b. College of Life Science, Shanghai Normal University, Shanghai, 200234, China;
c. Shenzhen Key Laboratory for Orchid Conservation and Utilization, National Orchid Conservation Center of China and the Orchid Conservation & Research Center of Shenzhen, Shenzhen, 518114, China;
d. Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, 510650, China;
e. College of Forestry, Central South University of Forestry & Technology, Changsha, Hunan, 410004, China;
f. State Environmental Protection Key Laboratory of Regional Eco-process and Function Assessment, Chinese Research Academy of Environmental Sciences, Beijing, 100012, China
The genus Ceratopteris is an aquatic or subaquatic annual plant, distributed in tropical and subtropical regions worldwide (Brongniart, 1821; Lloyd, 1974; Masuyama et al., 2002). However, because of the high degree of polymorphism within the genus, several reproductively isolated taxa are morphologically indistinguishable (Grant, 1981; Paris et al., 1989; Masuyama et al., 2002; Masuyama, 2008). This phenomenon strongly suggests that many cryptic species exist in Ceratopteris (Liao et al., 2011; Kinosian et al., 2020a). Consequently, the number of Ceratopteris species around the world remains unknown.
The genus Ceratopteris currently contains over 14 species names (IPNI 2021; Tropicos 2021), of which only four are widely accepted: Ceratopteris cornuta (P. Beauv.) Lepr., Ceratopteris richardii Brongn., Ceratopteris thalictroides (L.) Brongn., and Ceratopteris pteridoides (Hook.) Hieron (Brongniart, 1821, Brongniart, 1823, Hieronymus, 1905, Leprieur, 1830). C. thalictroides may be a cryptic species complex and the most polymorphic among the four species (Benedict, 1909; Lloyd, 1974). Masuyama and Watano (2010) demonstrated that C. thalictroides contains at least five taxa, including Ceratopteris froesii Brade., C. thalictroides, C. gaudichaudii var. gaudichaudii Brongn., C. gaudichaudii var. vulgaris Masuyama & Watano., and C. oblongiloba Masuyama & Watano (Brade, 1964, Brongniart, 1821, Masuyama and Watano, 2010). Liao et al. (2011) proposed two cryptic species of C. thalictroides (the south type and the north type species) in China. A new cryptic species of Ceratopteris, Ceratopteris shingii Y.H. Yan & R. Zhang, has been reported based on strong molecular and morphological evidence (Zhang et al., 2020b). Cytological results suggest that C. cornuta, C. richardii and C. pteridoides are diploids with 2n = 78; other Ceratopteris species are tetraploid with 2n = 154 or 156 (Smith et al., 1977; Hickok, 1979; Masuyama and Watano, 2005). The speciation of tetraploid Ceratopteris species is more complex than expected because of frequent hybridization. Ceratopteris is known to have cryptic allotetraploid taxa, but one of the diploid parents is likely extinct or has yet to be identified among known diploids (Hickok and Klekowski, 1974; Lloyd, 1974; Hickok, 1977, 1979; Adjie et al., 2007; Kinosian et al., 2020b).
From 2017 to 2019, we collected and confirmed living individuals belonging to two species of Ceratopteris. Samples from Guangdong Province were sequenced using RNA-Seq (Shen et al., 2018) and have been reported as diploids, with a chromosome number 2n = 78 (Zhang et al., 2019). However, the taxonomy of these samples has yet to be revised. Here we identified this specimen as Ceratopteris chunii. Another species collected from the Yangtze River area in central China and previously misidentified as C. pteridoides is here described as Ceratopteris chingii. We also confirm two new undescribed species through morphological observation, palynology, cytological study, and phylogenetic analysis.
2. Material and methods 2.1. Morphological description and scanning electron microscopy (SEM)Morphological features were described based on specimens measured with Vernier calipers. Micromorphological features were examined under a JSZ6 dissecting microscope (Jiangnan Novel, China) (Tables 1, 2).
| Taxon | Stipe width of sterile leaf (cm) | Stipe length of sterile leaf (cm) | Blade length of sterile leaf (cm) | Ratio of stipe length to blade length (cm) | Blade length of 1st pinna (cm) | Ratio of 1st pinna of blade to stipe length (cm) | Voucher (Herbarium) |
| Ceratopteris gaudichaudii var. vulgaris | 0.122 | 0.731 | 1.580 | 0.463 | 1.225 | 1.676 | C. Gaudichaud, 1549 (MNHN) |
| Ceratopteris gaudichaudii var. vulgaris | 0.148 | 4.040 | 12.704 | 0.318 | 4.816 | 1.192 | B.C. Stone, 4321 (NL) |
| Ceratopteris gaudichaudii var. vulgaris | 0.155 | 2.505 | 7.220 | 0.347 | 2.738 | 1.093 | M. Evans, 628 (US) |
| Ceratopteris gaudichaudii var. vulgaris | 0.121 | 3.608 | 8.553 | 0.422 | 2.742 | 0.760 | L. Raulerson, 15, 656 (US) |
| Ceratopteris gaudichaudii var. vulgaris | 0.142 | 3.147 | 6.221 | 0.506 | 2.631 | 0.836 | F.R. Fosberg, 35, 454 (US) |
| Ceratopteris chunii | 0.100 | 7.000 | 14.000 | 0.500 | 2.702 | 0.386 | Y.H. Yan, Fern09730 (CSH) |
| Ceratopteris chunii | 0.150 | 9.500 | 12.500 | 0.760 | 3.572 | 0.376 | F.G. Wang, YYH15464 (CSH) |
| Ceratopteris chunii | 0.150 | 9.000 | 12.000 | 0.750 | 3.177 | 0.353 | F.G. Wang, YYH15463 (CSH) |
| Ceratopteris chunii | 0.200 | 9.000 | 11.300 | 0.796 | 3.708 | 0.412 | F.G. Wang, YYH15462 (CSH) |
| Ceratopteris chunii | 0.100 | 10.500 | 12.500 | 0.840 | 4.410 | 0.420 | F.G. Wang, YYH15461 (CSH) |
| Taxon | Stipe width of fertile leaf (cm) | Stipe length of fertile leaf (cm) | Blade length of fertile leaf (cm) | Width of fertile leaf (cm) | Ratio of stipe length to blade length (cm) | Aspect ratio of fertile leaf (cm) | Voucher (Herbarium) |
| Ceratopteris pteridoides | 1.925 | 13.000 | 29.500 | 23.475 | 0.441 | 0.703 | R.M. Lloyd, s.n. (GH) |
| Ceratopteris pteridoides | 0.836 | 14.545 | 36.364 | 14.545 | 0.400 | 1.500 | S.R. Hill & C.N. Horn, 27, 110 (US) |
| Ceratopteris pteridoides | 0.182 | 18.182 | 34.545 | 26.727 | 0.526 | 0.612 | C.R. Sperling, 6543 (US) |
| Ceratopteris pteridoides | 0.867 | 4.321 | 14.708 | 12.775 | 0.294 | 0.813 | A.H. Curtiss, 3690 (US) |
| Ceratopteris pteridoides | 1.727 | 7.273 | 17.309 | 18.182 | 0.420 | 0.552 | M.Y. Rimachi, 9213 (US) |
| Ceratopteris pteridoides | 1.455 | 7.273 | 20.000 | 21.818 | 0.364 | 0.583 | M.Y. Rimachi, 9213 (US) |
| Ceratopteris pteridoides | 2.909 | 23.636 | 30.909 | 9.091 | 0.765 | 0.800 | M.Y. Rimachi, 10, 650 (US) |
| Ceratopteris pteridoides | 1.200 | 18.982 | 41.818 | 25.455 | 0.454 | 0.897 | M.Y. Rimachi, 10, 650 (US) |
| Ceratopteris pteridoides | 1.436 | 11.091 | 23.636 | 14.545 | 0.469 | 0.863 | M.H. Grayum, 8030 (US) |
| Ceratopteris pteridoides | 2.782 | 9.091 | 21.818 | 25.455 | 0.417 | 0.500 | T.C. Plowman, 6380 (US) |
| Ceratopteris pteridoides | 1.018 | 16.364 | 30.909 | 33.273 | 0.529 | 0.437 | J. Zainúm, 19 Bo127 (US) |
| Ceratopteris pteridoides | 1.091 | 14.545 | 23.636 | 18.182 | 0.615 | 0.500 | N.C. Fassett, 28, 557 (US) |
| Ceratopteris pteridoides | 0.727 | 8.891 | 20.000 | 19.200 | 0.445 | 0.579 | E.L. Ekman, H12109 (US) |
| Ceratopteris pteridoides | 0.545 | 14.545 | 21.818 | 21.818 | 0.667 | 0.333 | E.L. Ekman, H12109 (US) |
| Ceratopteris pteridoides | 0.836 | 10.909 | 29.091 | 20.000 | 0.375 | 0.909 | Gomez, 6766 (US) |
| Ceratopteris pteridoides | 2.878 | 12.887 | 26.632 | 18.782 | 0.484 | 0.732 | D.S. Conant, 948 (F) |
| Ceratopteris pteridoides | 1.261 | 13.068 | 34.014 | 23.678 | 0.384 | 0.885 | J. Popenoe, 1617 (NCU) |
| Ceratopteris pteridoides | 1.873 | 10.354 | 25.859 | 18.821 | 0.400 | 0.824 | S.G. Beck, 5523 (F) |
| Ceratopteris pteridoides | 1.506 | 16.202 | 31.452 | 25.592 | 0.515 | 0.596 | F.J. Roldán, 1686 (NY) |
| Ceratopteris pteridoides | 0.695 | 9.136 | 27.458 | 18.034 | 0.333 | 1.016 | T.M. Pedersen, 14, 800 (RMNH) |
| Ceratopteris pteridoides | 2.776 | 6.343 | 19.949 | 19.581 | 0.318 | 0.695 | S. McDaniel, 17329 (F) |
| Ceratopteris chingii | 1.400 | 5.000 | 24.000 | 17.500 | 0.208 | 1.086 | H. Shang, SG2911 (CSH) |
| Ceratopteris chingii | 2.100 | 6.000 | 23.000 | 25.000 | 0.261 | 0.680 | H. Shang, SG2912 (CSH) |
| Ceratopteris chingii | 1.200 | 4.500 | 23.500 | 22.000 | 0.191 | 0.864 | H. Shang, SG2912 (CSH) |
| Ceratopteris chingii | 0.500 | 3.000 | 20.000 | 14.000 | 0.150 | 1.214 | H. Shang, SG2912 (CSH) |
| Ceratopteris chingii | 1.700 | 5.000 | 25.000 | 20.000 | 0.200 | 1.000 | H. Shang, SG2913 (CSH) |
| Ceratopteris chingii | 1.800 | 9.000 | 37.000 | 30.000 | 0.243 | 0.933 | H. Shang, SG2914 (CSH) |
| Ceratopteris chingii | 2.300 | 10.000 | 40.000 | 24.000 | 0.250 | 1.250 | H. Shang, SG2914 (CSH) |
| Ceratopteris chingii | 2.400 | 9.000 | 40.000 | 30.000 | 0.225 | 1.033 | H. Shang, SG2914 (CSH) |
| Ceratopteris chingii | 2.200 | 8.000 | 37.000 | 21.000 | 0.216 | 1.381 | H. Shang, SG2915 (CSH) |
| Ceratopteris chingii | 0.900 | 6.000 | 19.000 | 14.000 | 0.316 | 0.929 | H. Shang, SG2916 (CSH) |
| Ceratopteris chingii | 0.400 | 9.000 | 24.000 | 11.000 | 0.375 | 1.364 | H. Shang, SG2917 (CSH) |
| Ceratopteris chingii | 0.400 | 5.000 | 16.000 | 13.000 | 0.313 | 0.846 | H. Shang, SG2918 (CSH) |
| Ceratopteris chingii | 1.000 | 12.000 | 30.000 | 14.000 | 0.400 | 1.286 | H. Shang, SG2919 (CSH) |
| Ceratopteris chingii | 1.600 | 10.000 | 36.000 | 24.000 | 0.278 | 1.083 | H. Shang, SG2920 (CSH) |
| Ceratopteris chingii | 1.300 | 7.000 | 34.000 | 22.000 | 0.206 | 1.227 | H. Shang, SG2920 (CSH) |
| Ceratopteris chingii | 2.419 | 6.452 | 24.194 | 24.194 | 0.267 | 0.733 | J.M. Wang, 3886 (NAS) |
| Ceratopteris chingii | 2.581 | 4.839 | 20.968 | 16.129 | 0.231 | 1.000 | J.M. Wang, 3886 (NAS) |
| Ceratopteris chingii | 3.065 | 8.065 | 40.323 | 32.258 | 0.200 | 1.000 | J.M. Wang, 3886 (NAS) |
| Ceratopteris chingii | 1.129 | 5.484 | 17.742 | 16.129 | 0.309 | 0.760 | Y. Zou, 01679 (PE) |
| Ceratopteris chingii | 1.774 | 4.839 | 20.968 | 19.355 | 0.231 | 0.833 | Y. Zou, 01679 (PE) |
| Ceratopteris chingii | 0.600 | 7.000 | 15.000 | 14.000 | 0.467 | 0.571 | Y.H. Yan, YYH15457 (CSH) |
| Ceratopteris chingii | 0.600 | 6.500 | 19.000 | 15.000 | 0.342 | 0.833 | Y.H. Yan, YYH15458 (CSH) |
| Ceratopteris chingii | 0.300 | 7.000 | 16.800 | 7.000 | 0.417 | 1.400 | Y.H. Yan, YYH15459 (CSH) |
Spores were picked from samples with an insect needle and fixed on a copper platform with conductive adhesive. The gold-coated spores were examined under a Quanta250 (FEI, Hillsboro, OR, USA) SEM. We observed 30 spores for each sample.
2.2. Chromosome countingRoot-tips were pretreated with P-dichlorobenzene at room temperature for 3 h and then fixed in ethanol: glacial acetic acid (3:1 v/v) for 1 h at 4 ℃. The fixed root-tips were dissociated in 3% cellulase: 2.5% pectinase (1:1 v/v) for 15 min at 25 ℃, and the chromosomes were stained with carbol fuchsin. The photomicrographs were imaged on an Axio Scope A1 microscope (Carl Zeiss, Zena, Germany) (Zhang et al., 2019).
2.3. Phylogenetic analysisSequences for five plastid DNA regions (rbcL, rpoC2, rbcL–atpB, trnL (UAA)–trnF (GAA), and trnW (CCA)–trnP (UGG)) were used for the phylogenetic analysis according to Masuyama et al. (2002), Wei et al. (2017) and Zhang et al. (2020b). For primers and sequence amplification, we followed protocols described in Masuyama et al. (2002) and Abid et al. (2020). We sampled 25 species of Ceratopteris including the presumed new species. Acrostichum speciosum was chosen for the outgroup based on Zhang et al. (2020b). A list of samples, location information, GenBank accession numbers, and voucher information are provided in Table S1.
Phylogenetic trees were constructed based on maximum parsimony (MP) in MEGA X v.10.0.5 (Kumar et al., 2018), maximum likelihood (ML) in IQ-TREE v.1.6.8 (Nguyen et al., 2015), and Bayesian inference (BI) in MrBayes v.3.2.6 (Ronquist et al., 2012). Each DNA sequence matrix was aligned using MAFFT v.7 (Katoh and Standley, 2013) with manual adjustments using BioEdit v.4.0.6.2 (Hall, 1999).
For MP analysis, 1000 replicates were performed, with up to 10 tree-bisection-reconnection (TBR) searches per replicate and a maximum of 100 trees held per TBR search. In BI analysis, ModelFinder v.1.6.8 (Kalyaanamoorthy et al., 2017) was used to select the best-fit model of nucleotide substitution based on the Bayesian information criterion (BIC). The HKY + F model was selected for the cpDNA data set. Markov Chain Monte Carlo analyses were run for five million generations in BI analysis, with sampling every 500 generations, and the first 25% discarded as burn-in. We confirmed that the runs had converged by verifying that the average standard deviation of the split frequencies was below 0.01. For ML analysis, ModelFinder v.1.6.8 (Kalyaanamoorthy et al., 2017) was used to select the best-fit model of nucleotide substitution based on the BIC. The HKY + F model was selected for the cpDNA data set and 5000 replicates were performed for bootstrap analyses.
In addition to the MP phylogenetic trees, all phylogenetic analyses were performed in PhyloSuite 1.2.1 (Zhang et al., 2020a).
3. Results 3.1. Taxonomic treatment 3.1.1. Ceratopteris chunii Y.H. Yan, sp. nov. (Fig. 1)Type: CHINA, Guangdong, Guangzhou, South China Botanical Garden, the Chinese Academy of Sciences (CAS), 23°11′N, 113°21′E, elev. 9 m, 20 August 2017. Yue-Hong Yan Fern09730 (holotype: CSH; isotypes: CSH, IBSC and PE).
Morphologically, Ceratopteris chunii and C. gaudichaudii var. vulgaris are similar, however, the sterile leaf of C. chunii has a longer stipe and shorter basal pinna than that of C. gaudichaudii var. vulgaris.
Subaquatic, annual, 7–27 cm tall, soft and juicy, green when young and brownish when old. Rhizomes: erect, dense cover root. Scales sparse on stipe apex and dense on the base, brown to hyaline. Leaves: clustered and dimorphic. Sterile leaves: 13–23 cm long; stipes 4–11 cm long, 0.1–0.3 cm wide at base, green and semicylindrical; laminae 2–4 cm, 2–3-pinnate, broadly ovate or ovate-triangular, deeply divided, apex acute, base rounded-cuneate, pinna 1 × 0.5 cm, alternate, ovate to oblong, base rounded-cuneate. Fertile leaves: 8–27 cm long; stipes 4–12 cm long at base, 0.1–0.3 cm wide, stipe same as in sterile leaves; laminae 4–10 cm, 2–3-pinnate, oblong or ovate-triangular, base rounded-cuneate or rounded-truncate, apex acuminate; entire, terminal segments linear, acute to attenuate; pinna 1 × 4 cm, alternate, ovate or narrowly triangular. Spores: tetrahedral-globose, 120–131 μm in diameter, granulate deposits, coarse ridges, and rodlets formed of coalescent particles on the surface. Chromosomes: 2n = 78, diploid.
Additional Specimens Examined —CHINA. Guangdong Province, Guangzhou City, South China Botanical Garden, CAS, 20 August 2017. Yue-Hong Yan Fern09680 (CSH); 30 June 2020, Fa-Guo Wang YYH15461–YYH15462 (PE and IBSC).
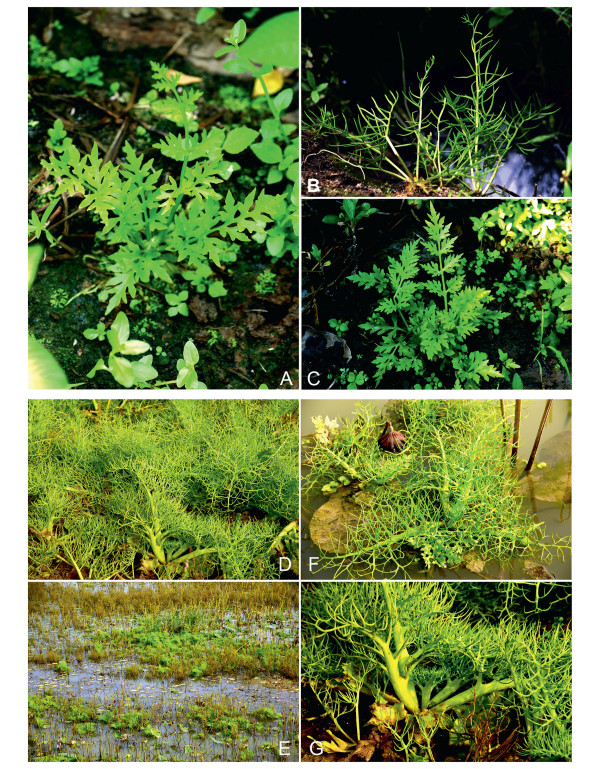
Habitat —Ceratopteris chunii is currently only found in Guangdong Province, China, where it grows in ponds, ditches, and rice fields at elevations of 9–21 m (Fig. 2A–C).

|
| Fig. 1 Illustrations of Ceratopteris chunii Y.H. Yan, sp. nov. and C. chingii Y.H. Yan & Jun H. Yu, sp. nov. (A) Blade of sterile leaf of C. chunii; (B) Habit of C. chunii; (C) Spores of C. chunii; (D) Habit of C. chingii; (E) Blade of sterile leaf of C. chingii; (F) Spores of C. chingii. |
|
| Fig. 2 Habitat of Ceratopteris. (A) Habitat of C. chunii; (B), (C) Young fertile leaf and sterile leaf of C. chunii. (D), (E) Habitat of C. chingii; (F) Young fertile leaf and sterile leaf of C. chingii; (G) Stipe of fertile leaf of C. chingii. |
Chinese name —Huan Yong Shui Jue (焕镛水蕨).
Etymology —In honor of Professor Huan-Yong Chen for his contribution to plant investigations in China and the establishment of South China Botanical Garden, CAS.
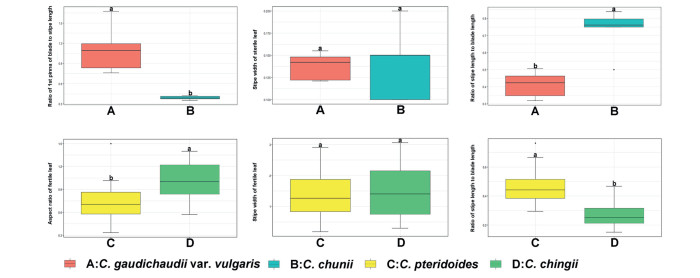
Morphological evidence —Morphological comparisons show that the stipe width of the sterile leaf of Ceratopteris chunii and C. gaudichaudii var. vulgaris is similar (P > 0.05; Fig. 3). However, C. chunii has shorter basal pinnae (P < 0.05) and longer stipes (P < 0.05). These are important diagnostic characters for separating C. chunii from C. gaudichaudii var. vulgaris.
|
| Fig. 3 Analysis of variance of the Ceratopteris morphometric data. (A) C. gaudichaudii var. vulgaris; (B) C. chunii; (C) C. pteridoides; (D) C. chingii. |
Spore evidence —The spores of Ceratopteris gaudichaudii var. vulgaris and C. thalictroides are similar. Spores of both have dense granulate deposits and rodlets on the surface (Dettmann and Clifford, 1991; Tryon and Lugardon, 1991); however, C. chunii spores have comparatively fewer granulate deposits and rodlets (Fig. 4A).

|
| Fig. 4 Spore morphology of Ceratopteris. (A) C. chunii; (B) C. chingii. |
Chromosome evidence —Ceratopteris gaudichaudii var. vulgaris is a tetraploid with 2n = 156 (Masuyama and Watano, 2010), whereas C. chunii is considered diploid with a chromosome number of 2n = 78 (Fig. S1A).
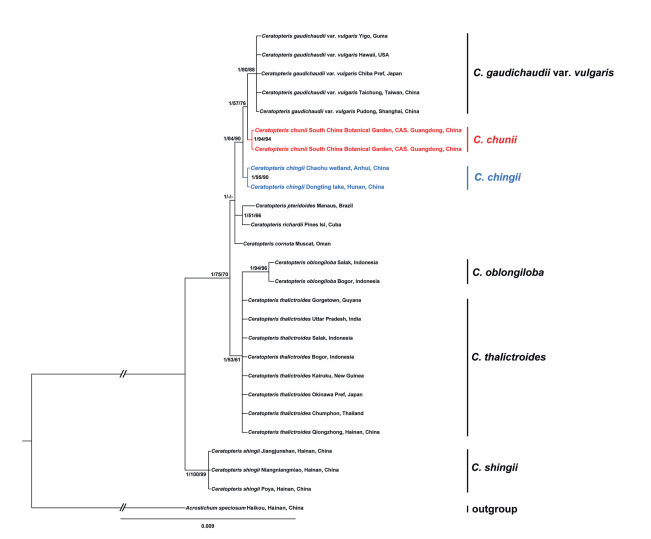
Phylogenetic evidence —The topologies of the phylogenetic trees from the MP, BI, and ML analyses were mostly identical, with support values shown in Fig. 5. The sampled populations of Ceratopteris chunii formed a monophyletic group with strong support (MPBS = 91%, MLBS = 94%, and BIPP = 1), suggesting that C. chunii is an independent lineage from other previously reported Ceratopteris groups.
|
| Fig. 5 Bayesian consensus tree of Ceratopteris based on the sequences of rbcL, ropC2, rbcL–atpB, trnL (UAA)–trnF (GAA) and trnW (CCA)–trnP (UGG) intergenicspacer regions. Numbers on the branches are support values (PPBI/BSML/BSMP). Dash (-) indicates nodes with BSMP or BSML < 50%. The branch length of the outgroups is shortened as indicated by "//". |
Type: CHINA, Anhui, Chaohu, Chaohu wetland, in some reed marshes, 31°38′N, 117°44′E, elev. 5 m, 16 October 2018. Hui Shang SG2911 (holotype: CSH; isotypes: CSH, IBSC and PE).
The morphological features of Ceratopteris chingii are similar to those of C. pteridoides except for the shorter stipe and the subelliptic shape of fertile leaf.
Subaquatic, annual, 10–41 cm tall, soft and juicy, green when young and brownish when old. Stipe, rachis, and costa all obviously expanded toward base. Rhizomes: floating or penetrating deep into the mud, branched. Scales sparse on stipe apex, brownish hyaline. Leaves: clustered and dimorphic. Sterile leaves: 1–10 cm long; stipes 1–7 cm, 0.3–2 cm wide at base; green, smooth, and hemicylindrical; lamina ovate-triangular, 2–7 cm, 2-pinnate, terminal pinna blunt, lobes triangular to broadly loriform. Fertile leaves: subelliptic, 10–33 cm long; stipes 3–12 cm, 0.3–3 cm wide at base; stipe same as in sterile leaves; lamina 4–15 cm, 3-pinnate, base rounded-cuneate or rounded-truncate, apex acuminate; entire, terminal segments linear, acute to attenuate; pinna 3 × 6 cm, alternate, ovate or narrowly triangular. Spores: tetrahedral-globose, 95–113 μm in diameter, smooth, granulate deposits and coarse ridges on sporangium surface. Chromosomes: 2n = 78, diploid.
Additional Specimens Examined —CHINA. Anhui and Hunan Provinces, Anhui Province Chaohu City, Chaohu wetland, in some reed marshes, 16 October 2018. Hui Shang SG2912–SG2920 (CSH); Hunan Province, Yueyang City, north of Dongting Lake National Nature Reserve at Caisang Lake, 12 October 2017, Xun Lin Yu YYH15457–YYH15458 (PE and IBSC).
Habitat —Ceratopteris chingii is currently found in Anhui and Hunan Provinces, China. It usually floats on the lakes or grows in wetlands, at elevations of 5–30 m (Fig. 2D–G).
Chinese name —Cu Geng Shui Jue (粗梗水蕨).
Etymology —In honor of Professor Ren-Chang Ching for his contribution to fern investigations in China and the establishment of Lushan Botanical Garden, CAS. The Chinese name preserves the name widely used in the past.
Morphological evidence —Morphological comparisons show that the stipe width of the fertile leaf of Ceratopteris chingii and C. pteridoides is similar (P > 0.05; Fig. 3). However, two important diagnostic characters separate C. chingii from C. pteridoides. The relative length of the stipe to blade is smaller in C. chingii (P < 0.05), indicating that the stipe is shorter. In addition, the aspect ratio indicates that the fertile leaf of C. chingii is subelliptical rather than triangular (P < 0.05).
Spores evidence —Ceratopteris chingii spores are relatively smooth with fewer granulate deposits on the surface (Fig. 4B) than those of C. pteridoides (Dettmann and Clifford, 1991; Tryon and Lugardon, 1991).
Chromosome evidence —Ceratopteris chingii chromosome number is 2n = 78 (Fig. S1B), and this species was identified as diploid.
Phylogenetic evidence —MP, BI, and ML phylogenetic analyses all showed that Ceratopteris chingii formed a monophyletic clade with support of MPBS = 87%, MLBS = 95%, and BIPP = 1, and was sister to the lineage including C. gaudichaudii var. vulgaris and C. chunii (Fig. 5).
4. DiscussionGenerally, plants in relatively stable environments (e.g., aquatic plants) show little morphological variation due to convergent evolution (Schneider and Meyer, 2017). Consequently, identifying morphologically cryptic species such as those in Ceratopteris is challenging. Liao et al. (2011) noted that there are two cryptic species within C. thalictroides complex throughout China, which agreed with previous studies (Masuyama et al., 2002). However, only two species (C. thalictroides and C. pteridoides) are recorded in China (Lin and Masuyama, 2013). Zhang et al. (2020b) described a new endemic species C. shingii in Hainan, China. In addition, Kinosian et al. (2020a) isolated a novel putative cryptic species from C. thalictroides using restriction-site associated DNA sequencing. Multiple lines of evidence indicate that there are many cryptic species in the genus Ceratopteris. Here, C. chunii and C. chingii are recognized as two new cryptic species of Ceratopteris in China.
Our phylogenetic analyses showed that Ceratopteris chunii from Guangdong Province formed a well-support monophyletic group closely related to C. gaudichaudii var. vulgaris (Fig. 5). Masuyama and Watano (2010) separated C. gaudichaudii var. vulgaris (type from Japan) from C. gaudichaudii (type from Guam) at the variety level due to their morphological differences, although they have the same rbcL gene sequence and chromosome number (2n = 156, tetraploid) (type from Japan). Although morphologically similar to C. gaudichaudii var. vulgaris, C. chunii has a sterile leaf with a longer stipe and shorter basal pinna (Fig. 3). SEM showed that C. chunii spores have dense granulate deposits and rodlets on the surface (Fig. 4A). Most importantly, C. chunii is diploid (2n = 78) (Fig. S1A) rather than tetraploid. Based on these cytological analyses, we recommend that C. chunii be separated from these relatives. C. chingii has long been misidentified as C. pteridoides (type from South America) in China. Our cytological studies showed that C. chingii and C. pteridoides are both diploid with 2n = 78 (Fig. S1B; Masuyama and Watano, 2005). The phylogenetic analyses indicated that C. chingii (type from Anhui and Hunan Provinces) and C. pteridoides were more distantly related; C. chingii formed a well-supported clade with MPBS = 87%, MLBS = 95%, and BIPP = 1 (Fig. 5). Furthermore, the leaf-shape is nearly triangular, and the stipe of the fertile leaf is longer in C. pteridoides, as recorded by Hieronymus (1905), whereas those of C. chingii are subelliptic and short (Fig. 3). Most importantly, the spore surface of C. chingii is relatively smooth with fewer granulate deposits (Fig. 4B). Overall, our results indicate that C. chingii is a new cryptic species in China.
The origin of the tetraploid complex of Ceratopteris thalictroides is a mystery, and all cryptic species in C. thalictroides are tetraploid (2n = 154, 156; Masuyama and Watano, 2005). Various hypotheses have been proposed to explore the origin and speciation mechanism of the C. thalictroides complex. Unfortunately, one of the diploid parents is likely extinct or has yet to be identified (Masuyama and Watano, 2005; Adjie et al., 2007; Kinosian et al., 2020a). Here, the discovery of C. chunii and C. chingii, two diploid Ceratopteris in Asia, provides a framework for testing hypotheses about the origins of the tetraploid C. thalictroides complex. To unravel the origins of tetraploid Ceratopteris, future work should use nuclear gene sequencing or whole-genome sequencing.
Author contributionsYue-Hong Yan, Yong-Bo Liu and Rui Zhang designed the experiments. Jun-Hao Yu drafted this paper; Field investigations and data analyses were performed by Jun-Hao Yu, Qiao-Ling Liu, Fa-Guo Wang, Xun-Lin Yu, and Xi-Ling Dai; Yue-Hong Yan, Rui Zhang, and Yongbo Liu revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interestThe author declares no conflict of interest.
AcknowledgmentsWe would like to thank Hui Shang, Yu-Feng Gu, Li-Jun Chen and Wen Shao for their assistance in data analyses. The research was funded by the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China (2019HJ2096001006), the Shanghai Municipal Administration of Forestation and City Appearance (grant number G192421), the Biological Resource Programme CAS (ZSZY-001-8), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA13020603) and the Basic Project of Ministry of Science and Technology of China under Grant (2015FY110200).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.10.002.
Abid, S., Kaliraj, L., Arif, M.H., et al., 2020. Molecular and morphological discrimination of Chrysanthemum indicum using allele-specific PCR and T-shaped trichome. Mol. Biol. Rep., 47: 7699-7708. DOI:10.1007/s11033-020-05844-2 |
Adjie, B., Masuyama, S., Ishikawa, H., et al., 2007. Independent origins of tetraploid cryptic species in the fern Ceratopteris thalictroides. J. Plant Res., 120: 129-138. DOI:10.1007/s10265-006-0032-5 |
Benedict, R.C., 1909. The Genus Ceratopteris: a preliminary revision. Bull. Torrey Bot. Club., 36: 463-476. DOI:10.2307/2479023 |
Brade, A.C., 1964. Filices novae Brasilienses VIII. Arq. Jard. Bot. Rio de Janeiro, 18: 31. |
Brongniart, A., 1821. Description dun nouveau genre de fougere, nomme Ceratopteris. Bull. ci. Soc. Philom. Paris, 8: 184-187. |
Brongniart, A., 1823. Dictionnaire classique d'histoire naturelle. Dict. Class. Hist. Nat., 3: 351. |
Dettmann, M.E., Clifford, H.T., 1991. Spore morphology of anemia, Mohria, and Ceratopteris (Filicales). Am. J. Bot., 78: 303-325. DOI:10.1002/j.1537-2197.1991.tb15194.x |
Grant, V., 1981. Plant Speciation, second ed. Columbia University Press
|
Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser., 41: 95-98. |
Hickok, L.G., Klekowski, E.J., 1974. Inchoate speciation in Ceratopteris: an analysis of the synthesized hybrid C. richardii x C. pteridoides. Evolution, 28: 439-446. |
Hieronymus, G., 1905. Plantae Lehmannianae in Guatemala, Columbia et Ecuador regionibusque finitimis collectae. Pteridophyta. Bot. Jahrb. Syst., 34: 561-582. |
Hickok, L.G., 1977. Cytological relationships between three diploid species of the fern genus Ceratopteris. Can. J. Bot., 55: 1660-1667. DOI:10.1139/b77-194 |
Hickok, L.G., 1979. A cytological study of intraspecific variation in Ceratopteris thalictroides. Can. J. Bot., 57: 1694-1700. DOI:10.1139/b79-207 |
IPNI, 2021. The International Plant Names Index. http://www.ipni.org
|
Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., et al., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods, 14: 587-589. DOI:10.1038/nmeth.4285 |
Katoh, K., Standley, D.M., 2013. MAFFT: multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kinosian, S.P., Pearse, W.D., Wolf, P.G., 2020. Cryptic diversity in the model fern genus Ceratopteris (Pteridaceae). Mol. Phylogenet. Evol., 152: 106938. DOI:10.1016/j.ympev.2020.106938 |
Kinosian, S.P., Pearse, W.D., Wolf, P.G., 2020. There and back again: reticulate evolution in Ceratopteris. Am. Fern J., 110: 193-210. |
Kumar, S., Stecher, G., Li, M., et al., 2018. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol., 35: 1547-1549. DOI:10.1093/molbev/msy096 |
Leprieur, F.M.R., 1830. Annals of the natural Sciences. Ann. Sci. Nat. (Paris)., 19: 103. |
Liao, Y.Y., Yang, X.Y., Motley, T.J., et al., 2011. Phylogeographic analysis reveals two cryptic species of the endangered fern Ceratopteris thalictroides (L.) Brongn. (Parkeriaceae) in China. Conserv. Genet., 12: 1357. DOI:10.1007/s10592-011-0236-7 |
Lin, Y., Masuyama, S., 2013. Ceratopteris. Flora of China 2-3, 180-181
|
Lloyd, R.M., 1974. Systematics of the genus Ceratopteris Brongn. (Parkeriaceae). II. Taxonomy. Brittonia, 26: 139-160. DOI:10.2307/2805883 |
Masuyama, S., Yatabe, Y., Murakami, N., et al., 2002. Cryptic species in the fern Ceratopteris thalictroides (L.) Brongn. (Parkeriaceae). I. Molecular analyses and crossing tests. J. Plant Res., 115: 87-97. DOI:10.1007/s102650200013 |
Masuyama, S., Watano, Y., 2005. Cryptic species in the fern Ceratopteris thalictroides (L.) Brongn. (Parkeriaceae). II. Cytological characteristics of three cryptic species. Acta Phytotax. Geobot., 56: 231-240. |
Masuyama, S., 2008. Cryptic species in the fern Ceratopteris thalictroides (Parkeriaceae). III. Referential diagnostic characters of three cryptic species. J. Plant Res., 121: 279-286. DOI:10.1007/s10265-008-0159-7 |
Masuyama, S., Watano, Y., 2010. Cryptic species in the fern Ceratopteris thalictroides (L.) Brongn. (Parkeriaceae). IV. Taxonomic revision. Acta Phytotax. Geobot., 61: 75-86. |
Nguyen, L.T., Schmidt, H.A., Von Haeseler, A., et al., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32: 268-274. DOI:10.1093/molbev/msu300 |
Paris, C.A., Wagner, F.S., Wagner, W.H., 1989. Cryptic species, species delimitation, and taxonomic practice in the homosporous ferns. Am. Fern J., 79: 46-54. DOI:10.2307/1547159 |
Ronquist, F., Teslenko, M., Van Der Mark, P., et al., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Schneider, R.F., Meyer, A., 2017. How plasticity, genetic assimilation and cryptic genetic variation may contribute to adaptive radiations. Mol. Ecol., 26: 330-350. DOI:10.1111/mec.13880 |
Shen, H., Jin, D.M., Shu, J.P., et al., 2018. Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience, 7: 116. |
Smith, A.R., Love, A., Love, D., et al., 1977. Cytotaxonomical atlas of the Pteridophyta. Syst. Bot., 2: 86. DOI:10.2307/2418510 |
Tropicos, 2021. Tropicos. org. Missouri botanical garden. http://www.tropicos.org
|
Tryon, A.F., Lugardon, B., 1991. Spores of the Pteridophyta: Surface, Wall Structure, and Diversity Based on Electron Microscope Studies. Springer Verlag New York Inc. vol. 33, 126
|
Wei, R., Yan, Y.H., Harris, A.J., et al., 2017. Plastid phylogenomics resolve deep relationships among eupolypod II ferns with rapid radiation and rate heterogeneity. Genome Biol. Evol., 9: 1646-1657. DOI:10.1093/gbe/evx107 |
Zhang, D., Gao, F., Jakovlić, I., et al., 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour., 20: 348-355. DOI:10.1111/1755-0998.13096 |
Zhang, R., Wang, F.G., Zhang, J., et al., 2019. Dating whole genome duplication in Ceratopteris thalictroides and potential adaptive values of retained gene duplicates. Int. J. Mol. Sci., 20: 19-26. |
Zhang, R., Yu, J.H., Shao, W., et al., 2020. Ceratopteris shingii, a new species of Ceratopteris with creeping rhizomes from Hainan, China. Phytotaxa, 449: 23-30. DOI:10.11646/phytotaxa.449.1.3 |



