF (Table 2). These suggest that carotenoid pigment (its component lutein esters) play pivotal role in fruit coloration in YF, RF, ORF and least in PF.
| Components | Carotenoid compounds (μg/g) in each sample | |||
| YF | PF | RF | ORF | |
| Lutein dimyristate | 31.70 ± 1.34b | 22.20 ± 0.91c | 35.63 ± 1.04a | 37.00 ± 0.72a |
| Lutein myristate | 31.70 ± 0.69b | 21.13 ± 0.35c | 34.20 ± 0.91a | 29.37 ± 0.94b |
| Lutein | 35.27 ± 0.28a | 23.33 ± 0.95c | 27.67 ± 0.47b | 24.60 ± 0.36c |
| Lutein palmitate | 9.50 ± 0.30b | 6.92 ± 0.37c | 12.10 ± 0.85a | 11.87 ± 0.20a |
| Zeaxanthin dimyristate | 6.99 ± 0.07a | 3.92 ± 0.25d | 5.65 ± 0.16b | 4.74 ± 0.07c |
| β-Carotene | 3.71 ± 0.07c | 2.48 ± 0.10d | 4.27 ± 0.07a | 3.98 ± 0.09b |
| Lutein laurate | 3.79 ± 0.12b | 2.21 ± 0.11d | 4.42 ± 0.14a | 3.16 ± 0.02c |
| β-Cryptoxanthin | 1.38 ± 0.05c | 1.68 ± 0.23b | 1.45 ± 0.07c | 2.34 ± 0.03a |
| Lutein dipalmitate | 1.86 ± 0.20bc | 1.32 ± 0.07c | 2.72 ± 0.25a | 2.17 ± 0.16ab |
| Lutein dilaurate | 1.78 ± 0.01b | 1.27 ± 0.08c | 2.67 ± 0.20a | 2.01 ± 0.03b |
| Zeaxanthin palmitate | 1.51 ± 0.14b | 1.02 ± 0.08c | 1.98 ± 0.13a | 1.79 ± 0.07ab |
| α-Carotene | 1.94 ± 0.06a | 0.75 ± 0.05c | 1.68 ± 0.07b | 1.71 ± 0.01b |
| Zeaxanthin-laurate-palmitate | 1.79 ± 0.03a | 0.82 ± 0.02c | 1.39 ± 0.09b | 1.08 ± 0.13c |
| Zeaxanthin | 1.81 ± 0.03a | 0.75 ± 0.03c | 0.73 ± 0.05c | 0.96 ± 0.05b |
| Volaxanthin palmitate | 1.00 ± 0.01a | 1.00 ± 0.02a | 1.00 ± 0.03a | 0.90 ± 0.02a |
| Violaxanthin myristate | 0.95 ± 0.06ab | 1.07 ± 0.04a | 0.99 ± 0.25a | 0.84 ± 0.02b |
| Zeaxanthin-myristate-palmitate | 0.98 ± 0.03a | 0.51 ± 0.02c | 0.86 ± 0.05ab | 0.75 ± 0.07b |
| 5, 6 epoxy-lutein-caprate-palmitate | 0.34 ± 0.03b | 0.41 ± 0.02b | 0.45 ± 0.03b | 0.69 ± 0.07a |
| α-Cryptoxanthin | 0.81 ± 0.02a | 0.50 ± 0.01c | 0.81 ± 0.03a | 0.65 ± 0.04b |
| Lutein caprate | 0.77 ± 0.02b | 0.38 ± 0.02d | 0.91 ± 0.03a | 0.54 ± 0.02c |
| Violaxanthin | 0.54 ± 0.04a | 0.60 ± 0.04a | 0.35 ± 0.03b | 0.34 ± 0.02b |
| Neoxanthin | 0.41 ± 0.02a | 0.34 ± 0.01b | 0.21 ± 0.01d | 0.26 ± 0.01c |
| Violaxanthin-myristate-laurate | 0.23 ± 0.02ab | 0.24 ± 0.01a | 0.24 ± 0.02a | 0.20 ± 0.00b |
| Violaxanthin dimyristate | 0.21 ± 0.03a | 0.20 ± 0.04a | 0.20 ± 0.01a | 0.17 ± 0.06a |
| Violaxanthin-myristate-palmitate | 0.17 ± 0.02a | 0.16 ± 0.01a | 0.15 ± 0.01a | 0.17 ± 0.01a |
| Antheraxanthin dipalmitate | 0.11 ± 0.04a | N/A | 0.12 ± 0.06a | 0.12 ± 0.03a |
| Violaxanthin dipalmitate | 0.05 ± 0.01c | 0.04 ± 0.01d | 0.08 ± 0.02b | 0.09 ± 0.01a |
| Lutein oleate | 0.03 ± 0.00b | 0.02 ± 0.00c | 0.06 ± 0.01a | 0.06 ± 0.00a |
| Violaxanthin dibutyrate | 0.07 ± 0.00b | 0.08 ± 0.00a | 0.06 ± 0.00c | 0.04 ± 0.00d |
| Violaxanthin dilaurate | 0.04 ± 0.01a | 0.03 ± 0.00b | 0.04 ± 0.01a | 0.04 ± 0.00b |
| Capsanthin | 0.02 ± 0.00a | 0.01 ± 0.00b | 0.02 ± 0.00a | 0.02 ± 0.00a |
| Violaxanthin laurate | 0.02 ± 0.00a | 0.02 ± 0.00a | 0.02 ± 0.00a | 0.01 ± 0.00a |
| Antheraxanthin | N/A | 0.27 ± 0.04 | N/A | N/A |
| Total | 141.48 | 95.68 | 143.13 | 132.67 |
| Yellow, purple, red and orange colored fruits of Coffea arabica are denoted as YF, PF, RF and ORF, respectively. Means ± standard error with a common alphabet in a row indicate no significant difference with Duncan Multiple Range Test at 0.05 probability level. N/A represents not applicable. | ||||
β-carotene is reported to be responsible for orange pigment whereas α-carotene causes yellow pigment in fruits and vegetables (Clevidence et al., 2000). The highest amount of α-carotene was observed in YF while the highest content of β-carotene was in RF, which correlate well with the colors of the fruits. Antheraxanthin was detected only in PF while antheraxanthin dipalmitate was found only in ORF, RF and YF. In sum, the total carotenoid detected in the current study followed RF (143.13 μg/g) > YF (141.48 μg/g) > ORF (132.67 μg/g) > PF (95.68 μg/g), respectively. This trend suggests that carotenoid is the major pigment in YF, RF and ORF whereas PF coloration is largely determined by anthocyanin.
4. DiscussionFruit pigmentation is a major quality trait that influences consumer preference and correlates directly with nutritional composition and health benefit (Yuan et al., 2015; Zhang et al., 2020; Jiang et al., 2021). As a result, fruit coloration and its pigmentations have been a major focus of research among scholars (Petropoulos et al., 2019). To date, anthocyanins, betalains, chlorophylls and carotenoids have been reported as the major pigmentations regulating coloration in crops (Zhao et al., 2017; Petropoulos et al., 2019). The present study used fully matured and ripe fruits from four Coffea arabica varieties (Fig. 1) to conduct transcriptome profiling to elucidate mechanisms of fruit coloration. We identified key DEGs, enriched biosynthesis pathways and TFs implicated in fruit coloration in coffee.
The KEGG pathway enrichment analysis revealed a number of enriched pathways (Table S2) which may explain the basis for the influence of fruit coloration on quality and nutritional value. For instance, fructose and mannose metabolism; vitamin B6 metabolism, together with valine, leucine and isoleucine degradation have been documented to regulate nutritional composition of many fruits (Bernstein et al., 2020). From the KEGG pathway, two pathways known to regulate fruit coloration were detected: carotenoid biosynthesis and phenylpropanoid/flavonoid/anthocyanin biosynthesis (Table S2) of which the former was more prominent than the latter. Carotenoids are known to be responsible for yellow, orange and red coloration of fruits and vegetables (García-Gómez et al., 2021); while it has also been reported to form chlorophyll–carotenoid complexes through its component, β-carotenoid found to bound either chlorophylls or xanthophylls leading to absorption of light in the orange or red-light spectrum, yielding green, purple or blue coloration (Wieruszewski, 2002). On the carotenoid biosynthetic pathway, we detected 32 DEGs encoding for 14 known enzymes (Fig. 6). The major precursor on this biosynthetic pathway is phytoene synthase (PSY) which is a major regulator of the carotenoid biosynthesis pathway by converting geranyl–geranyl diphosphate (GGPP) to phytoene which further converts into phytofluence and ζ-carotene by phytoene desaturase (PDS) (Yuan et al., 2015). According to recent studies, the transcriptional control of various genes encoding PSY is another key contributor to carotenoid production (Welsch et al., 2000; Toledo-Ortiz et al., 2010; Liu et al., 2015; Fu et al., 2016; Mercadante, 2019; Wan et al., 2019). De-etiolation of Arabidopsis thaliana seedlings is associated with a dramatic burst in carotenoid production, which occurs conterminously with a sharp upregulation of PSY gene expression and protein levels, as well as a jump in PSY enzyme activity (Welsch et al., 2000; Toledo-Ortiz et al., 2010; Liu et al., 2015; Wan et al., 2019). The phytochrome family of photoreceptors regulates the PSY gene expression by stimulating the PSY transcription factors under red (R) and far-red (FR) light. Genes linked to PSY are reported to be the primary bottlenecks in carotenogenesis (Yuan et al., 2015). Specifically, the phytochrome-interacting factor 1 (PIF1) and other transcription factors of the PIF family down-regulate the accumulation of carotenoids by inhibiting the expression of the enzyme that catalyzes the key rate-limiting step in the PSY pathway (Welsch et al., 2000; Toledo-Ortiz et al., 2010). Thus, the low expression (down-regulation) of 1 PSY gene (gene-LOC113688784) may partly account for the differential coloration in the fruits which was expressed in order of YF > ORF > RF > PF. This however requires functional validation to deepen our understanding of carotenoid biosynthesis and accumulation in C. arabica (Liu et al., 2015).
Subsequently, lycopene epsilon-cyclase (LYCE) is involved in the conversion of lycopene to δ-carotene, α-carotene and α-cryptoxanthin. One LYCE gene (gene-LOC113696970) expressed 2.21 times higher in RF than YF. This corroborates with earlier report that lycopene is one of the naturally occurring red colored carotenoids (Boileau et al., 2002). In contrast, naturally occurring β-carotene is orange in color. In our study, no gene was differentially involved in the conversion of lycopene/γ-carotene to β-carotene; however, two beta-carotene isomerases (BCIS) (gene-LOC113704159 and gene-LOC113704210) were expressed higher in ORF than YF. This is in consonance with the report that β-carotene occurs as an orange pigment, while α-carotene is a yellow pigment, which can be found in fruits and vegetables, but the yellow colored fruits contain low or trace amounts of β-carotene (Clevidence et al., 2000). The 32 genes linked to 14 known enzymes in carotenoid biosynthetic pathway with differential expression among the four fruits could be further studied for further functional validation.
Another pigment which has received much attention is anthocyanin (Stanley and Yuan, 2019). This pigment is synthesized through the flavonoid pathway which is considered as a branch of the phenylpropanoid pathway. On this broad and complex pathway, the biosynthesis of anthocyanins starts with the lysis of phenylalanine ammonia catalyzed via the enzyme PAL (Zhang et al., 2014) which was encoded by 3 of 4 DEGs (gene-LOC113707625, gene-LOC113720652, and gene-LOC113725801) in the present study. These genes had higher expression in either PF, RF or ORF than YF which may have contributed to reduction in anthocyanin biosynthesis and accumulation in YF leading to weak anthocyanin coloration. This enzyme, PAL and several others (Supplementary Fig. S5) suggest that anthocyanin is not a major pigment in YF. Specifically, two enzymes, BZI and 3AT involved in last steps of the anthocyanin biosynthetic pathway, were linked to 5 genes which were expressed higher in either PF, RF or ORF than YF.
The study revealed that MYB, bHLH and NAC TFs may regulate carotenoid biosynthesis negatively (Fig. 8A, B and F). The trend for MYB agrees with the R2R3-MYB repression of the conversion of α- and β-branch of carotenoid by negatively modulating the expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulata (Zhu et al., 2017b). Similarly, when bHLH TF (Solyc02g067380.2.1) expression was repressed markedly, SlPRE2 downregulated SlPSY1, SlPDS and SlZDS in the carotenoid biosynthetic pathway in tomato (Zhu et al., 2017a). The trend observed for NAC TF contrasts the report that CpNAC1 positively regulates carotenoid biosynthesis during papaya fruit ripening (Fu et al., 2016). There are numerous studies on the upregulation of MYB and bHLH TFs on anthocyanins biosynthesis; however, the actual mechanism in C. arabica is yet to be unraveled. Therefore, these together with other major TFs (bHLH, MYB, AP2/ERF, NAC, MADS, AUX/IAA and WRKY) would be foundational for future studies to elucidate their modulating roles in fruit coloration in C. arabica via functional genomics such as CRISPR/Cas9 technology (Zhi et al., 2020).
The composition and concentration of carotenoids determine the nutritional value and fruit color in most crops. We further conducted quantitative profiling of carotenoid constituents among the four fruit samples (Table 2). The major component consisted of lutein esters (e.g., lutein dimyristate, myristate, palmitate, laurate, dipalmitate and dilaurate) (Table 2) which have been reported in several crops (Mercadante, 2019). Lutein esters as pigment like its sister compound, has primarily been used in food and supplement manufacturing as a colorant due to its yellow-red color (Mercadante, 2019). It absorbs blue light which appears yellow at low concentration and orange-red at high concentrations. This may account for the higher price of yellow colored coffee fruits than either red, orange or purple colored fruits. Cumulatively, the total carotenoids in RF was very close to that of YF indicating that difference in anthocyanins may have contributed to color differentiation between red and yellow (Wan et al., 2019). Most of the key genes involved in anthocyanin biosynthetic pathway expressed higher in RF relative to YF.
The findings presented in this study offer valuable insights into the transcriptional regulation of fruit coloration among the four Coffea arabica varieties. The numerous genes identified in this study could be further screened for functional validation experiments such CRISPR/Cas9, overexpression and gene silencing approaches to elucidate their actual roles in fruit coloration. It is also recommended to conduct metabolome profiling among the four different varieties to identify metabolites responsible for fruit coloration for possible functional biomarker development.
5. ConclusionDifferentially expressed genes and key biosynthesis pathways involved in Coffea arabica fruit color formation were identified using transcriptome profiling in this study. The DEGs were found to be involved in the biosynthesis of carotenoids, and anthocyanins/phenylpropanoid/flavonoids in the KEGG enrichment analyses, indicating that these biosynthesis pathways are important in Coffea arabica fruit coloration. These findings shed light on the formation and variation of fruit color in C. arabica. Further study into the inherent roles of the candidate genes and TFs identified in this study may be required to decipher their regulatory mechanisms in C. arabica fruit color formation and variation.
Author contributionsConceptualization, Faguang Hu, Xinping Luo and Hongming Liu; Methodology, Faguang Hu; Formal analysis, Hongming Liu, Xingfei Fu, Guiping Li; Investigation, Yanan Li, Yang, Xiaofang Zhang, Ruirui Wu; Resources, Yulan Lv; Data curation, Xiaofei Bi; Writing-original draft preparation, Faguang Hu, Xiaofei Bi, Hongming Liu; Writing-review and editing, Xinping Luo, Rui Shi; Supervision, Jiaxiong Huang; Project administration, Faguang Hu; Funding acquisition, Faguang Hu. All authors have read and agreed to the content of the manuscript.
Data availabilityThe RNA-sequencing trimmed data used in this manuscript has been submitted to the NCBI SRA database under accession number: PRJNA743796.
Declaration of competing interestThe authors declare no conflict of interest.
AcknowledgementThis study was financially supported by the Yunnan Science and Technology Introducing project (International Science and Technology Cooperation): Construction Project of Coffee Scientific and Technological Demonstration Districts in Mountainous Areas of Northern Laos (2019IB013), High-end Foreign Experts Program of Yunnan Thousand Talents Program (2019013), and Yunnan provincial key programs (2019ZG00901, 202002AA10007).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.11.005.
Allan, A.C., Hellens, R.P., Laing, W.A., 2008. MYB transcription factors that colour our fruit. Trends Plant Sci., 13: 99-101. DOI:10.1016/j.tplants.2007.11.012 |
Ben-Meir, H., Zuker, A., Weiss, D., Vainstein, A., 2002. Molecular control of floral
pigmentation: anthocyanins. In: Breeding for Ornamentals: Classical and Molecular Approaches, pp. 253-272.
|
Bernstein, L.E., Burns, C., Drumm, M., et al., 2020. Impact on isoleucine and valine supplementation when decreasing use of medical food in the nutritional management of methylmalonic acidemia. Nutrients, 12: 473. DOI:10.3390/nu12020473 |
Boileau, T.W.M., Boileau, A.C., Erdman, J.W., 2002. Bioavailability of all-trans and cisisomers of lycopene. In: Proceedings of the Experimental Biology and Medicine;
Society for Experimental Biology and Medicine, vol. 227, pp. 914e919.
|
Cheng, B., Furtado, A., Henry, R.J., 2017. Long-read sequencing of the coffee bean transcriptome reveals the diversity of full-length transcripts. GigaScience, 6: 1-13. DOI:10.1093/gigascience/gix086 |
Cheng, B., Furtado, A., Henry, R.J., 2018. The coffee bean transcriptome explains the accumulation of the major bean components through ripening. Sci. Rep., 8: 11414. DOI:10.1038/s41598-018-29842-4 |
Cheng, B., Furtado, A., Smyth, H.E., et al., 2016. Influence of genotype and environment on coffee quality. Trends Food Sci. Technol., 57: 20-30. DOI:10.1016/j.tifs.2016.09.003 |
Clevidence, B., Paetau, I., Smith, J.C., 2000. Bioavailability of Carotenoids from
vegetables. HortScience, 35, 585-588.
|
Cui, X., Deng, J., Huang, C., et al., 2021. Transcriptomic analysis of the anthocyanin biosynthetic pathway reveals the molecular mechanism associated with purple color formation in Dendrobium nestor. Life, 11: 1-19. DOI:10.3390/life11020113 |
Dai, J., Gupte, A., Gates, L., et al., 2009. A comprehensive study of anthocyanin-containing extracts from selected blackberry cultivars: extraction methods, stability, anticancer properties and mechanisms. Food Chem. Toxicol., 47: 837-847. DOI:10.1016/j.fct.2009.01.016 |
Davis, A.P., Tosh, J., Ruch, N., et al., 2011. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data; implications for the size, morphology, distribution and evolutionary history of Coffea. Bot. J. Linn. Soc., 167: 357-377. DOI:10.1111/j.1095-8339.2011.01177.x |
Denoeud, F., Carretero-Paulet, L., Dereeper, A., et al., 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science, 345: 1181-1184. DOI:10.1126/science.1255274 |
Dubos, C., Stracke, R., Grotewold, E., et al., 2010. MYB transcription factors in Arabidopsis. Trends Plant Sci., 15: 573-581. DOI:10.1016/j.tplants.2010.06.005 |
Espley, R.v., Hellens, R.P., Putterill, J., et al., 2007. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J., 49: 414-427. DOI:10.1111/j.1365-313X.2006.02964.x |
Falcone Ferreyra, M.L., Rius, S.P., Casati, P., 2012. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci., 3: 1-15. DOI:10.3389/fpls.2012.00222 |
Fenilli, T.A.B., Reichardt, K., Dourado-Neto, D., et al., 2007. Growth, development, and fertilizer-15N recovery by the coffee plant. Sci. Agric., 64: 541-547. DOI:10.1590/S0103-90162007000500012 |
Fraser, P.D., Pinto, M.E.S., Holloway, D.E., et al., 2008. Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J., 24: 551-558. DOI:10.1111/j.1365-313x.2000.00896.x |
Fu, C.C., Han, Y.C., Fan, Z.Q., et al., 2016. The papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J. Agric. Food Chem., 64: 5454-5463. DOI:10.1021/acs.jafc.6b01020 |
Furtado, M.B., Wilmanns, J.C., Chandran, A., et al., 2015. A novel conditional mouse model for NKX2-5 reveals transcriptional regulation of cardiac ion channels. Differentiation, 91: 29-41. DOI:10.1016/j.diff.2015.12.003 |
García-Gómez, B.E., Salazar, J.A., Nicolás-Almansa, M., et al., 2021. Molecular bases of fruit quality in prunus species: an integrated genomic, transcriptomic, and metabolic review with a breeding perspective. Int. J. Mol. Sci., 22: 1-38. |
Geleta, M., Herrera, I., Monzón, A., et al., 2012. Genetic diversity of Arabica coffee (Coffea arabica L.) in Nicaragua as estimated by simple sequence repeat markers. Sci. World J., 2012: 939820. DOI:10.1100/2012/939820 |
Guo, W.H., Tu, C.Y., Hu, C.H., 2008. Cis-trans isomerizations of β-carotene and lycopene: a theoretical study. J. Phys. Chem. B, 112: 12158-12167. DOI:10.1021/jp8019705 |
Hao, Y., Oh, E., Choi, G., et al., 2012. Interactions between HLH and BHLH factors modulate light-regulated plant development. Mol. Plant, 5: 688-697. DOI:10.1093/mp/sss011 |
Heberle, H., Meirelles, V.G., da Silva, F.R., et al., 2015. A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf., 16: 169. DOI:10.1186/s12859-015-0611-3 |
Holton, T.A., Cornish, E.C., 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell, 7: 1071-1083. DOI:10.1105/tpc.7.7.1071 |
Ikeda, M., Mitsud, N., Ohme-Takagi, M., 2013. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic BHLH system. Plant Signal. Behav., 8: e23448. DOI:10.4161/psb.23448 |
Inbaraj, B.S., Lu, H., Hung, C.F., et al., 2008. Determination of carotenoids and their esters in fruits of Lycium barbarum Linnaeus by HPLC-DAD-APCI-MS. J. Pharmaceut. Biomed. Anal., 47: 812-818. DOI:10.1016/j.jpba.2008.04.001 |
International Coffee Organization Trade Statistics. 2021. Available online. http://www.ico.org/trade_statistics.asp?section=Statistics. (Accessed 24 May 2021).
|
Ivamoto, S.T., Reis, O., Domingues, D.S., et al., 2017. Transcriptome analysis of leaves, flowers and fruits perisperm of Coffea arabica L. reveals the differential expression of genes involved in raffinose biosynthesis. PLoS One, 12: e0169595. DOI:10.1371/journal.pone.0169595 |
Jian, W., Cao, H., Yuan, S., et al., 2019. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res, 6: 22. DOI:10.1038/s41438-018-0098-y |
Jiang, S., Sun, Q., Zhang, T., et al., 2021. MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4-like in apple fruit. J. Plant Physiol., 257: 153353. DOI:10.1016/j.jplph.2020.153353 |
Kanehisa, M., Goto, S., 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 28: 27-30. DOI:10.1093/nar/28.1.27 |
Kanehisa, M., Sato, Y., Kawashima, M., et al., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res., 32: 277-280. DOI:10.1093/nar/gkv1070 |
Khoo, H.E., Azlan, A., Tang, S.T., et al., 2017. Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res., 61: 1361779. DOI:10.1080/16546628.2017.1361779 |
Knevitt, D., 2016. Characterising Chlorogenic Acid Biosynthesis in Coffee. Doctoral
thesis, University of East Anglia.
|
Koseki, M., Goto, K., Masuta, C., et al., 2005. The star-type color pattern in petunia hybrid 'red star' flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Psysiol., 46: 1879-1883. DOI:10.1093/pcp/pci192 |
Kumar, V., Yadav, S.K., 2013. Overexpression of CsANR increased flavan-3-ols and decreased anthocyanins in transgenic tobacco. Mol. Biotechnol., 54: 426-435. DOI:10.1007/s12033-012-9580-1 |
Lai, B., Li, X. -J., Hu, B., et al., 2014. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS One, 9: 1-12. DOI:10.1371/journal.pone.0086293 |
Langfelder, P., Horvath, S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf., 9: 10.1186/1471-2105-9-559. DOI:10.1186/1471-2105-9-559 |
Li, J., Li, G., Gao, S., et al., 2010. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome a signaling. Plant Cell, 22: 3634-3649. DOI:10.1105/tpc.110.075788 |
Licausi, F., Ohme-Takagi, M., Perata, P., 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol., 199: 639-649. DOI:10.1111/nph.12291 |
Liu, L., Shao, Z., Zhang, M., et al., 2015. Regulation of carotenoid metabolism in tomato. Mol. Plant, 8: 28-39. DOI:10.1016/j.molp.2014.11.006 |
Lloyd, A., Brockman, A., Aguirre, L., et al., 2017. Advances in the MYB-BHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and Co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol., 58: 1431-1441. DOI:10.1093/pcp/pcx075 |
Lohse, M., Nagel, A., Herter, T., et al., 2015. Mercator: a Fast and simple Web server for genome scale functional annotation of plant sequence data. Plant Cell Environ., 37: 1250-1258. DOI:10.1111/pce.12231 |
Lopes, C.T., Max, F., Farzana, K., et al., 2010. Cytoscape Web: an interactive web-based network browser. Bioinformatics, 26: 2347-2348. DOI:10.1093/bioinformatics/btq430 |
Lu, S., Zhang, Y., Zhu, K., et al., 2018. The Citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol., 176: 2657-2676. DOI:10.1104/pp.17.01830 |
Machemer, K., 2011. Interplay of MYB factors in differential cell expansion, and consequences for tomato fruit development. Plant J., 68: 337-350. DOI:10.1111/j.1365-313x.2011.04690.x |
Malien-Aubert, C., Dangles, O., Amiot, M.J., 2001. Color stability of commercial anthocyanin-based extracts in relation to the phenolic composition. Protective effects by intra- and intermolecular copigmentation. J. Agric. Food Chem., 49: 170-176. DOI:10.1021/jf000791o |
Mercadante, A.Z., 2019. Carotenoid esters in foods: physical, chemical and biological
properties. In: Food chemistry, function and analysis, vol. 13. Royal Society of
Chemistry, UK. https://www.worldcat.org/title/carotenoid-esters-in-foodsphysical-chemical-and-biological-properties/oclc/1112235716.
|
Mekbib, Y., Saina, J.K., Tesfaye, K., et al., 2020. Chloroplast genome sequence variations and development of polymorphic markers in Coffea arabica. Plant Mol. Biol. Rep., 38: 491-502. DOI:10.1007/s11105-020-01212-3 |
Mishra, M.K., Slater, A., 2012. Recent advances in the genetic transformation of coffee. Biotechnol. Res. Int.: 1-17. DOI:10.1155/2012/580857 |
Mitsis, T., Efthimiadou, A., Bacopoulou, F., et al., 2020. Transcription factors and evolution: an integral part of gene expression (review). World Acad. Sci. J., 2: 3-8. DOI:10.3892/wasj.2020.32 |
Mofatto, L.S., Carneiro, F. de A., Vieira, N.G., et al., 2016. Identification of candidate genes for drought tolerance in coffee by high-throughput sequencing in the shoot apex of different Coffea arabica cultivars. BMC Plant Biol., 16: 10.1186/s12870-016-0777-5. DOI:10.1186/s12870-016-0777-5 |
Neto, A.P., Favarin, J.L., de Almeida, R.E.M., et al., 2011. Changes of nutritional status during a phenological cycle of coffee under high nitrogen supply by fertigation. Commun. Soil Sci. Plant Anal., 42: 2414-2425. DOI:10.1080/00103624.2011.607731 |
Petropoulos, S.A., Sampaio, S.L., di Gioia, F., et al., 2019. Grown to be blue—antioxidant properties and health effects of colored vegetables. Part I: root vegetables. Antioxidants, 8: 617. DOI:10.3390/antiox8120617 |
Rashid, M.I., Fareed, M.I., Rashid, H., et al., 2019. Flavonoids and their biological
secrets. In: Ozturk, M., et al. (Eds.), Plant and Human Health: Phytochemistry
and Molecular Aspects. Springer, pp. 579-605.
|
Reis, A.R., Favarin, J.L., Gallo, L.A., et al., 2009. Nitrate reductase and glutamine synthetase activity in coffee leaves during fruit development. Rev. Bras. Ciência do Solo, 33: 315-324. DOI:10.1590/S0100-06832009000200009 |
Riechmann, J.L., Heard, J., Martin, G., et al., 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science, 290: 2105-2110. DOI:10.1126/science.290.5499.2105 |
Rodríguez-Villaló, A., Gas, E., Rodríguez-Concepció, M., 2009. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J., 60: 424-435. DOI:10.1111/j.1365-313X.2009.03966.x |
Ruiz-Sola, M. Á., Rodríguez-Concepción, M., 2012. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book, 10: e0158. DOI:10.1199/tab.0158 |
Ságio, S.A., Lima, A.A., Barreto, H.G., et al., 2013. Physiological and molecular analyses of early and late Coffea arabica cultivars at different stages of fruit ripening. Acta Physiol. Plant., 35: 3091-3098. DOI:10.1007/s11738-013-1342-6 |
Sant'Ana, G.C., Pereira, L.F.P., Pot, D., et al., 2018. Genome-wide association study reveals candidate genes influencing lipids and diterpenes contents in Coffea arabica L. Sci. Rep., 8: 465. DOI:10.1038/s41598-017-18800-1 |
Stanley, L., Yuan, Y.W., 2019. Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front. Plant Sci., 10: 1017. DOI:10.3389/fpls.2019.01017 |
Takos, A.M., Jaffé, F.W., Jacob, S.R., et al., 2006. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol., 142: 1216-1232. DOI:10.1104/pp.106.088104 |
Tanaka, Y., Sasaki, N., Ohmiya, A., 2008. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J., 54: 733-749. DOI:10.1111/j.1365-313X.2008.03447.x |
Toledo-Ortiz, G., Huq, E., Rodríguez-Concepción, M., 2010. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A., 107: 11626-11631. DOI:10.1073/pnas.0914428107 |
Tran, H.T.M., Ramaraj, T., Furtado, A., et al., 2018. Use of a draft genome of coffee (Coffea arabica) to identify SNPs associated with caffeine content. Plant Biotechnol. J., 16: 1756-1766. DOI:10.1111/pbi.12912 |
Ullah, I., Magdy, M., Wang, L., et al., 2019. Genome-wide identification and evolutionary analysis of TGA transcription factors in soybean. Sci. Rep., 9: 11186. DOI:10.1038/s41598-019-47316-z |
Usadel, B., Poree, F., Nagel, A., et al., 2009. A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ., 32: 1211-1229. DOI:10.1111/j.1365-3040.2009.01978.x |
Varaud, E., Brioudes, F., Szécsi, J., et al., 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the BHLH transcription factor BIGPETALp. Plant Cell, 23: 973-983. DOI:10.1105/tpc.110.081653 |
Verweij, W., Spelt, C.E., Bliek, M., et al., 2016. Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell, 28: 786-803. DOI:10.1105/tpc.15.00608 |
Wan, H., Yu, C., Han, Y., et al., 2019. Determination of flavonoids and carotenoids and their contributions to various colors of rose cultivars (Rosa spp.). Front. Plant Sci., 10: 123. DOI:10.3389/fpls.2019.00123 |
Wang, C.C., Chang, S.C., Inbaraj, B.S., et al., 2010. Isolation of carotenoids, flavonoids and polysaccharides from Lycium barbarum L. and evaluation of antioxidant activity. Food Chem., 120: 184-192. DOI:10.1016/j.foodchem.2009.10.005 |
Welsch, R., Beyer, P., Hugueney, P., et al., 2000. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta, 211: 846-854. DOI:10.1007/s004250000352 |
Welsch, R., Maass, D., Voegel, T., et al., 2007. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol., 145: 1073-1085. DOI:10.1104/pp.107.104828 |
Wieruszewski, J.B., 2002. Astaxanthin Bioavailability, Retention Efficiency and Kinetics in Atlantic Salmon (Salmo Salar) as Influenced by Pigment Concentration
and Method of Administration (Kinetics Only). Ottawa.
|
Xu, Q., He, J., Dong, J., et al., 2018. Genomic survey and expression profiling of the MYB gene family in watermelon. Hortic. Plant J., 4: 1-15. DOI:10.1016/j.hpj.2017.12.001 |
Yahia, E.M., de Jesús Ornelas-Paz, J., Emanuelli, T., et al., 2017. Chemistry, stability,
and biological actions of carotenoids In: Fruit and Vegetable Phytochemicals:
Chemistry and Human Health, second ed., vol. 1. Wiley Blackwell,
pp. 285-345.
|
Yan, H., Kerr, W.L., 2013. Total phenolics content, anthocyanins, and dietary fiber content of apple pomace powders produced by vacuum-belt drying. J. Sci. Food Agric., 93: 1499-1504. DOI:10.1002/jsfa.5925 |
Ye, J., Hu, T., Yang, C., et al., 2015. Transcriptome profiling of tomato fruit development reveals transcription factors associated with ascorbic acid, carotenoid and flavonoid biosynthesis. PLoS One, 10: e0130885. DOI:10.1371/journal.pone.0130885 |
Yuan, H., Zhang, J., Nageswaran, D., et al., 2015. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res., 2: 1-11. DOI:10.3109/00207454.2015.1036421 |
Yuyama, P.M., Reis Júnior, O., Ivamoto, S.T., et al., 2016. Transcriptome analysis in Coffea eugenioides, an arabica coffee ancestor, reveals differentially expressed genes in leaves and fruits. Mol. Genet. Genom., 291: 323-336. DOI:10.1007/s00438-015-1111-x |
Zhang, Y., Chen, G., Dong, T., et al., 2014. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple bok choy (Brassica rapa var. chinensis). J. Agric. Food Chem., 62: 12366-12376. DOI:10.1021/jf503453e |
Zhang, H., Zhang, S., Zhang, H., et al., 2020. Carotenoid metabolite and transcriptome dynamics underlying flower color in marigold (Tagetes erecta L.). Sci. Rep., 10: 16835. DOI:10.1038/s41598-020-73859-7 |
Zhao, Y., Dong, W., Wang, K., et al., 2017. Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Front. Plant Sci., 8: 1552. DOI:10.3389/fpls.2017.01552 |
Zhi, J., Liu, X., Li, D., et al., 2020. CRISPR/Cas9-mediated SlAN2 mutants reveal various regulatory models of anthocyanin biosynthesis in tomato plant. Plant Cell Rep., 39: 799-809. DOI:10.1007/s00299-020-02531-1 |
Zhou, D., Shen, Y., Zhou, P., et al., 2019. Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res., 6: 80. DOI:10.1038/s41438-019-0162-2 |
Zhu, Z., Chen, G., Guo, X., et al., 2017. Overexpression of SlPRE2, an atypical BHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato. Sci. Rep., 7: 5786. DOI:10.1038/s41598-017-04092-y |
Zhu, F., Luo, T., Liu, C., et al., 2017. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulata. New Phytol., 216: 178-192. DOI:10.1111/nph.14684 |
Zhuang, H., Lou, Q., Liu, H., et al., 2019. Differential regulation of anthocyanins in green and purple turnips revealed by combined de novo transcriptome and metabolome analysis. Int. J. Mol. Sci., 20: 4387. DOI:10.3390/ijms20184387 |



