b. School of Ecology and Environmental Science & Yunnan Key Laboratory for Plateau Mountain Ecology and Restoration of Degraded Environments, Yunnan University, Kunming 650091, PR China;
c. Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining 810001, PR China;
d. The Germplasm Bank of Wild Species, Institute of Tibetan Plateau Research at Kunming, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, PR China;
e. Yunnan Lijiang Forest Ecosystem National Observation and Research Station, Kunming Institute of Botany, Chinese Academy of Sciences, Lijiang 674100, Yunnan, PR China
A species is the fundamental unit of classification in the biological sciences. However, defining species and understanding speciation represent important and controversial challenges for evolutionary biologists. Currently, there are between 20 (Wilkins, 2009) to 60 (Zhou and Yang, 2011) different species concepts, each emphasizing different criteria for delimiting a species (e.g., the Biological Species Concept emphasizes reproductive isolation, whereas the Phylogenetic Species Concept emphasizes evolutionary descent). Consequently, the species concept used to define a species affects where species boundaries are drawn and the number of species recognized (de Queiroz, 2005a).
In recent years, several unified species concepts have been proposed to define a species as a separately evolving metapopulation lineage (de Queiroz, 1998, 2005b, 2007). The integrative species concept emphasizes the importance of multiple lines of evidences in delimiting species, especially for stable species and species undergoing speciation (Liu, 2016). The gen-morph species concept has modified the morphological-biological species concept to define a species as a group of natural populations that is genetically compatible, with gene flow within the group, but isolated from any other such groups and that, accordingly, shows continuous variation within the group, but discontinuous (or statistically discontinuous) variation between groups in at least two independent morphological characteristics (Hong, 2016, 2020). Regardless of which species concept is used to define a species, researchers advocate the use of multiple lines of evidence to delimit species (de Queiroz, 2007; Hong, 2016, 2020; Liu, 2016).
Because speciation is a process, and most species concepts focus on the different cut-offs in this process (Liu, 2016), the use of multiple methods to define a species has been proposed to increase accuracy. For example, the Paeonia delavayi Franch. complex (Ranunculaceae), which has traditionally included one to four species, has been revised into two species without any varieties (P. delavayi and P. ludlowii (Stern & Taylor) D.Y. Hong) based on several morphological traits (Hong, 2016, 2020). This revision is fully supported by phylogenetic analysis of Paeonia based on chloroplast sequence data and single-copy nuclear markers (Zhou et al., 2014). Similarly, researchers have used multiple morphological traits, ecological niche and molecular data to delimit Orinus (Poaceae) species, identifying a new undescribed species and dividing the genus into three species instead of the six species identified earlier (Su et al., 2015). These studies demonstrate that integrating multiple lines of evidence is an efficient approach to delimiting, describing and discovering species.
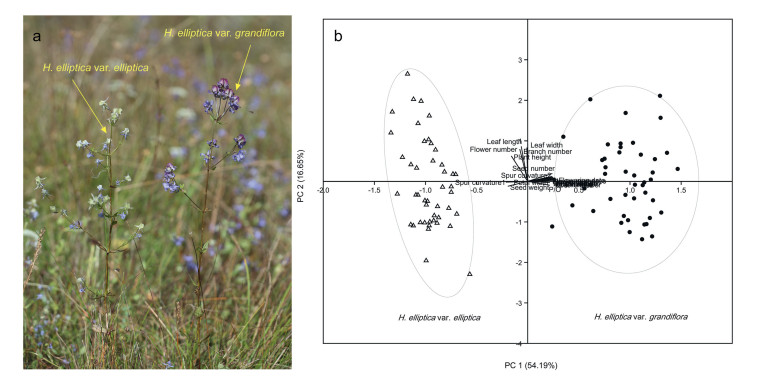
Halenia Borkh. (spurred gentian), a genus of Gentianaceae that includes about 100 species, originated in East Asia and migrated to America, where it diversified (von Hagen and Kadereit, 2003). In China, only two species in this genus are known to occur, H. corniculata (L.) Cornaz and H. elliptica D. Don (Ho and Prigle, 1995). H. elliptica is widely distributed in western China, and produces seeds via outcrossing and autonomous selfing (Yang et al., 2018). Two varieties of H. elliptica have been identified based on flower size (Ho and Prigle, 1995), spur length and curvature: H. elliptica var. elliptica and H. elliptica var. grandiflora (Fig. 1a). Our previous work suggested that flower size of both varieties showed a clinal change and no obvious gap existed between them, indicating the two varieties could be merged (Wang et al., 2011). However, hand-pollinated hybridization between the two varieties did not produce any seed in our preliminary experiment, indicating reproductive isolation. Here, we used multiple lines of evidence to evaluate the taxonomic treatment of H. elliptica var. elliptica and H. elliptica var. grandiflora. For this purpose, we determined morphological and mating system differences, the degree of reproductive isolation, and phylogenetic relationships between the two varieties.
|
| Fig. 1 (a) Morphology of the two Halenia elliptica varieties in the sympatric population. (b) Principle component analysis (PCA) of 21 traits of the two varieties of H. elliptica. PC 1 explains 54.19% of the total variance, with major contributions from initial flowering date of plant, flower openness, petal height, petal width at the base, spur length, width at the base of spur, distance from the top to the base of spur, pistil height, stamen height, pollen number, ovule number, pollen/ovule ratio and seed number. PC 2 explains 16.65% of the total variance, with major contributions from plant height, total flower number, number of lateral branches, length and width of the third leaf on the main branch from the bottom. |
We compared Halenia elliptica var. elliptica and H. elliptica var. grandiflora morphology, flowering phenology, mating system, pollinator preference, and hybrid pollination at one site in the alpine meadow area around Lijiang Alpine Botanical Garden, where the two varieties occur sympatrically (26°59′56″N, 100°11′59″E, 2622 m, Yunnan province, P. R. China). All field observations, measurements and experiments were performed in this population in 2020. It should be noted that trait differences between the two varieties with allopatric distribution were not considered because geographical isolation may have been sufficient to cause reproductive isolation between the two varieties.
2.2. Morphological traitsA total of 45 plants of each variety were identified according to flower morphology of the top flower and subsequently labelled every three days. For each variety, we recorded the initial flowering date, plant height and number of lateral branches. We also measured the length and width of the third pair of leaves from the bottom of the main branch because the first and second pairs of leaves had wilted when we performed our measurements.
We determined the durations of pollen shedding and stigma receptivity by recording key time points in the flowering process twice daily. We considered the absence of obvious pollen grains on the anther as the time point for completion of pollen shedding. In accordance with the flowering process of many gentians (Petanidou et al., 2001; Duan et al., 2005, 2010; Meng et al., 2012), we considered stigmas no longer receptive after turning brown. In addition, we determined seed number per fruit and seed weight from fruit collected from the top flower on each plant when the capsule was ripe but not dehiscent. Seed number per fruit was determined after air-drying fruits and the total weight of all seeds in each fruit was weighed using a digital balance (minimum to 0.1 mg).
For each variety of Halenia elliptica we characterized 21 floral traits from one newly opened flower adjacent to the top flower from each plant (Table 1). Flower openness, petal height and width, spur base width, and spur length were measured with a digital caliper. We measured spur length with a piece of string. To evaluate the curvature of the spur, we measured from the top of the spur to the top of the petal, and from the top of the spur to its base (Fig. S1). To determine the total number of pollen grains, ovule number, and the pollen: ovule ratio, we first fixed flowers in standard FAA solution (38% formalin–acetic acid–70% alcohol = 5:5:90 by volume). Anthers were squashed in a centrifuge tube with 5 ml of 70% alcohol and one drop of detergent. Pollen grains were counted under a microscope in ten replicates of 10 μL. Ovule number was determined under a stereoscope.
| Trait | H. e. var. elliptica | H. e. var. grandiflora | Data transformation | t-test | |||
| N | Mean ± SD | N | Mean ± SD | ||||
| Initial flowering date of plant | 45 | 240.64 ± 1.88 | 45 | 284.4 ± 2.18 | Log10 | P < 0.01 | |
| Plant height (cm) | 45 | 71.17 ± 11.57 | 45 | 58.46 ± 8.05 | Log10 | P < 0.01 | |
| Total flower number | 45 | 111.16 ± 64.07 | 45 | 87 ± 47.15 | Log10 | P < 0.05 | |
| Number of lateral branches | 45 | 6.67 ± 1.61 | 45 | 6.73 ± 1.51 | Log10 | P = 0.84 | |
| Length of the third leaf on the main branch from the bottom (cm) | 45 | 3.77 ± 0.76 | 45 | 3.51 ± 0.75 | Log10 | P = 0.11 | |
| Width of the third leaf on the main branch from the bottom (cm) | 45 | 1.86 ± 0.37 | 45 | 1.89 ± 0.36 | Log10 | P = 0.67 | |
| Length/width ratio of leaf | 45 | 2.03 ± 0.2 | 45 | 1.85 ± 0.20 | Log10 | P < 0.01 | |
| Flower openness (cm) | 45 | 0.86 ± 0.1 | 45 | 1.13 ± 0.12 | Sqrt | P < 0.01 | |
| Petal height (cm) | 45 | 0.76 ± 0.04 | 45 | 0.95 ± 0.09 | Sqrt | P < 0.01 | |
| Petal width at the base (cm) | 45 | 0.46 ± 0.06 | 45 | 0.57 ± 0.06 | Sqrt | P < 0.01 | |
| Spur length (cm) | 45 | 0.92 ± 0.07 | 45 | 1.34 ± 0.14 | Sqrt | P < 0.01 | |
| Width at the base of spur (cm) | 45 | 0.17 ± 0.03 | 45 | 0.27 ± 0.03 | Sqrt | P < 0.01 | |
| Distance from the top of spur to the top of petal (cm) | 45 | 0.96 ± 0.09 | 45 | 0.70 ± 0.14 | Sqrt | P < 0.01 | |
| Distance from the top of spur to the base of spur (cm) | 45 | 0.85 ± 0.06 | 45 | 1.02 ± 0.11 | Sqrt | P < 0.01 | |
| Pistil height (cm) | 45 | 0.46 ± 0.05 | 45 | 0.81 ± 0.08 | Sqrt | P < 0.01 | |
| Stamen height (cm) | 45 | 0.42 ± 0.04 | 45 | 0.64 ± 0.07 | Sqrt | P < 0.01 | |
| Pollen number | 45 | 12, 662 ± 1753 | 45 | 37, 034 ± 6187 | Log10 | P < 0.01 | |
| Ovule number | 45 | 10.00 ± 1.99 | 45 | 19.40 ± 3.41 | Log10 | P < 0.01 | |
| Pollen/ovule ratio | 45 | 1298 ± 231 | 45 | 1938 ± 340 | Log10 | P < 0.01 | |
| Seed number | 45 | 8.33 ± 2.01 | 45 | 17.89 ± 3.81 | Log10 | P < 0.01 | |
| Seed weight (mg) | 45 | 1.35 ± 0.29 | 45 | 0.9 ± 0.21 | Sqrt | P < 0.01 | |
We also examined whether chromosome number differed between the two varieties. A previous study reported that the chromosome number of Halenia elliptica var. elliptica is 2n = 2x = 22 (Yuan and Kupfer, 1993), which is shared by most Halenia species (Rice et al., 2015). We thus only examined the chromosome number of H. elliptica var. grandiflora. Seeds of H. elliptica var. grandiflora were germinated into seedlings in the laboratory, and root tips with a length of 0.5 cm were collected and pretreated with a high-pressure nitrous oxide (N2O) at a pressure of 6 standard atmosphere pressure for 2 h at room temperature using a gas pressure chamber (Kato, 1999; Andres and Kuraparthy, 2013). They were then fixed in ethanol: glacial acetic acid (3:1 in volume) for at least 5 min at 4 ℃. Each root tip was squashed in a drop of 45% acetic acid. Slides with well-spread and intact chromosomes were stored at −80 ℃ for over 30 min. Slides were air-dried before chromosomes were counted and photographed using an Olympus BX63 fluorescence microscope (Olympus Corp, Tokyo, Japan).
2.3. Flowering phenologyOur preliminary observations suggested that flowering phenology of the two varieties differed. To quantify these differences in flowering phenology, we set up five plots (1m × 1m) for each variety. Because the longevity of a single flower is about four days, we observed the total number of flowers in each plot every week from August 15 to October 24. The relative flowering phenology was calculated by dividing the total number of flowers with the number of flowers on each time point in each plot (Savolainen et al., 2006).
2.4. Breeding systems and hand pollinated hybridizationTo compare breeding systems of the two varieties, we performed the following treatments for each. (1) Autonomous selfing was examined by excluding pollinators from flower buds with fine nylon nets. (2) The contribution of pollinator-mediated seed production was examined by emasculating flower buds and leaving them open for pollination. (3) Apomixis was examined by emasculating flower buds and netting. (4) We hand-pollinated receptive plants with emasculated and netted flower buds using pollen from the same plant. (5) We hand-pollinated receptive plants with emasculated and netted flower buds using pollen grains from plants of the same variety but about 10 m away. (6) We hand-pollinated receptive plants with emasculated and netted flower buds using pollen grains from plants belonging to the other variety. (7) As a control, flower buds were left for open pollination. All labelled flowers in these treatments were on different plants, but with similar position on the plants so that the position effect on seed production could be excluded (Zhang et al., 2011). Although we planned experiments for 60 flowers per treatment, unexpected damage reduced the sample size to 40 ripe dehiscent fruits per treatment. Seed number per fruit was determined in the laboratory.
2.5. Pollinator preferenceTo examine pollinator preference for the two varieties of Halenia elliptica, we firstly identified potential pollinators when both varieties were in anthesis. We pre-selected three to five plants before formal observation, and recorded all insects that visited these flowers. If an insect touched the anthers and stigmas when it visited the flowers, we considered it to be a potential pollinator. Formal observations were performed on five sunny days, with a total observation time of 20 h. We classified the pollinators according to pollinator functional groups, i.e., bees (including bumblebees and honeybees), butterflies, moths and flies. Visitation rates were calculated for the dominant pollinator functional group for each variety.
To identify the fidelity of pollinators, we tracked dominant pollinators and recorded their visitation bouts when the two varieties flowered simultaneously. Pollinators were tracked and recorded until the pollinator shifted to other plant species or we were unable to follow it. We hypothesized that pollinators did not prefer either variety of Halenia elliptica, and that pollinators would visit H. elliptica var. elliptica and H. elliptica var. grandiflora with the same probability (0.5). To score pollinator fidelity, we gave 0.5 to a pollinator when it was observed visiting one variety and then shifted to visit another plant of the same variety. If the pollinator shifted from one variety to visit a plant of the other variety, we afforded that pollinator a score of −0.5. For each pollinator, the probability was summed and then divided by the number of shifts among plants within one bout. Finally, the average score for a variety was calculated by first summing the scores of one pollinator functional group and then dividing by the total number of insects. Theoretically, if no visitation preference occurred, the average score approached 0. If there was a visitation preference, the average score would approach 0.5 for one variety and −0.5 for the alternative variety.
2.6. Phylogenetic relationshipWe collected leaves from 12 individuals in each of three populations of Halenia elliptica var. elliptica and four populations of H. elliptica var. grandiflora (Table S1); leaves were stored in paper bags with silica gel inside. Voucher specimens have been deposited at the Herbarium of Kunming Institute of Botany (KUN), Chinese Academy of Sciences. We randomly selected a total of 14 individuals (two per population) for analysis. Total genomic DNA was extracted using DNeasy™ Tissue Kit (Qiagen, Hilden, Germany). PCR primers and the amplification protocol for the nuclear ribosomal Internal Transcribed Spacer (nrITS) region and rpl16 intron followed von Hagen and Kadereit (2001) and Small et al. (1998), and the sequencing protocol was done by Tsingke Biotechnology Co., Ltd. MEGA X (Kumar et al., 2018) was used to align and manually adjust sequences. The newly obtained sequences of the two varieties were deposited in GenBank under accession numbers MZ097596-MZ097609 for ITS1, MZ097610-MZ097623 for ITS2 and MZ090593-MZ090606 for rpl16, respectively. Sequences of ITS1, ITS2 and rpl16 were concatenated for phylogenetic analysis following von Hagen and Kadereit (2003).
The final data set included an earlier published data set of Halenia (von Hagen and Kadereit, 2003) and the newly generated sequences. Phylogenetic placement and relationship of the two varieties of H. elliptica were analyzed by means of partitioned Bayesian inference and Maximum Likelihood (ML) analyses. We used Veratrilla baillonii Franch., Swertia bimaculata (Sieb. et Zucc.) Hook. f. et Thoms. ex C.B. Clarke and S. tetraptera Maxim. as outgroup taxa according to von Hagen and Kadereit (2003). Bayesian inference was performed in MrBayes v.3.2.1 (Ronquist and Huelsenbeck, 2003). The best fit models of sequence evolution for the ITS1, ITS2 and rpl16 regions were determined to be TIM3 + G, TIM3 + G and TVM + I, respectively, by the Akaike Information Criterion (Akaike, 1974) using jModelTest v.2.1.10 (Guindon and Gascuel, 2003; Darriba et al., 2012). Two independent runs with one cold and three incrementally heated Monte Carlo Markov chains (MCMCs) were run for 1, 000, 000 generations, with trees sampled every 100 generations. Model parameters were unlinked across partitions. After discarding the first 2500 trees out of the 10, 001 trees as burn-in, the remaining trees were used to build a 50% majority rule consensus tree. Maximum likelihood analysis was performed with RAxML v.8.2.10 (Stamatakis, 2014) via a graphical interface raxmlGUI v.2.0 beta (Edler et al., 2021), implementing the "ML + rapid bootstrap" option with MR-based Bootstrapping criterion (Silvestro and Michalak, 2012). A GTRGAMMA substitution model was specified for each data partition with the model parameters calculated separately.
2.7. Variations in chloroplast genomesThe fresh leaves of two Halenia elliptica varieties were collected from the sympatric population in the study site, and stored in liquid nitrogen. Total genomic DNA was extracted with the Ezup plant genomic DNA prep kit (Sangon Biotech, Shanghai, China). After generation of libraries with an average insert size of 350 bp, sequencing was performed to generate paired-end reads using the Illumina HiSeq 2500 platform at the Grandomics Co., China.
After quality control, clean reads were used to assemble the chloroplast genome with GetOrganelle software (Jin et al., 2020). Bandage v.0.81 was used to check the assembled contigs. The whole chloroplast sequence data of the two varieties have been deposited in the Genome Warehouse in National Genomics Data Center (CNCB-NGDC Members and Partners, 2021), Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers GWHBAVI00000000 and GWHBAVJ00000000, and are publicly accessible at https://bigd.big.ac.cn/gwh. Plastid Genome Annotator (Qu et al., 2019) was used to annotate the assembled plastomes into inverted repeats regions (IRs), large single copy (LSC) and short single copy (SSC) regions. Chloroplast genome sequences were compared and visualized by Mummer v.4.0.0rc1 (Kurtz et al., 2004) and R package circlize (Gu et al., 2014). We compared the numbers of SNP/InDels in the two varieties, and then used previous published data to compare the number of SNP/InDels in H. elliptica var. elliptica and other species of Gentianaceae (Zhang et al., 2020a).
2.8. Statistical analysisInitial flowering phenology of plants was transformed to numeric data by calculating the days from January 1, 2020 (Sletvold et al., 2015), which were subsequently transformed to meet the assumptions of comparisons (Table 1). We first employed independent t-tests to compare the morphological traits between the two varieties, and then a principle component analysis (PCA) was performed to examine the contribution of 21 morphological traits to the main differences between the two varieties.
Seed number per fruit subjected to different treatments was log transformed, and then generalized linear models were employed to analyze the effects of treatment, variety and their interactions on seed production. In addition, we used one simple t-test to compare the score of pollinator fidelity with 0 and 0.5. All analyses were performed using IBM SPSS Statistics v.20.
3. Results 3.1. Morphological traitsExcept for three traits (number of lateral branches, length and width of the third leaf on the main branch from the bottom), the morphological traits we measured for the two Halenia elliptica varieties differed significantly (Table 1). Halenia elliptica var. elliptica flowered earlier, and produced larger plants, more flowers and longer leaves than H. elliptica var. grandiflora. Furthermore, H. elliptica var. elliptica had smaller flowers and fewer but larger seeds. Importantly, the spur of H. elliptica var. grandiflora was more curved than that of H. elliptica var. elliptica, as the spur length H. elliptica var. elliptica was smaller and the distance from the top to the base of spur was shorter, although the distance was greater from the top of spur to the top of petal.
PCA showed that the two varieties could be separated clearly based on the 21 morphological traits measured (Fig. 1b). Specifically, PC 1 and PC 2 explain 54.19% and 16.65% of the total variance (Fig. 1b; Table S2). Several traits contributed significantly to PC1, including initial flowering date of plant, flower openness, petal height, petal width at the base, spur length, width at the base of spur, distance from the top to the base of the spur, pistil height, stamen height, pollen number, ovule number, pollen/ovule ratio and seed number (Fig. 1b; Table S2). Distinct traits contributed significantly to PC 2, including plant height, total flower number, number of lateral branches, length and width of the third leaf on the main branch from the bottom (Fig. 1b; Table S2).
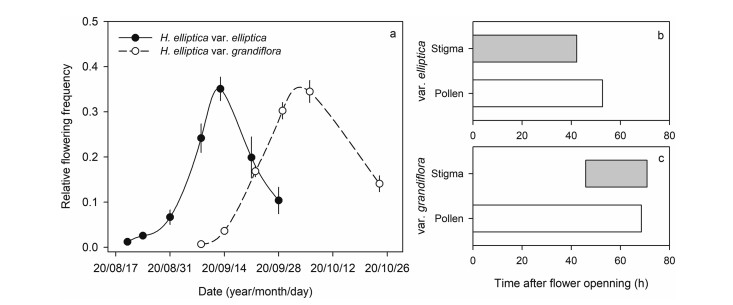
Our field observations also indicated that Halenia elliptica var. elliptica flowered earlier than H. elliptica var. grandiflora, with peak flowering phenology about 18 days earlier (Fig. 2a). When the single flower of H. elliptica var. elliptica opened, the four anthers began to shed pollen and the two stigma lobes opened immediately, both of which lasted about two days (Fig. 2b). In contrast, the four anthers of H. elliptica var. grandiflora began to shed pollen when flower opened, but the two stigma lobes began to open after about two days (Fig. 2c). Since the four stamens differed in height, we compared the lowest and highest stamens and pistil height within one flower using paired sample t-test to evaluate the potential ability of autonomous selfing in each of the two varieties. The results indicated that pistil height was higher than the lowest stamen in H. elliptica var. elliptica (T = 11.40, df = 44, P < 0.01), but did not differ significantly with the highest stamen (T = 0.49, df = 44, P = 0.626). For H. elliptica var. grandiflora, pistil height was significantly higher than both the lowest (T = 8.65, df = 44, P < 0.01) and highest (T = 21.56, df = 44, P < 0.01) stamens. Taken together, these findings indicate that flowers of H. elliptica var. grandiflora exhibit incomplete protandry and complete herkogamy, whereas H. elliptica var. elliptica lacks both of these traits.
|
| Fig. 2 Flowering phenology of the two Halenia elliptica varieties (a) and duration of pollen shedding and stigma receptivity of H. elliptica var. elliptica (b) and H. elliptica var. grandiflora (c) after flower opened. |
Chromosomes of Halenia elliptica var. grandiflora were generally small, and were counted as 2n = 22 (Fig. S2), which is same as other Halenia species (Rice et al., 2015).
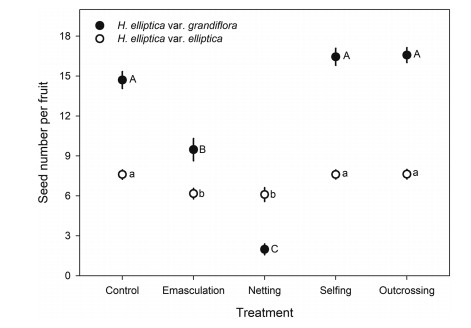
3.2. Breeding system and hand hybrid pollinationHand-pollinated hybridization with pollen grains from the alternate variety failed to produce any seed in either variety of Halenia elliptica, indicating complete post-pollination reproductive isolation. In addition, flowers subjected to emasculation and netting did not produce any seed in either variety, suggesting that apomixis did not occur. We, therefore, did not consider these two treatments in the statistical analysis on seed production. According to our generalized linear models, seed production per fruit was significantly affected by treatment, variety, and their interactions (Table S3). Specifically, flowers of H. elliptica var. elliptica subjected to hand-selfing and outcrossing produced similar seed number, and both flowers subjected to emasculation without netting and netting without emasculation produced seeds successfully, although significantly fewer than the control (Fig. 3). Flowers of H. elliptica var. grandiflora subjected to hand-selfing and outcrossing produced similar numbers of seeds, and emasculation without netting produced fewer seeds than the control (Fig. 3). However, flowers subjected to netting without emasculation produced almost no seeds. These results suggest that H. elliptica var. elliptica exhibits a mixed mating system and can produce seeds via pollinators and autonomous selfing. In contrast, H. elliptica var. grandiflora relies solely on pollinators to produce seeds, and thus exhibits an outcrossing mating system, although this variety was fully self-compatible.
|
| Fig. 3 Seed number per fruit of flowers subjected to different treatments. Comparisons were performed using a generalized linear model. Values labelled with different letters (uppercase letters and lowercase letters were for different varieties) indicate that the difference is significant at the 0.05 level. |
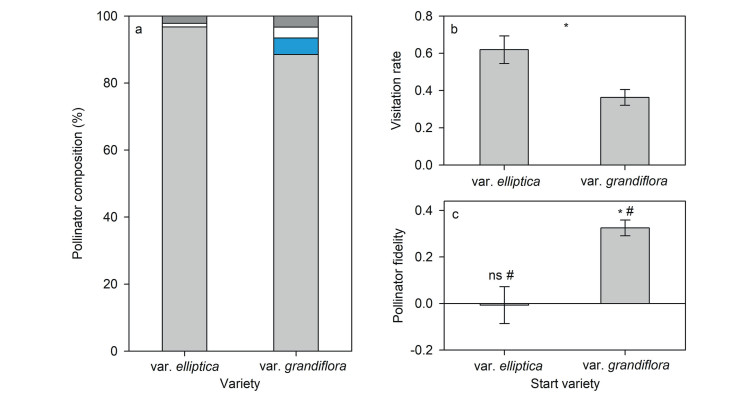
During the observation period, we recorded 92 and 61 types of insects visiting Halenia elliptica var. elliptica and H. elliptica var. grandiflora, respectively. Collectively, four pollinator functional groups were identified, namely bees, moths, butterflies, and flies, with bees the most abundant pollinators for both varieties (96.7% for H. elliptica var. elliptica and 88.5% for H. elliptica var. grandiflora) (Fig. 4a). Thus, we only considered the dominant bees in subsequent analyses. Bees visited H. elliptica var. elliptica at a significantly higher rate than they visited H. elliptica var. grandiflora (Fig. 4b). Bees that started visitations on H. elliptica var. elliptica showed no preference for either H. elliptica variety (−0.007 ± 0.079, Mean ± SE) (Fig. 4c). In contrast, bees that started visitations on H. elliptica var. grandiflora showed a high but not strong preference to H. elliptica var. grandiflora (0.325 ± 0.034).
|
| Fig. 4 Pollinator component (a), pollinator visitation rate (b) and pollinator fidelity (c) of the two varieties of Halenia elliptica. Different colored bars indicate bees, moths, butterflies, and flies. Moths were not observed visiting H. elliptica var. elliptica (a). Visitation rate of bees to H. elliptica var. elliptica was significantly higher than that to H. elliptica var. grandiflora, as indicated by an asterisk (b). Bees that started visitations on H. elliptica var. elliptica showed no preference for either H. elliptica variety, i.e., not significantly different than 0 (ns) but significantly higher than −0.5 (#). Bees that started visitations on H. elliptica var. grandiflora showed a high but not strong preference for H. elliptica var. grandiflora, i.e., significantly higher than 0 (*) but less than 0.5 (#) (c). |
The lengths of the alignments of the ITS1 + 2 region and rpl16 were 477 bp and 763 bp, respectively. A total of 80 and 26 parsimony informative sites were found in the nrITS data and the rpl16 region, respectively. For all the newly generated sequences of the two varieties, a total of 12 base substitutions and two different length variants (1 and 7 bp, respectively) were found, of which only four substitutions were informative and present in the ITS1 + 2 region only (Table S4). Additionally, no intra-population sequence variation was observed for the three regions; thus, we used one individual from each population for further phylogenetic analyses.
The phylogenetic analyses by both ML and Bayesian methods yielded an identical topology with slightly different support values (Fig. S3). The relationships among the taxa of Halenia were well resolved and supported, and H. elliptica was placed as the basal lineage, in complete accord with the previous inferences reported by von Hagen and Kadereit (2003). Furthermore, the monophyly of H. elliptica was strongly supported (BP = 100%, PP = 1.00), which was further divided into two monophyletic clades corresponding to the two varieties identified, respectively (Fig. S3).
It is worth noting that one population (XGLL) was identified as Halenia elliptica var. grandiflora based on morphological traits. However, the nrITS sequences of this population showed nucleotide additivity at four sites that were variable between H. elliptica var. elliptica (GL) and H. elliptica var. grandiflora (XZD) (Fig. S4, Table S4), potentially indicating this population might be a hybrid swarm. Therefore, we did not consider this population when we constructed the phylogeny of Halenia.
3.5. Variations in chloroplast genomesThe chloroplast genomes of the two Halenia elliptica varieties shared the typical circular quadripartite structure and similar GC content. The H. elliptica var. grandiflora chloroplast genome was larger (153, 358 bp) than that of H. elliptica var. elliptica (153, 260 bp), which can be explained mainly by changes in the LSC region (82, 815 vs 82, 689 bp) (Fig. S4). Using H. elliptica var. elliptica as a sequence reference, we found 52 SNPs and 32 InDels in H. elliptica var. grandiflora (Fig. S5). The overall SNP transition rate, SNP transversion rate, InDel insert rate, and InDel deletion rate were 5.95%, 55.95%, 20.24%, and 17.86%, respectively. Of the SNPs detected, 30 located in the LSC and 12 in the SSC regions (Fig. S4), where the SNP transition rates were 80% and 20%, whereas the SNP transversion rates were 55.32% and 23.40%, respectively. In addition, the LSC region contributed significantly to InDels, with an InDel insert rate of 76.47% and an InDel deletion rate of 86.67%.
When we compared the chloroplast genomes of the two Halenia elliptica varieties, we found that the number of SNPs/InDels unique to H. elliptica var. grandiflora was similar to the difference in SNPs/InDels between our H. elliptica var. elliptica sequences and the published data of H. elliptica (Table S5). Similarly, we found that the difference in the number of SNPs/InDels in H. elliptica var. elliptica from our study and in H. corniculata (L.) Cornaz from a previous study was greater than that between the two varieties in the present study (Table S5). Differences in the number of SNPs/InDels increased significantly when H. elliptica var. elliptica in our results were compared with species of other genera (Swertia L., Veratrilla (Baill.) Franch. and Gentiana (Tourn.) L.) in Gentianaceae (Table S5).
4. DiscussionDefining species has long been controversial, leading to the recent unification of species concepts (de Queiroz, 2005b; Hong, 2016, 2020; Liu, 2016). However, unified species concepts require more lines of evidence to delimit species. Here, multiple lines of evidence demonstrate that two varieties of Halenia elliptica should be treated as two species.
Morphological traits are the simplest and most important criteria in describing species and discovering new species. For example, over the past two decades, the number of new species discovered in China has exceeded 177 per year, totaling 3543, 80% of which were described using morphological traits (Du et al., 2020). The gen-morph species concept requires that at least two traits be distinct or statistically discontinuous (Hong, 2020). In the present study, we found that 18 of 21 traits measured showed significant differences between the two varieties of Halenia elliptica, indicating that varieties can be well discriminated. Vegetative traits, such as plant height, flower number and leaf size, might vary greatly depending on abiotic stresses and available resources for plants. Generally, plant height decreases at the high elevations (Zhang et al., 2020b), and/or under drought stress (Guo et al., 2016; Meng et al., 2017). Flowering phenology is also affected by environmental factors, among which temperature and photoperiod might be dominant (Elzinga et al., 2007; Tooke and Battey, 2010). Therefore, these traits might not be operational in identifying species. Flower traits, however, which can be more reliably used to discriminate between taxa, were also significantly different between the two varieties of H. elliptica. Specifically, we found that the spur of H. elliptica var. grandiflora is longer and more curved than that of H. elliptica var. elliptica (Ho and Prigle, 1995); in addition, the pistils and stamens of H. elliptica var. grandiflora were higher than those of H. elliptica var. elliptica. Furthermore, H. elliptica var. grandiflora had more pollen grains and ovules, higher pollen: ovule ratio and smaller seeds than H. elliptica var. elliptica. The gen-morph species concept posits that when varieties show distinct differences in more than two traits they can be treated as separate species (Hong, 2020).
Molecular markers are also powerful tools that can be used to delimit species (Hollingsworth et al., 2009; Li et al., 2011; Li and Zeng, 2015). Previous studies have used multiple molecular markers to identify new or cryptic species and resolve controversial taxonomic relationships. For example, taxonomists had suggested that Orychophragmus Bunge (Brassicaceae) consisted of two to seven species before molecular analysis recovered nine species, two of which are new species (Hu et al., 2015). Similarly, previous research indicated that the Cycas segmentifida complex (Cycadaceae), consists of eight until multiple molecular markers were used to revise the complex into only two species (Feng et al., 2016). Our phylogenetic analysis based on nrITS and rpl16 identified the two varieties of H. elliptica as two independent evolutionary lineages. However, one population (XGLL) may be a hybrid swarm, potentially resulting from gene exchanges between the two varieties in the early stage of splitting.
The chloroplast genome, for which gene content and organization is exceptionally conserved, can resolve phylogenetic relationships at different taxonomic levels (Jansen et al., 2007; Moore et al., 2010; Li et al., 2019). Comparison of the complete chloroplast genomes of the two Halenia elliptica varieties varied; furthermore, the number of variations between the two varieties was similar to that between populations of H. elliptica var. elliptica but far fewer than those between species of Gentianaceae. These findings suggest H. elliptica var. elliptica and H. elliptica var. grandiflora should be treated as two genetic lineages.
Reproductive isolation has been widely accepted as the only necessary criterion for delimiting species (Coyne et al., 1988; Mayr, 1992, 2000), especially for research on speciation (Butlin and Stankowski, 2020; Gao and Rieseberg, 2020). However, although reproductive isolation is generally strong in plant species, it might not be complete, with the degree of reproductive isolation often varying greatly (Rieseberg et al., 2006). In the Orychophragmus violaceus (L.) O.E. Schulz species complex (Brassicaceae), which consists of four species, inter-specific post-mating reproductive isolation shows great variation, ranging from 40% to 100% (Hu et al., 2015), and directional reproductive isolation has also been found (Hu et al., 2018). Further, in the three species of Arctic Draba L. (Brassicaceae), intra-specific pollination among populations generates more than 90% infertile hybrids, suggesting high cryptic diversity in Arctic flora (Grundt et al., 2006). In the two varieties of Halenia elliptica, the variation in flowering phenology (Fig. 2a) could contribute to a certain degree of pre-mating reproductive isolation. Additionally, the lack of seed production in inter-specific hand-pollinated plants suggest strong post-mating reproductive isolation. Post-mating reproductive isolation is generally frequent in the contact zones of the plants with different ploidy levels (Ramsey and Schemske, 1998; Castro et al., 2020). However, our results suggest that both varieties of H. elliptica could be diploid. Although further investigation is required to reveal the specific mechanisms that underlie post-mating reproductive isolation between H. elliptica var. grandiflora and H. elliptica var. elliptica, according to the Biological Species Concept these two varieties should be elevated to species status.
5. ConclusionIn conclusion, we found that the two varieties of Halenia elliptica represent two distinct evolutionary lineages based on multiple lines of evidence, including morphological traits, phylogenetic relationships, chloroplast genome, mating system and reproductive isolation. We recommend that the two varieties of H. elliptica be revised into two species, respectively as H. elliptica D. Don and Halenia grandiflora (Hemsl.) M.Y. Chen (text for taxonomic treatment follows). We further conclude that an evolutionary shift of mating system from outcrossing to mixed mating may have contributed to speciation of H. grandiflora and H. elliptica (Widmer et al., 2009; Baack et al., 2015), along with changes in flowering phenology that may have reduced pollen exchange between the two gene pools. Furthermore, changes in floral traits of H. elliptica, including reduction of flower size, protandry, herkogamy, allocation to pollen and ovule, and pollen: ovule ratio, support the evolution of selfing syndrome (Sicard and Lenhard, 2011; Cutter, 2019). The principle caveat of our conclusion is that we did not take population demographic history into consideration, and thus cannot exclude the possibility that the observed sympatry of the two species in the study site might have resulted from secondary contact. If the two species originated from allopatric speciation, post-mating reproductive isolation might have resulted from reinforcement selection against maladaptive hybridization in our study site (Butlin, 1987; Servedio and Noor, 2003). This issue could be resolved by examining the population genetic structure and demographic history in future. Lastly, the findings of our study highlight the importance of multiple lines of evidence in discovering, delimiting and revising species.
Taxonomic treatments of Halenia elliptica var. elliptica and H. elliptica var. grandiflora.
(1) Key to H. elliptica and H. grandiflora in China
1a. Corolla including spurs 1–1.5 cm in diam., spur curvature low, stigma height same as the four anthers ….….….….….….….….….….….….….….….….….….….….…. H. elliptica
1b. Corolla including spurs ca. 2.5 cm in diam., spur curvature high, stigma higher than four anthers ….….….….….….….….….….….….….….….….….….….….….….… H. grandiflora
(2) Taxonomic revision of H. elliptica var. elliptica and H. elliptica var. grandiflora
2a Halenia elliptica D. Don, London Edinburgh Philos. Mag. & J. Sci. 8: 77. 1836. (TYPE: INDIA. Choor et Kaderkanta, Royle s. n., syntypes CAL, DD, not seen; Kumaon, R. Brown, syntype K, not seen)
卵萼花锚【luǎn è huā máo】
Herbal, biennial. Plants (7–) 15–90 cm tall. Stems erect, subquadrangular, striate, simple or branched from base and/or above base. Basal leaves petiole flattened, 1–1.5(–3) cm; leaf blade spatulate, elliptic, or sometimes suborbicular, 2– 3 × 0.5–1.5 cm, base narrowed to cuneate, apex acute to rounded, veins 3–5. Stem leaves sessile or short petiolate; leaf blade oblong, elliptic, ovate-lanceolate, or ovate, 1.5– 7 × 0.5–3.5 cm, base rounded to truncate, apex acute to rounded, veins 5. Pedicel 0.5–3.5 cm. Calyx lobes elliptic to ovate, apex acuminate. Corolla blue to purple, campanulate, less than 8 mm, basal spurs less than 10 mm; lobes elliptic to ovate and curvature low, apex obtuse and apiculate. Filaments 3–5 mm; anthers ovoid, ca. 1 mm, pistil 4–5 mm. Ovules 8–12. Seeds light brown, ca. 2 × 1 mm. 2n = 22.
Phenology: Flowering from July to September, fruiting from August to October.
Habit and distribution: Beside streams in valleys, grassland slopes, scrub, forest margins, forests; 700–4100 m. Gansu, Guizhou, Hubei, Hunan, Liaoning, Inner Mongolia, Qinghai, Shanxi, Xinjiang, Xizang, Yunnan [Bhutan, India, Kyrgyzstan, Myanmar, Nepal, Sikkim].
Specimens examined: CHINA. Gansu: Tianshui, on the grassland near river in poplar forest, alt. 1600 m, 15 Aug. 1951, J.M. Liu 10466 (KUN 0554017). Lanzhou, Yuzhong County, Xinglongshan Natural Reserve, in the slope forest, alt. 2408 m, 8 Aug. 2019, L. Zhang, X.Y. Fan, J.Q. Guo & H.R. Yang, QTP-LJQ-1437-3030 (KUN 1477053). Zhangye, Sunan County, Dahe town, alt. 2699 m, 8 Aug. 2020, L.T. Li, D.S. He & J.B. Jiang QTP-LJQ-CHNO027-3065 (KUN 1510019). Shaanxi: Lueyang County, Sigou Mountain, Xiajiagou, on the slope grassland, alt. 1000 m, 11 Sept. 1952, K.J. Fu 5772 (KUN 0554058). Xitaishan, on the slope grassland, alt. 2880 m, 7 Aug. 1957, K.J. Fu 10206 (KUN 0554059). Xinjiang: Yining, Zhaosu County, northern foot of the Tenshan Mountains, Aug. 2005, N. Abdusalik FK35608 (KUN 0891550). Xizang: Changdu, Basu County, in the alpine shrubs, alt. 4827 m, 15 Jul. 2018, D.G. Zhang Deng5151 (KUN 1352044). Ningxia: Zhongwei, Haiyuan County, forest farm, on the slope grassland, alt. 2459 m, 17 Aug. 2019, L. Zhang, X.Y. Fan, J.Q. Guo & H.R. Yang QTP-LJQ-1338-3067 (KUN 1476870). Qinghai: Menyuan County, Xianmi temple, Zharigou, on the slope, alt 2600 m, 30 Jul. 1958, B.Q. Zhong 10011 (KUN 0554048). Nangqian County, Forest farm, in the forest, alt. 3800 m, 30 Jul. 1972, Tibetan Medicine Team 1165 (KUN 0554051). Huzhu County, Daban Mountains, on the slope grassland, alt. 2700 m, 23 Aug. 1965, B.Z. Guo 6648 (KUN 0554050).
2b. Halenia grandiflora (Hemsl.) M.Y. Chen, comb. & stat. nov.
Basionym: Halenia elliptica var. grandiflora Hemsl. in F. B. Forbes & Hemsl., J. Linn. Soc., Bot. 26: 141. 1890. (TYPE: CHINA. Prov. Hupeh, 1885–1888, A. Henry 4982, holotype NY00178945!)
大花花锚【dà huā huā máo】
Herbal, biennial. Plants (7–) 15–90 cm tall. Stems erect, subquadrangular, striate, simple or branched from base and/or above base. Basal leaves petiole flattened, 1–1.5(–3) cm; leaf blade spatulate, elliptic, or sometimes suborbicular, 2–3 × 0.5–1.5 cm, base narrowed to cuneate, apex acute to rounded, veins 3–5. Stem leaves sessile or short petiolate; leaf blade oblong, elliptic, ovate-lanceolate, or ovate, 1.5–7 × 0.5–3.5 cm, base rounded to truncate, apex acute to rounded, veins 5. Pedicel 0.5–3.5 cm. Calyx lobes elliptic to ovate, apex acuminate. Flowers often hermaphroditic and occasionally female. Corolla blue to purple, campanulate, more than 9 mm, basal spurs more than 10 mm; lobes elliptic to ovate and curvature obvious, apex obtuse and apiculate. Filaments 5–8 mm; anthers ovoid, ca. 1 mm or aborted, pistil 7–10 mm. Ovules 15–23. Seeds light brown, ca. 1.3 × 1.0 mm. 2n = 22.
Phenology: Flowering from September to October, fruiting from October to November.
Habit and distribution: Beside streams, grassland slopes; 1300–2500 m. Gansu, Guizhou, Hubei, Qinghai, Shaanxi, Sichuan, Yunnan.
Specimens examined: CHINA. Yunnan: Wuding County, Guanpo, in the forest, alt. 2530 m, 11 Nov. 2002, H. Li, S.X. Yang, R. Li, Y.H. Ji & B.X. Liu 1133 (KUN 1305709). Lijiang, Baisha, on grassland along the road, alt. 2500 m, 31 Aug. 1958, W.T. Wang s. n. (KUN 0553924). Lijiang, Xusongcun, on grassland, 18 Sept. 1939, Z.G. Zhao 30642 (PE 01399585). Lijiang, Yulong Snow Mountains, 28 Aug. 1940, R.C. Ching 30996 (PE 01399589). Heqing County, Lianping, Fengchuiling, on grassland slope, alt. 2700 m, 20 Aug. 1929, R.C. Ching 23806 (KUN 0553762). Kunming, Xishan, on slope, 21 Sept. 1955, Anonymous collector 51041 (PE 01399608). Kunming, Motianling, on slope, 12 Sept. 1957, B.Y. Qiu 55066 (PE01399610). Dali, Cangshan Mountains, on grassland slope, alt. 2100 m, 26 Aug. 1976, Y.Q. He et al. 087 (WUK 0346642). Luquan County, Emao Town, Emao village, on wet grassland, alt. 2400 m, 29 Oct. 1952, P.Y. Mao 01494 (WUK0244892). Songming County, Taposhao, swamp, alt. 2400, 14 Oct. 1940, Y.P. Chang 0191 (PE 01399564). Zhenxiong County, Niuchang, Huashan, on wild land, alt. 1850 m, 28 Sept. 1972, Northeast Yunnan Team 1100 (KUN 0554016).
Author contributionsY.W.D. and L.H.M. designed the research. J.F.W., R.J.L. and L.L.W. performed the field experiments. D.R.J. performed phylogenetic analysis. Z.L.Z. performed data analysis of chloroplast genome. M.Y.C. proposed the initial idea. Y.W.D., L.H.M. and D.R.J. wrote the manuscript.
Declaration of competing interestWe declare no conflict of interest.
AcknowledgementsWe are grateful to the staff at the Lijiang Alpine Botanical Garden (Lijiang Forest Ecosystem Research Station) for their kind help and support during the field experiments. We thank Mr. Kai-Wen Jiang and Dr. Bin Tian from the Southwest Forest University for their suggested revisions of the taxonomic treatment. Financial support for this research was provided by the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), National Key R & D Program of China (2017YFC0505200), National Natural Science Foundation of China (31460096 and 31590823) and State Key Laboratory of Phytochemistry and Plant Resources in West China (P2020-KF04).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.09.004.
Akaike, H., 1974. New look at statistical model identification. IEEE Trans. Automat. Contr., 19: 716-723. DOI:10.1109/TAC.1974.1100705 |
Andres, R.J., Kuraparthy, V., 2013. Development of an improved method of mitotic metaphase chromosome preparation compatible for fluorescence in situ hybridization in cotton. J. Cotton Sci., 17: 149-156. |
Baack, E., Melo, M.C., Rieseberg, L.H., et al., 2015. The origins of reproductive isolation in plants. New Phytol., 207: 968-984. DOI:10.1111/nph.13424 |
Butlin, R., 1987. A new approach to sympatric speciation. Trends Ecol. Evol., 2: 310-311. DOI:10.1016/0169-5347(87)90085-1 |
Butlin, R.K., Stankowski, S., 2020. Is it time to abandon the biological species concept?. No. Natl. Sci. Rev., 7: 1400-1401. DOI:10.1093/nsr/nwaa109 |
Castro, M., Loureiro, J., Husband, B.C., et al., 2020. The role of multiple reproductive barriers: strong post-pollination interactions govern cytotype isolation in a tetraploid-octoploid contact zone. Ann. Bot., 126: 991-1003. DOI:10.1093/aob/mcaa084 |
Coyne, J.A., Orr, H.A., Futuyma, D.J., 1988. Do we need a new species concept?. Syst. Biol., 37: 190-200. DOI:10.2307/2992276 |
Cutter, A.D., 2019. Reproductive transitions in plants and animals: selfing syndrome, sexual selection and speciation. New Phytol., 224: 1080-1094. DOI:10.1111/nph.16075 |
Darriba, D., Taboada, G.L., Doallo, R., et al., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods, 9: 772. DOI:10.1038/nmeth.2109 |
de Queiroz, K., 1998. The general lineage concept of species, species criteria, and the process of speciation - A conceptual unification and terminological recommendations. In: Howard, D.J., Berlocher, S.H. (Eds), Endless Forms. Oxford University Press, New York, 57-75
|
de Queiroz, K., 2005. Ernst Mayr and the modern concept of species. Proc. Natl. Acad. Sci. U.S.A., 102: 6600-6607. DOI:10.1073/pnas.0502030102 |
de Queiroz, K., 2005. A unified concept of species and its consequences for the future of taxonomy. Proc. Calif. Acad. Sci., 56: 196-215. |
de Queiroz, K., 2007. Species concepts and species delimitation. Syst. Biol., 56: 879-886. DOI:10.1080/10635150701701083 |
Du, C., Liao, S., Boufford, D.E., et al., 2020. Twenty years of Chinese vascular plant novelties, 2000 through 2019. Plant Divers., 42: 393-398. DOI:10.1016/j.pld.2020.08.004 |
Duan, Y.W., He, Y.P., Liu, J.Q., 2005. Reproductive ecology of the Qinghai-Tibet Plateau endemic Gentiana straminea (Gentianaceae), a hermaphrodite perennial characterized by herkogamy and dichogamy. Acta Oecol., 27: 225-232. DOI:10.1016/j.actao.2005.01.003 |
Duan, Y.-W., Dafni, A., Hou, Q.-Z., et al., 2010. Delayed selfing in an alpine biennial Gentianopsis paludosa (Gentianaceae) in the Qinghai-Tibetan plateau. J. Integr. Plant Biol., 52: 593-599. DOI:10.1111/j.1744-7909.2010.00951.x |
Edler, D., Klein, J., Antonelli, A., et al., 2021. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol., 12: 373-377. DOI:10.1111/2041-210x.13512 |
Elzinga, J.A., Atlan, A., Biere, A., et al., 2007. Time after time: flowering phenology and biotic interactions. Trends Ecol. Evol., 22: 432-439. DOI:10.1016/j.tree.2007.05.006 |
Feng, X., Liu, J., Gong, X., 2016. Species delimitation of the Cycas segmentifida complex (Cycadaceae) resolved by phylogenetic and distance analyses of molecular data. Front. Plant Sci., 7: 134. |
Gao, L., Rieseberg, L.H., 2020. While neither universally applicable nor practical operationally, the biological species concept continues to offer a compelling framework for studying species and speciation. Natl. Sci. Rev., 7: 1398-1400. DOI:10.1093/nsr/nwaa108 |
Grundt, H.H., Kjolner, S., Borgen, L., et al., 2006. High biological species diversity in the arctic flora. Proc. Natl. Acad. Sci. U.S.A., 103: 972-975. DOI:10.1073/pnas.0510270103 |
Gu, Z., Gu, L., Eils, R., et al., 2014. Circlize implements and enhances circular visualization in R. Bioinformatics, 30: 2811-2812. DOI:10.1093/bioinformatics/btu393 |
Guindon, S., Gascuel, O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol., 52: 696-704. DOI:10.1080/10635150390235520 |
Guo, W., Yang, J., Sun, X.-D., et al., 2016. Divergence in eco-physiological responses to drought mirrors the distinct distribution of Chamerion angustifolium cytotypes in the Himalaya-Hengduan Mountains region. Front. Plant Sci., 7: 1329. |
Ho, T.N., Prigle, J.S., 1995. Gentianaceae. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds). Flora of China. Science Press & Missouri Botanical Garden, Beijing & St. Louis
|
Hollingsworth, P.M., Forrest, L.L., Spouge, J.L., et al., 2009. A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A., 106: 12794-12797. DOI:10.1073/pnas.0905845106 |
Hong, D.-Y., 2016. Biodiversity pursuits need a scientific and operative species concept. Biodivers. Sci., 24: 979-999. DOI:10.17520/biods.2016203 |
Hong, D.-Y., 2020. Gen-morph species concept—a new and integrative species concept for outbreeding organisms. J. Systemat. Evol., 58: 725-742. DOI:10.1111/jse.12660 |
Hu, H., Al-Shehbaz, I.A., Sun, Y., et al., 2015. Species delimitation in Orychophragmus (Brassicaceae) based on chloroplast and nuclear DNA barcodes. Taxon, 64: 714-726. DOI:10.12705/644.4 |
Hu, H., Zeng, T., Wang, Z., et al., 2018. Species delimitation in the Orychophragmus violaceus species complex (Brassicaceae) based on morphological distinction and reproductive isolation. Bot. J. Linn. Soc., 188: 257-268. |
Jansen, R.K., Cai, Z., Raubeson, L.A., et al., 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A., 104: 19369-19374. DOI:10.1073/pnas.0709121104 |
Jin, J.-J., Yu, W.-B., Yang, J.-B., et al., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 241. DOI:10.1186/s13059-020-02154-5 |
Kato, A., 1999. Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem., 74: 160-166. DOI:10.3109/10520299909047968 |
Kumar, S., Stecher, G., Li, M., et al., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol., 35: 1547-1549. DOI:10.1093/molbev/msy096 |
Kurtz, S., Phillippy, A., Delcher, A.L., et al., 2004. Versatile and open software for comparing large genomes. Genome Biol., 5: R12. DOI:10.1186/gb-2004-5-2-r12 |
Li, D., Zeng, C.-X., 2015. Prospects for plant DNA barcoding. Biodivers. Sci., 23: 297-298. DOI:10.17520/biods.2015135 |
Li, D.-Z., Gao, L.-M., Li, H.-T., et al., 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A., 108: 19641-19646. DOI:10.1073/pnas.1104551108 |
Li, H.-T., Yi, T.-S., Gao, L.-M., et al., 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants, 5: 461-470. DOI:10.1038/s41477-019-0421-0 |
Liu, J., 2016. The integrative species concept" and "species on the speciation way. Biodivers. Sci., 24: 1004-1008. DOI:10.17520/biods.2016222 |
Mayr, E., 1992. Species concepts and their application. In: Ereshefsky, M. (Ed), The unit of evolution: Essays on the nature of species. MA: MIT Press, Cambridge, 15-25
|
Mayr, E., 2000. The Biological Species Concept. In: Wheeler, Q.D., Meier, R. (Eds), Species Concepts and Phylogenetic Theory: A Debate. Columbia University Press, New York
|
CNCB-NGDC Members and Partners, 2021. Database resources of the national genomics data center, China national center for bioinformation in 2021. Nucleic Acids Res., 49: D18-D28. DOI:10.1093/nar/gkaa1022 |
Meng, L.-H., Wang, Y., Luo, J., et al., 2012. Pollination ecology and its implication for conservation of an endangered perennial herb native to the East-Himalaya, Megacodon stylophorus (Gentianaceae). Plant Ecol. Evol., 145: 356-362. DOI:10.5091/plecevo.2012.684 |
Meng, L.-H., Yang, J., Guo, W., et al., 2017. Differentiation in drought tolerance mirrors the geographic distributions of alpine plants on the Qinghai-Tibet Plateau and adjacent highlands. Sci. Rep., 7: 42466. DOI:10.1038/srep42466 |
Moore, M.J., Soltis, P.S., Bell, C.D., et al., 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U.S.A., 107: 4623-4628. DOI:10.1073/pnas.0907801107 |
Petanidou, T., Ellis-Adam, A., Den Nijs, H.C.M., et al., 2001. Differential pollination success in the course of individual flower development and flowering time in Gentiana pneumonanthe L. (Gentianaceae). Bot. J. Linn. Soc., 135: 25-33. DOI:10.1111/j.1095-8339.2001.tb02365.x |
Qu, X.-J., Moore, M.J., Li, D.-Z., et al., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. DOI:10.1186/s13007-019-0435-7 |
Ramsey, J., Schemske, D.W., 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst., 29: 467-501. DOI:10.1146/annurev.ecolsys.29.1.467 |
Rice, A., Glick, L., Abadi, S., et al., 2015. The chromosome counts database (CCDB) - a community resource of plant chromosome numbers. New Phytol., 206: 19-26. DOI:10.1111/nph.13191 |
Rieseberg, L.H., Wood, T.E., Baack, E.J., 2006. The nature of plant species. Nature, 440: 524-527. DOI:10.1038/nature04402 |
Ronquist, F., Huelsenbeck, J.P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
Savolainen, V., Anstett, M.C., Lexer, C., et al., 2006. Sympatric speciation in palms on an oceanic island. Nature, 441: 210-213. DOI:10.1038/nature04566 |
Servedio, M.R., Noor, M.A.F., 2003. The role of reinforcement in speciation: theory and data. Annu. Rev. Ecol. Evol. Syst., 34: 339-364. DOI:10.1146/annurev.ecolsys.34.011802.132412 |
Sicard, A., Lenhard, M., 2011. The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann. Bot., 107: 1433-1443. DOI:10.1093/aob/mcr023 |
Silvestro, D., Michalak, I., 2012. raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol., 12: 335-337. DOI:10.1007/s13127-011-0056-0 |
Sletvold, N., Moritz, K.K., Ågren, J., 2015. Additive effects of pollinators and herbivores result in both conflicting and reinforcing selection on floral traits. Ecology, 96: 214-221. DOI:10.1890/14-0119.1 |
Small, R.L., Ryburn, J.A., Cronn, R.C., et al., 1998. The tortoise and the hare: choosing between noncoding plastome and nuclear ADH sequences for phylogeny reconstruction in a recently diverged plant group. Am. J. Bot., 85: 1301-1315. DOI:10.2307/2446640 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Su, X., Wu, G., Li, L., et al., 2015. Species delimitation in plants using the Qinghai-Tibet plateau endemic Orinus (Poaceae: tridentinae) as an example. Ann. Bot., 116: 35-48. DOI:10.1093/aob/mcv062 |
Tooke, F., Battey, N.H., 2010. Temperate flowering phenology. J. Exp. Bot., 61: 2853-2862. DOI:10.1093/jxb/erq165 |
von Hagen, K.B., Kadereit, J.W., 2001. The phylogeny of Gentianella (Gentianaceae) and its colonization of the southern hemisphere as revealed by nuclear and chloroplast DNA sequence variation. Org. Divers. Evol., 1: 61-79. DOI:10.1078/1439-6092-00005 |
von Hagen, K.B., Kadereit, J.W., 2003. The diversification of Halenia (Gentianaceae): ecological opportunity versus key innovation. Evolution, 57: 2507-2518. DOI:10.1554/02-742 |
Wang, L.-L., Zhao, M.-F., Wang, Y., et al., 2011. A preliminary study on geographical variations in floral traits of Halenia elliptica (Gentianaceae) based on herbaria. Plant Divers. Resour., 33: 503-508. |
Widmer, A., Lexer, C., Cozzolino, S., 2009. Evolution of reproductive isolation in plants. Heredity, 102: 31-38. DOI:10.1038/hdy.2008.69 |
Wilkins, J.S., 2009. Species: A history of the idea. University of California Press, Berkeley
|
Yang, M.-L., Wang, L.-L., Zhang, G.-P., et al., 2018. Equipped for migrations across high latitude regions? Reduced spur length and outcrossing rate in a biennial Halenia elliptica (Gentianaceae) with mixed mating system along a latitude gradient. Front. Genet., 9: 223. DOI:10.3389/fgene.2018.00223 |
Yuan, Y.-M., Kupfer, P., 1993. Karyological studies of Gentianopsis Ma and some related genera of Gentianaceae from China. Cytologia, 58: 115-123. DOI:10.1508/cytologia.58.115 |
Zhang, C., Hu, L., Wang, Y., et al., 2011. Effects of the position on floral traits and reproductive success of Comastoma pulmonarium (Gentianaceae). Plant Divers. Resour., 33: 495-502. |
Zhang, X., Sun, Y., Landis, J.B., et al., 2020. Plastome phylogenomic study of Gentianeae (Gentianaceae): widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol., 20: 340. DOI:10.1186/s12870-020-02518-w |
Zhang, C., An, Y.-M., Jaeschke, Y., et al., 2020. Processes on reproductive ecology of plant species in the Qinghai-Xizang Plateau and adjacent highlands. Chinese J. Plant Ecol., 44: 1-21. DOI:10.17521/cjpe.2019.0296 |
Zhou, C.F., Yang, G., 2011. Esistence and Definition of Species. Beijing: Science Press.
|
Zhou, S.-L., Zou, X.-H., Zhou, Z.-Q., et al., 2014. Multiple species of wild tree peonies gave rise to the 'king of flowers', Paeonia suffruticosa Andrews. Proc. R. Soc. B-Biol. Sci., 281: 20141687. DOI:10.1098/rspb.2014.1687 |



