b. College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China;
c. Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Menglun, Mengla, Yunnan 666303, China;
d. The Innovative Academy of Seed Design, Chinese Academy of Sciences, Menglun, Mengla, Yunnan 666303, China
Rice (Oryza sativa L.), as one of the most important food crops worldwide, fulfills half of the food demand for the world's population (Qi et al., 2012). With the incessantly growing population and shortage of cultivated land, food security is becoming increasingly important (Alexandratos and Bruinsma, 2012). Researchers and breeders have conducted extensive studies to improve rice yield by utilizing heterosis in rice. Hybrid rice has become one of the best strategies to overcome the current challenges for rice production, which accounted for more than half of China's main rice production area (Yuan, 1986; Wang, 2020). The yield of hybrid rice has improved by 20–30% compared with the best semi-dwarf inbred varieties since the 1970s (Wang and Wang, 2017).
The three-line hybrid system, including sterile lines, maintainer lines and restorer lines, is the predominant strategy for rice hybridization. However, the unitary cytosterility system may make hybrid crops more vulnerable to destructive diseases or insects. Due to the high cost of seeds produced by complex breeding procedures, such as long cycles and low efficiency, the planting area and yield improvement per unit area of three-line hybrid rice are relatively limited (Yuan, 1990, 1998). Yuan proposed the development of two-line and even one-line hybrids in rice breeding (Yuan, 1997). The utilization of heterosis in rice breeding will be easier to operate and more efficient compared with the three-line systems.
Nongken 58, a male sterile rice variety altered fertility depending on day length and variable temperature, was first discovered in 1973. Since then, the development of two-line hybrid rice has been exploited in China (Shi, 1985). Recently, one-line hybrid using parthenogenetic line has also been tentatively proposed to best utilize heterosis. Hybrid seeds could be obtained by asexual reproduction without double fertilization. Unfortunately, the seed setting rate of these lines is relatively low, and most of the embryos are tetraploid. There remains much work to be done to realize the practical application of one-line hybrid (Wang et al., 2019; Khanday et al., 2019), and the two-line approach is still the main way to produce hybrid seeds at present.
The two-line system, which is performed without a maintainer line, is convenient, less time-consuming and great potential to reduce the price of hybrid seeds. Hybrid seed production without cytoplasmic sterility is a simpler method that could effectively avoid the potential threat of negative cytoplasmic effects (Yuan, 1990). Photosensitive genic male sterility (PGMS) and thermosensitive genic male sterility (TGMS) are two main types of resources used in two-line system. TGMS is considered more useful than PGMS under tropical conditions, where the day length differences are marginal (Virmani, 1996). Therefore, breeders prefer to use TGMS lines in hybrid rice via this system. The fertility of these lines is mainly controlled by recessive nuclear genes, which allows them to be used in hybrid rice production. Several recessive TGMS genes, including tms (Kadirimangalam et al., 2019; Khlaimongkhon et al., 2019), tms4 (Dong et al., 2000), tms5 (Wang et al., 2003; Jiang et al., 2006), tms8 (Hussain et al., 2012) and tms9-1 (Qi et al., 2014), have been extensively studied. In addition to these recessive genes, the dominant genes in PingXiang and the 8987 male-sterile line have also been proven to play a critical role in TGMS (Huang et al., 2007; Deng and Zhou, 1994).
Most TGMS lines are sterile when the temperature is high (>25 ℃) but fertile at lower temperatures, which severely restricts their applicable region. In field practice, TGMS rice sterile at a lower temperature and its fertility controlled by recessive genes will have wide application potential. Reverse thermosensitive genic male sterile (RTGMS) lines ideally fulfill these requirements (Jia et al., 2001; Liu et al., 2010). RTGMS rice materials, which are fertile at high environmental temperatures and sterile at low temperatures, were first discovered in China in the 1980s. In particular, J207S was fertile or sterile when the temperature was higher or lower than 31 ℃, respectively (Jia et al., 2001). The fertility transition of G20S occurred at approximately 29.5 ℃ (Liu et al., 2010). Genetic analysis showed that the fertility of J207S and G20S was also controlled by recessive genes, which were separately identified as rtms1 and tms6(t) via molecular mapping. Both genes were mapped to an overlapping region on chromosome 10, and rtms1 contained tms6(t) (Jia et al., 2001; Liu et al., 2010). However, cloning and functional analyses of RTGMS genes are largely scarce.
In this study, a RTGMS line, Diannong S-1 xuan (DNS-1X), was identified. The critical temperature for the fertility transition detected in the natural environment showed that the pollen was sterile below 28–30 ℃. Phenotype analysis confirmed that fertility was controlled by a recessive gene. Moreover, we fine mapped the candidate region containing the rtms1-D gene by BSA and molecular mapping techniques. Our results will broaden the utilization of TGMS rice resources and provide insights into analysis of the function of RTGMS genes.
2. Materials and methods 2.1. Plant materialsDiannong S-1 was kindly provided by Yunnan Agriculture University and has been described previously (Rong, 1994; Jiang et al., 1997). Yunjing 26 (YJ26), a japonica rice variety, was provided by the Japonica Rice Breeding Center of Yunnan Academy of Agricultural Sciences. Diannong S-1 xuan (DNS-1X) is a new and stable RTGMS sterile line which was bred by purification and selection of Diannong S-1 for several generations. The F1 and F2:3 genetic populations and BC2F4:5 near-isogenic lines (NILs) derived from a cross between the donor YJ26 and recipient DNS-1X were used in this study. All materials were planted twice per year in a paddy field at the breeding bases of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan Province, China. The heading dates of the two crop seasons were in early May and late October. Breeding practice showed that the RTGMS sterile line DNS-1X presented a sterile phenotype in these two periods. NILs were sown every 10 days to investigate fertility conversion in 2020.
2.2. Identification of pollen fertility and phenotypic characterizationThe materials planted at the Sanya breeding base (Sanya, Hainan) and Xishuangbanna breeding base (Jinghong, Yunnan) were investigated for the indicated times. During heading, florets in the upper middle section of flowering primary panicles, twenty per plant, were sampled and fixed with 75% alcohol. Anthers of three florets were randomly selected from the sampled florets and squashed in 0.01 g/mL iodine and iodine-potassium solution (I2-KI) for staining the pollen grains. The pollen grains that were round, dark and uniformly stained were considered fertile. In contrast, the pollen grains that exhibited unstained spherical or shriveled shapes or pale coloration were considered sterile. Investigation of pollen fertility was carried out under a Carl Zeiss microscope (Carl Zeiss, Germany) at 10 × magnification. The pollen fertility of each plant was measured for three repetitions. Moreover, the morphology of anthers and pollen grains was photographed by an OLYMPUS SZX16 stereoscope microscope (Olympus, Japan) and a ZEISS EVO LS10 scanning electron microscope (Carl Zeiss, Germany), respectively.
2.3. Determination of agronomic traits of NILsThe seeds of the NILs and receptor parent YJ26 were soaked for 36 h, induced to germinate in a growth chamber at 28 ℃, and subsequently sown on seedbeds; the seedlings were transplanted at the four-leaf stage with 5 rows per plot and 12 plants per row. The density was 25 cm × 20 cm between plants. The pollen fertility of 10 individuals randomly selected from each population was investigated at the heading stage and at the mature stage. Agronomic traits, including plant height, panicle length, number of primary branches, number of secondary branches, effective tiller number, grain number per panicle, grain length and grain width, were measured.
2.4. Histological characterization of DNS-1XFor histological analysis, the fertile (YJ26) and sterile (BC2F5 sterile NILs) anthers of eight panicle development stages were sampled and fixed in 50% (v/v) FAA before made into semithin transverse sections (Feng et al., 2001), all anther samples were collected in early May of 2021. The fertility of NILs was determined by I2-KI staining the mature pollen grains, ensure that the NILs was sterile. After the samples were vacuum infiltrated, semithin sections were performed as described previously (Zhao et al., 2020). Images were photographed using a Nikon Eclipse E100 microscope (Nikon, Japan).
2.5. BSAHealthy leaves of 100 fertile and sterile individuals were collected from the F2 population. The materials were quickly frozen with liquid nitrogen and then stored at −80 ℃. The genomic DNA of all samples was extracted according to a previously described method (Murray and Thompson, 1980). Three mixed DNA pools (R01-DNS-1X, R02-fertile pool, and R03-sterile pool) were constructed for BSA. The FP (fertile pool) and SP (sterile pool) were made by mixing equal amounts of DNA from fertile and sterile samples, respectively. BSA was performed by OE Biotech Co., Ltd (Shanghai, China), and the analysis was performed according to a previous report (Takagi et al., 2013). The SNP-index of the two extreme trait pools and parents was calculated by genome sequencing. 1 Mb was used as the window, and a 10 kb walk was used to count the average value of all SNP-index when positioning. The coverage depths of the highbulk and lowbulk were tested by binomial distribution, and the 95% and 99% confidence intervals were calculated statistically. The fitting line (shown in red) outside of the confidence line on the chromosome (where the SNP index was close to 1) was considered to be the possible phenotypically linked region.
2.6. Identification of genotype by SSR markersGenomic DNA was extracted from fresh leaves as described above. Based on the results of BSA, a total of 191 reported SSR markers on chromosome 10 were used to test the parental polymorphisms (Sasaki, 2005). Four markers with polymorphisms between the parents were used to investigate the genotypes of 792 F2 individuals (DNS-1X × YJ26). For fine mapping, several new SSR markers were designed based on the reference genome of Nippobare, and markers with polymorphisms were used. PCRs with a total volume of 20 μL contained 10 μL 2 × Taq Master Mix (Novoprotein Scientific Inc.), 1 μL forward primer (10 mol/μL), 1 μL reverse primer (10 mol/μL), 2 μL DNA samples and 6 μL ddH2O. The PCR cycling conditions were as following: 95 ℃ for 5 min; 26 cycles of 30 s at 95 ℃, 30 s at 60 ℃, and 30 s at 72 ℃; and finally, 72 ℃ for 2 min. Polyacrylamide gel electrophoresis (PAGE) with silver staining was used to detect the PCR products.
2.7. Identification of the critical temperature for the fertility transitionTo determine the critical temperature for the fertility transition, we collected meteorological data, including temperature, sunshine hours and relative humidity in 2020, from the National Forest Ecosystem Research Station at Xishuangbanna, a location near the experimental breeding base, and further analyzed the correlation between the fertility transition and these environmental factors. The influence of meteorological factors on pollen fertility from 1 to 26 days before heading was evaluated (Rangaswamy, 1993).
2.8. Statistical analysisSPSS v.20 software (IBM) was used to analyze agronomic trait and fertility data. The χ2 test was applied to determine the segregation ratio of fertility. Student's t-tests were used to determine significant differences, with ∗ and ∗∗ indicating significant differences at P < 0.05 and P < 0.01, respectively.
3. Results 3.1. Pollen fertility of DNS-1X under different temperaturesWe first investigated the effect of temperature on the pollen fertility of DNS-1X, and found that pollen fertility was indeed affected by ambient temperature. From late February to early March in Sanya, Hainan, DNS-1X initiated panicle development at an average maximum temperature of 26 ℃. The anthers were shrunken and the filaments were abnormally long, resulting in complete male sterility (Fig. 1A–E, G). The pollen grains were irregular and unstained with I2-KI solution (Fig. 1J, K), resulting in no seeds setting in the panicles (Fig. 1A, B). However, the average maximum temperature was 35 ℃ in June in Xishuangbanna, and the initiation of panicle development presented normal and full anthers, short filaments, spherical and darkly stained pollen grains and normal seed set (Fig. 1A–E, F, H, I). These results indicated that DNS-1X was a potential RTGMS line.

|
| Fig. 1 Phenotype and morphology comparison of anthers and pollen fertility of DNS-1X under high- and low-temperature conditions. (A) Morphology of sterile and fertility restorer plant from Hainan and Xishuangbanna, respectively. Scale bars, 20 cm. Stereoscope microscope of fertile and sterile plants spikelet (B and C), and anther (D and E) during heading date. Scale bars, 5 cm (B) and 2 mm (D and E). The morphology of anthers (F and G), pollens (I and J) and the I2-KI (H and K) staining of fertile and sterile pollens. Plants were grown at 35 ℃ (F, I, H) or 26 ℃ (G, J, K). Scale bars, 200 μm, 35 × (F and G); 10 μm, 2000 × (I and J); and 100 μm, 100 × (H and K). |
To determine the genetic basis of sterility genes, we investigated the fertility of the F2 population derived from a cross between YJ26 as the donor and DNS-1X as the recipient. The pollen fertility and natural seed-setting rates of 1040 F2 individuals both showed obvious bimodal distributions (Fig. 2). There was a significant positive correlation between pollen fertility and the seed setting rate (r = 0.71). The χ2 tests showed that the break point occurred at 37.5% pollen fertility and a 28.13% seed setting rate and that the fertile and sterile types were fitted to a ratio of 3:1 (P > 0.05) (Table 1), suggesting that the sterility of DNS-1X was controlled by a single recessive gene. We further investigated the phenotypes of sterile and fertile NILs (BC2F4) in which there was no significant difference in plant height and grain size. However, the number of effective tillers of fertile NILs was significantly lower than that of sterile NILs, and the primary branching and number of grains per panicle of fertile plants were significantly higher than those of sterile plants (Table 2). The results indicated that the sterility gene in DNS-1X affected fertility conversion as well as some key agronomic traits.

|
| Fig. 2 The distribution of pollen fertility in F2 population and seed-setting rate. Distribution of pollen fertility (A) and seed-setting rate of 1040 plants in the F2 population (B) in a cross between DNS-1X and YJ26 grown in 2019. |
| Type | Total | Fertility | Sterility | Ratio | χ2 (3:1) | P value (0.05) | Correlation | |||||||
| Pollen | 1040 | 788 | 252 | 3.13:1 | 0.29 | 0.57 | 0.71∗∗ | |||||||
| Seed | 1040 | 755 | 285 | 2.65:1 | 3.08 | 0.07 | ||||||||
| Note: The fertile and sterile pollens of in F2 progenies segregated in the ratio of 3:1. The asterisk indicates significant difference (∗∗P<0.01). | ||||||||||||||
| Traits | Fertile | Sterile | ||||||||||||||||||||||||
| Plant height (cm) | 66.06±4.51 | 65.30±6.17 | ||||||||||||||||||||||||
| Effective tiller | 11.30±3.60∗∗ | 18.45±6.44 | ||||||||||||||||||||||||
| Panicle length (cm) | 16.38±1.57 | 15.57±1.26 | ||||||||||||||||||||||||
| Primary branching | 8.20±1.06∗ | 7.40±1.27 | ||||||||||||||||||||||||
| The secondary branch | 25.00±6.77 | 22.40±6.72 | ||||||||||||||||||||||||
| Number of grain per ear | 108.35±24.19∗ | 86.00±24.12 | ||||||||||||||||||||||||
| Grain length (mm) | 6.41±0.47 | 6.40±1.59 | ||||||||||||||||||||||||
| Grain width (mm) | 2.95±0.12 | 2.91±0.71 | ||||||||||||||||||||||||
| Grain length to width ratio | 2.23±0.13 | 2.11±0.53 | ||||||||||||||||||||||||
| The asterisk indicates significant difference (∗P<0.05, ∗∗P<0.01). | ||||||||||||||||||||||||||
Employing histological semithin transverse sectioning, we cytologically characterized male reproductive development in fertile (YJ26) and NILs sterile plants (Fig. 3). According to the previous classification of pollen growth, we tentatively characterized the sections into eight stages (Feng et al., 2001). During early development, from the early premeiosis stage to the young microspore stage, MMCs of both fertile and sterile anthers went through normal meiosis and tetrad period (Fig. 3A–D and I–L). Normal anther parietal cells including epidermis, endothecium, middle layer, and band-type-shape tapetum could all be found both in fertile (Fig. 3A–D) and sterile anthers (Fig. 3I–L). Obvious differences were observed during the start of the young microspore stage, in which tapetal layer cells of fertile pollen became degrading (Fig. 3E). In contrast, the sterile anthers still had visible tapetum (Fig. 3M). From the vacuolated pollen stage to the mature pollen stage, the tapetum of fertile anther gradually degenerated and vacuolated microspores went through mitosis, turning to mature pollen grains with fully accumulated nutrients on the surface (Fig. 3F–H). However, the sterile anther middle layer cells were still visible with intact tapetum. Meanwhile, pollens were shrunken and lacked starch granules (Fig. 3N–P). In conclusion, the pollen abortion of DNS-1X occurred in the young microspore stage.

|
| Fig. 3 Histological comparison of fertile and sterile anthers at eight developmental stages. A to H, transverse section analysis of fertile anthers; I to P, transverse section analysis of sterile anthers; A and I, the early premeiosis stage; B and J, the MMC stage; C and K, the late meiosis stage; D and L, the tetrad stage; E and M, the young microspore stage; F and N, the vacuolated pollen stage; G and O, the pollen mitosis stage; H and P, the mature pollen stage; BP: bicellular; DMsp: degraded microspore; E: epidermis; En: endothecium; MC: meiotic cell; MMC: microspore mother cell; ML: middle layer; MP: mature pollen; Msp: microspore; SC: sporogenous cell; SPC: secondary parietalcell; T: tapetum; Tds: tetrads; F: fertile anther of YJ26; S: sterile anther of BC2F5. Bars = 40 μm. |
We selected individuals with extreme phenotypes as two mixed pools (fertile pool and sterile pool) from the F2 population for BSA. Three mixed DNA pools (R01-parent DNS-1X, R02-fertile pool, and R03-sterile pool) were constructed and used for DNA sequencing. A total of 55.51 Gb of clean data were filtered from the raw data: quantification of the data showed a Q20 ≥ 97.71% and a Q30 ≥ 94.06%, and the GC content was 43.18%–43.82% across all three mixed samples (Table S1). The read depth of the parental pool (R01) was 43.5 × , and the depths of the two offspring pools (R02) and (R03) were 47.7 × and 42.5 × , respectively (Table S2). When the pooled samples were aligned to the reference genome, we identified approximately 1.09 million, 1.30 million, and 1.32 million single nucleotide polymorphisms (SNPs) in the three pools (R01–R03); among them, there were 0.22 million, 0.24 million and 0.25 million CDS mutations. There were more than 0.19 million, 0.24 million, and 0.24 million indels in the genomes of the pools; among them, 0.22 million, 0.24 million and 0.25 million CDS mutations were identified (Fig. S1). The transition of G > A and C > T was the main type of SNP mutation in each sample (Figs. S2 and S3). The ΔSNP-index and ΔInDel-index were calculated and visualized (Fig. 4A,B). There was a 9.99 Mb region on chromosome 10 that exceeded the threshold value 99%. This region contains 1163 genes. This result indicated that the genes responsible for fertility in DNS-1X might be located within this 9.99 Mb region on chromosome 10.
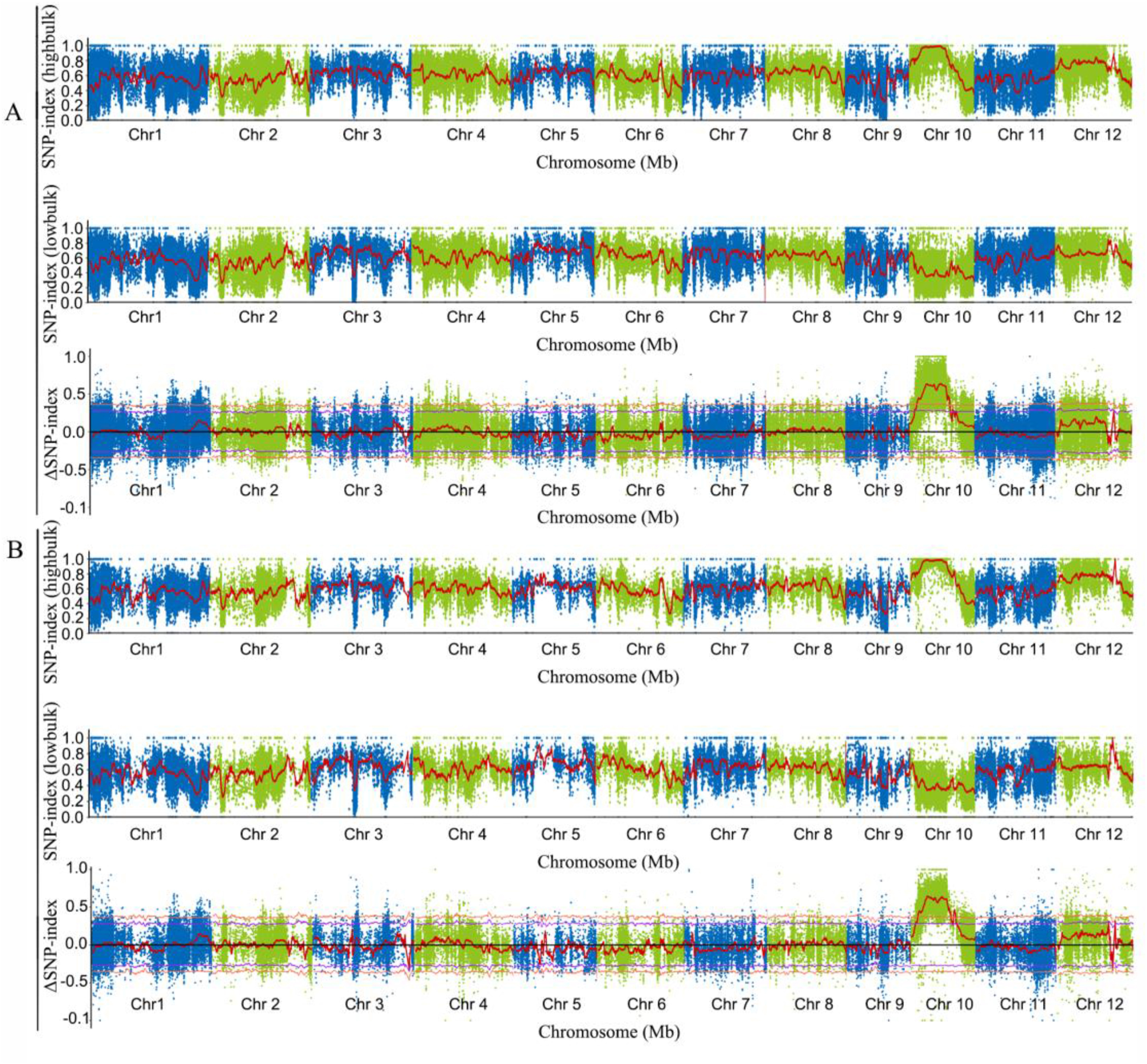
|
| Fig. 4 Bulked segregation analysis (BSA) of SNP. (A) SNP-index; (B) InDel-index; Note: Green and blue dots represent SNP-index or ΔSNP-index value; The red line: the SNP-index or ΔSNP-index after fitted; The purple line: the 95% confidence line; The orange yellow line: the 99% confidence line; SNP-index (highbulk): the linkage map of mutant offspring; SNP-index (lowbulk): the linkage map of wild offspring; ΔSNP-index: the linkage map of this population. |
To further narrow the candidate region, 191 SSR markers were selected to screen for polymorphisms in the putative region. Among them, 4 SSR markers (RM25213, RM25271, RM25289 and RM25308) showing polymorphisms were used to perform genetic linkage analysis and mapping of 829 F2 individuals. The sterility gene was mapped between RM25271 and RM25289 (Fig. 5A). For fine mapping of the sterility gene, we developed new SSR markers to screen recombinants in F3:4 segregating populations (Table S3). By comparing the phenotype and genotype of recombinants, we narrowed the sterile gene of DNS-1X to a 215 kb interval between markers DN32-3 and RM25289 on chromosome 10 (Fig. 5B) that contained 30 candidate genes (Table S4). The fine mapping region was narrower than that for the two previously reported reverse thermosensitive genic male sterile genes rtms1 and tms6(t) (Jia et al., 2001; Liu et al., 2010); all three regions overlapped on chromosome 10. We designated the newly identified region rtms1-D (Fig. 5C).
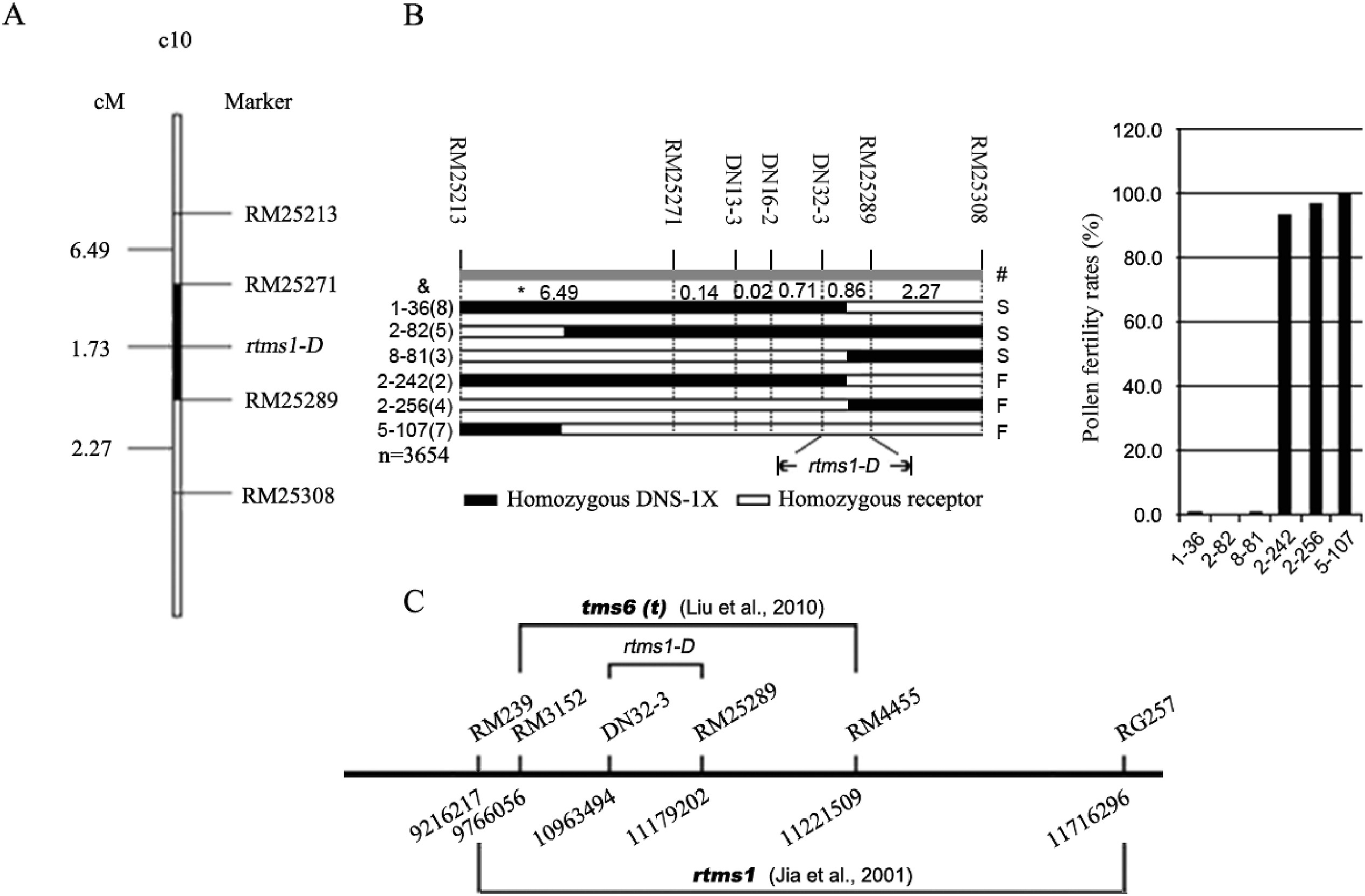
|
| Fig. 5 Fine mapping of the rtms1-D gene. (A) The result of mapping in F2 population. (B) Verification of homozygous recombinants in F3 and F4. &, ∗, and | represent the plant number, genetic distance (cM) of markers on the chromosome, and phenotype, respectively; the data in the brackets indicate the plants number of sterile or fertile genotype; S: sterile plant; F: fertile plant. (C) Comparison of the mapping region with the previously reported regions. |
A previous study showed that the fertility transition of TGMS lines was mainly attributed to temperature, and sometimes light, humidity, solar radiation, rainfall and other factors (Liu et al., 1997). Therefore, we analyzed the correlations between temperature, sunshine hours and humidity with the fertility of DNS-1X. The results demonstrated that the fertility of DNS-1X was obviously affected by temperature but not sunshine hours or relative humidity (Table 3). In addition, understanding the critical thermosensitive period of fertility alternation will help to manipulate the optimal sowing time for TGMS line reproduction and hybrid seed production (Latha and Thiyagarajan, 2010). Panicle development from first bract primordium differentiation to pollen maturation took approximately 26 days (Rangaswamy, 1993). To determine the critical temperature, the distribution of pollen fertility at both the fertile and sterile phases and the corresponding maximum temperature were charted. The results showed that the pollen was completely sterile when the temperature was lower than 28 ℃, whereas approximately 80% of the pollen was fertile when the temperature was higher than 30 ℃ (Fig. 6). We suggested that the critical temperature for fertility conversion was 28–30 ℃, implying that DNS-1X will be suitable for two-line seed production and for planting in a wider area.
| Parameter | Mean maximum temperature | Mean minimum temperature | Mean temperature | Relative humidity | Sunshine hours |
| Pollen fertility rates | 0.56∗∗ | 0.64∗∗ | 0.61∗∗ | 0.24 | −0.18 |
| The asterisk indicates significant difference (∗∗P<0.01). | |||||
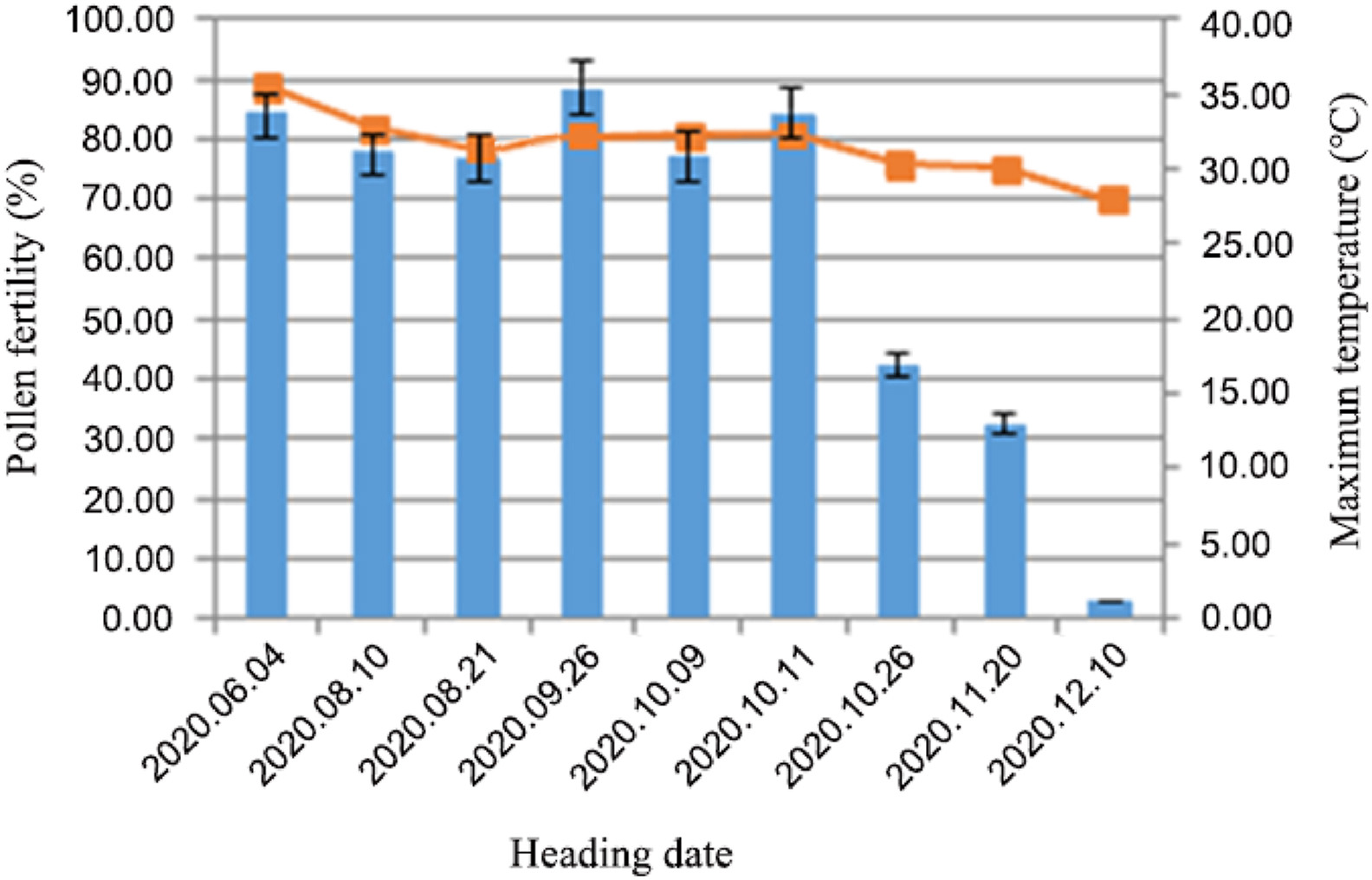
|
| Fig. 6 The observations of critical sterility and fertility temperatures in 2020. |
Heterosis, with a priority for male sterility, has been extensively applied in crop breeding. Its advantage lies in the fact that the hybrids are superior to their parental inbred lines, with an increased growth rate, a larger body size and higher yields (Shull et al., 1908; Kempe and Gils, 2011). TGMS lines have shown great potential in two-line hybrid rice breeding due to their great potential genetic characteristics. Previous studies demonstrated that tms1 was mapped to chromosome 8 (Wang et al., 1995), and tms2 was located in a 1.7 cM interval between RFLP markers R643A and R1440 on chromosome 7 (Yamaguchi et al., 1997). Subudhi et al. (1997) mapped tms3 between OPAC3640 and OPAA7550 on chromosome 6. tms5 was located in a 181 kb region on chromosome 2 between markers RMAN7 and RMAN54 (Jiang et al., 2006; Wang et al., 2003). These TGMS lines will facilitate the production of two-line hybrids.
In addition to TGMS, reverse TGMS will broaden the use of TGMS rice. In 2001, a RTGMS gene rtms1 in rice J207S was identified on chromosome 10 between markers RM239 and RG257 with genetic distances of 3.6 cM and 4.0 cM, respectively (Jia et al., 2001). Moreover, molecular mapping of the gene that controls the fertility transition of the sterile line G20S revealed a recessive gene tms6(t), which was fine mapped to an interval of 1455 kb on chromosome 10; this gene is closely linked with molecular markers RM3152 and RM4455 at distances of 3.0 cM and 1.1 cM, respectively (Liu et al., 2010). Although they overlap on chromosome 10, rtms1 and tms6(t) have different critical temperatures for fertility conversion.
In the current study, we reported a new RTGMS line, DNS-1X, which was sterile at low temperatures but fertile at high temperatures, presenting the same characteristics as rtms1 and tms6(t). Other environmental cues, such as sunshine hours and humidity, have no effect on the sterility of DNS-1X. Histological analyses showed the pollen abortion of DNS-1X occurred in the young microspore stage. The critical temperature for fertility conversion for DNS-1X is 28–30 ℃, indicating that DNS-1X has great potential in hybrid breeding. Especially in the low latitude plateau of Yunnan, China, hybrid rice seeds can be produced in large areas at middle and high altitudes, while male sterile lines can be produced in low hot valleys and at low altitudes in the south. The sterility gene was mapped on chromosome 10 between DN32-3 and RM25289 at a distance of 215 kb. This candidate region was narrower than that reported for rtms1 and tms6(t), indicating that the chromosome segment is closely related to fertility. As previous reports have not cloned or analyzed the functions of specific candidate genes, it could not be determined whether these three genes are the same gene. However, our phenotype observations further suggested that fertility is tightly controlled by a single recessive gene. DNS-1 was sterile at 24 ℃, but the inheritance pattern presented single recessive gene segregation (Rong, 1994). In the process of breeding the sterile line DNS-1X, we found that the fertility transformation of its recipient DNS-1 presented continuous segregation. After continuous selection for several generations, the sterile line DNS-1X with stable fertility and a narrow fertile transformation critical temperature of 28–30 ℃ was bred, implying that the genes mediating the fertility of DNS-1 might be gene clusters and that different combinations presented different fertility transitions. However, the underlying mechanism will be elucidated with further cloning and analysis of the function of rtms1-D.
Although numerous P/TGMS and RTGMS loci have been found and located in specific candidate regions, the gene sequences and underlying mechanisms are still largely unknown. Recent studies have demonstrated that TGMS traits are mediated at the transcriptional level. TMS5 encodes the gene Os02g0214300 (RNase ZS1), which causes the TGMS trait through a loss of RNase ZS1 function. RNase ZS1 processes the mRNAs of three ubiquitin fusion ribosomal protein L40 (UbL40) genes into multiple fragments in vitro and in vivo. The tms5 mutants showed increased levels of UbL40 mRNAs under high temperature (>28 ℃), and overexpression of UbL40 mRNAs caused defective pollen production and male sterility (Zhou et al., 2014). Moreover, the pollen fertility in rice is also regulated by long noncoding RNAs and phased small-interfering RNAs, as well as the material composition of the pollen coat (Fan et al., 2016; Jiang et al., 2020; Ding et al., 2012; Chen et al., 2007; Wang et al., 2020; Xue et al., 2018). Whether RTGMS line fertility is regulated at the transcriptional level needs to be further investigated.
Identifying a new gene locus regulating RTGMS and uncovering its underlying molecular mechanism will greatly promote large scale applications of two-line hybrid systems in rice breeding. Thus, further study is needed to investigate the candidate rtms1-D genes and their roles in regulating RTGMS. Together with the previously reported RTGMS lines, our reported line will provide beneficial support for using these RTGMS resources for rice breeding.
Author contributionsPX and DQY conceived the project and designed the experiments. XZ, GMC, ZHW, JY, FJW and FW performed the experiments. XZ, JPW and PX wrote and revised the manuscript. All authors interpreted and discussed the data.
Declaration of competing interestThe authors declare no conflict of interest.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.05.002.
AcknowledgementsThe authors gratefully acknowledge the Crop protection and breeding base and Public Technology Service Center of the Xishuangbanna Tropical Botanical Garden of CAS for providing research base and facilities. This work was financially supported by Science and technology projects of Yunnan Province, China (Grant No. 202003AD150007), Strategic Leading Science and Technology Programme of Chinese Academy of Sciences (Grant Nos. XDA24030301 and XDA24040308), Natural Science Foundation of Yunnan, China (Grant No. 2018FA 023), and National Natural Science Foundation of China (Grant No. 31902110).
Alexandratos, N., Bruinsma, J., 2012. World Agriculture towards 2030/2050. In: The 2012 Revision. FAO, Rome ESA Working Paper No. 12-03
|
Chen, R., Zhao, X., Shao, Z., et al., 2007. Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell, 19: 847-861. DOI:10.1105/tpc.106.044123 |
Deng, X., Zhou, K., 1994. Study on fertility transition and inheritance of the lower temperature thermo-sensitive dominant male-sterile rice"8987". Sichuan Agric. Univ., 12: 376-382. |
Ding, J., Lu, Q., Ouyang, Y., et al., 2012. A long noncoding RNA regulates photoperiod-sensitive male sterility,an essential component of hybrid rice. Proc. Natl. Acad. Sci. U. S. A, 109: 2654-2659. DOI:10.1073/pnas.1121374109 |
Dong, N.V., Subudhi, P.K., Luong, P.N., et al., 2000. Molecular mapping of a rice gene conditioning thermosensitive genic male sterility using AFLP, RFLP and SSR techniques. Theor. Appl. Genet., 100: 727-734. DOI:10.1007/s001220051345 |
Fan, Y., Yang, J., Mathioni, S.M., et al., 2016. PMS1T, producing Phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. U. S. A, 113: 15144-15149. DOI:10.1073/pnas.1619159114 |
Feng, J., Lu, Y., Liu, X., et al., 2001. Pollen development and its stages in rice (Oryza sativa L.). Chin. J. Rice Sci., 15: 21-28. |
Huang, T., Wang, Y., Ma, B., et al., 2007. Genetic analysis and mapping of genes involved in fertility of pingxiang dominant genic male sterile rice. J. Genet. Genomics., 34: 616-622. DOI:10.1016/S1673-8527(07)60070-8 |
Hussain, A.J., Ali, J., Siddiq, E.A., et al., 2012. Mapping of tms8 gene for temperature-sensitive genic male sterility (TGMS) in rice (Oryza sativa L.). Plant Breed., 131: 42-47. DOI:10.1111/j.1439-0523.2011.01897.x |
Jia, J.H., Zhang, D.S., Li, C.Y., et al., 2001. Molecular mapping of the reverse thermo-sensitive genic male-sterile gene (rtms1) in rice. Theor. Appl. Genet., 103: 607-612. DOI:10.1007/PL00002916 |
Jiang, D., Lu, S., Zhou, H., et al., 2006. Mapping of the rice (Oryza sativa L.) thermo-sensitive genic male sterile gene tms5 with EST and SSR markers. Chin. Sci. Bull., 51: 417-420. DOI:10.1007/s11434-006-0417-9 |
Jiang, P., Lian, B., Liu, C., et al., 2020. 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat. Commun., 11: 5191. DOI:10.1038/s41467-020-19034-y |
Jiang, Y., Rong, Y., Tao, G., et al., 1997. Breeding and performance of japonica TGMS line Diannong S-1 of a new resource. Hybrid. Rice, 5: 30. |
Kadirimangalam, S.R., Hifzur, R., R, S., et al., Fine mapping and expression analysis of thermosensitive genic male sterility gene (tms) in rice (Oryza sativa L.). Plant Gene. |
Kempe, K., Gils, M., 2011. Pollination control technologies for hybrid breeding. Mol. Breed., 27: 417-437. DOI:10.1007/s11032-011-9555-0 |
Khanday, I., Skinner, D., Yang, B., et al., 2019. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature, 565: 91-95. DOI:10.1038/s41586-018-0785-8 |
Khlaimongkhon, S., Chakhonkaen, S., Pitngam, K., et al., 2019. Molecular markers and candidate genes for thermo-sensitive genic male sterile in rice. Rice Sci., 26: 147-156. DOI:10.1016/j.rsci.2018.08.006 |
Latha, R., Thiyagarajan, K., 2010. Fertility alteration behaviour of thermosensitive genic male sterile lines in rice Oryza sativa L. Electron. J. Plant Breed., 1: 1118-1125. |
Liu, X., Li, X., Zhang, X., et al., 2010. Genetic analysis and mapping of a thermosensitive genic male sterility gene, tms6(t), in rice (Oryza sativa L.). Genome, 53: 119-124. DOI:10.1139/G09-092 |
Liu, Y., He, H., Shun, Y., et al., 1997. Light and temperature ecology of photo-thermo sensitive genic male sterile rice and its application in plant breeding. Proc. Int. Symp.: 49-58. |
Murray, M.G., Thompson, W.F., 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res., 8: 4321-4326. DOI:10.1093/nar/8.19.4321 |
Qi, P., Lin, Y.S., Song, X.J., et al., 2012. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res., 22: 1666-1680. DOI:10.1038/cr.2012.151 |
Qi, Y., Liu, Q., Zhang, L., et al., 2014. Fine mapping and candidate gene analysis of the novel thermo-sensitive genic male sterility tms9-1 gene in rice. Theor. Appl. Genet., 127: 1173-1182. DOI:10.1007/s00122-014-2289-8 |
Rangaswamy, M., 1993. Guidelines and techniques of Hybrid seed production in Rice. Dir. Rice Res
|
Rong, Y., 1994. A new type of japonica TGMS lines development at the Yunnan Agriculture University. Hybrid. Rice, 1: 43-44. |
Sasaki, T., 2005. The map-based sequence of the rice genome. Nature, 436: 793-800. DOI:10.1038/nature03895 |
Shi, M., 1985. The discovery and study of the photosensitive recessive male-sterile rice(Oryza sativa L. Subsp. Japonica). Sci. Agric. Sin.: 44-48. |
Shull, G.H., Harber, C.S., N, Y., 1908. The composition of a field of maize. Am. Breeder’s Assoc.: 296-301. DOI:10.1093/jhered/os-4.1.296 |
Subudhi, P.K., Borkakati, R.P., Virmani, S.S., et al., 1997. Molecular mapping of a thermosensitive genetic male sterility gene in rice using bulked segregant analysis. Genome, 40: 188-194. DOI:10.1139/g97-027 |
Takagi, H., Abe, A., Yoshida, K., et al., 2013. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J., 74: 174-183. DOI:10.1111/tpj.12105 |
Virmani, S.S., 1996. Hybrid rice. Adv. Agron., 57: 377-462. |
Wang, B., Fang, R., Zhang, J., et al., 2020. Rice LecRK5 phosphorylates a UGPase to regulate callose biosynthesis during pollen development. J. Exp. Bot., 71: 4033-4041. DOI:10.1093/jxb/eraa180 |
Wang, B., Wang, H., 2017. IPA1: a new “green revolution” gene?. Mol. Plant, 10: 779-781. DOI:10.1016/j.molp.2017.04.011 |
Wang, B., Xu, W.W., Wang, J.Z., et al., 1995. Tagging and mapping the thermo-sensitive genic male-sterile gene in rice (Oryza sativa L.) with molecular markers. Theor. Appl. Genet., 91: 1111-1114. DOI:10.1007/bf00223928 |
Wang, C., Liu, Q., Shen, Y., et al., 2019. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol., 37: 283-286. DOI:10.1038/s41587-018-0003-0 |
Wang, F., 2020. Achievements and prospects of hybrid rice breeding—review of 50 Years'research on hybrid rice by rice research institute of Guangdong Academy of agricultural Sciences. Guangdong Agric. Sci., 47: 1-11. |
Wang, Y.G., Xing, Q.H., Deng, Q.Y., et al., 2003. Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5. Theor. Appl. Genet., 107: 917-921. DOI:10.1007/s00122-003-1327-8 |
Xue, Z., Xu, X., Zhou, Y., et al., 2018. Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat. Commun., 9: 604. DOI:10.1038/s41467-018-03048-8 |
Yamaguchi, Y., Ikeda, R., Hirasawa, H., et al., 1997. Linkage analysis of thermosensitive genic male sterile gene,tms-2 in rice(Oryza sativa L.). Breed Sci., 47: 371-373. DOI:10.1270/jsbbs1951.47.371 |
Yuan, L., 1998. Exploiting crop heterosis by two-line system hybrids: current status and future prospects. High Technol. Lett, 4: 90-95. |
Yuan, L., 1997. Current status and development prospects in two-line hybrid rice research in China. Res. Agric. Mod., 18: 2-4. |
Yuan, L., 1990. Progress of two-line system Hybrid rice breeding. Sci. Agric. Sin., 23: 1-6. |
Yuan, L., 1986. Hybrid rice in China. Chin. J. Rice Sci., 1: 8-18. |
Zhao, J., Long, T., Wang, Y., et al., 2020. RMS2 encoding a GDSL lipase mediates lipid homeostasis in anthers to determine rice male fertility. Plant Physiol., 182: 2047-2064. DOI:10.1104/pp.19.01487 |
Zhou, H., Zhou, M., Yang, Y., et al., 2014. RNase ZS1 processes Ubnull mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun., 5: 4884. DOI:10.1038/ncomms5884 |



