b. Northwest A&F University, Yangling, 712100, China;
c. Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China (Ministry of Education), Southwest Forestry University, Kunming, 650224, China;
d. Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Science, Mengla, 666303, China;
e. Kunming Institute of Botany, Chinese Academy of Sciences, 132 Lanhei Road, Kunming, 650204, China;
f. University of Chinese Academy of Sciences, Beijing, 100101, China
Castor bean (Ricinus communis L., Euphorbiaceae) is an important non-edible oilseed crop whose seed oil is used extensively in industry. The main composition of its seed oil is ricinoleic acid (12-hydroxyoctadec-cis-9-enoic acid; 18:1-OH), which is considered an ideal and unique feedstock for biodiesel production (Akpan et al., 2006; Ogunniyi, 2006; Scholz and Silva, 2008). Due to an increased demand for production of castor bean seed oils in many countries, breeding and improvement of varieties have drawn great attention from breeders (Qiu et al., 2010). Thus, elucidating the molecular mechanisms that underlie the regulation of growth, development and seed oil accumulation is critical. The completion of the castor bean genome (Chan et al., 2010) provides an opportunity to identify and characterize genes that may regulate seed oil accumulation. Genes that may play a role in regulating seed oil accumulation include members of the B3 gene superfamily.
The B3 genes belong to a superfamily that encodes plant-specific transcription factors (TFs) found from unicellular green algae to eudicots (Swaminathan et al., 2008). B3 TFs are characterized by a common B3 domain approximately 110 amino acids in length, which has sequence-specific DNA binding activity initially identified in maize VIVIPAROUS1 (VP1) and Arabidopsis ABSCISIC ACID INSENSITIVE 3 (ABI3) (McCarty et al., 1989; Giraudat et al., 1992; Suzuki et al., 1997). Structural features and protein sequence homology have been used to divide the B3 gene superfamily into five major classes, including the ABI3/VP1 (Suzuki et al., 1997), HSI (High-level expression of sugar inducible gene) (Tsukagoshi et al., 2005), ARF (Auxin Response Factor) (Ulmasov et al., 1997), RAV (Related to ABI3/VP1) (Kagaya et al., 1999) and REM (Reproductive Meristem) (Franco-Zorrilla et al., 2002) subfamilies. B3 TFs have been characterized in various species, including bryophyte (Physcomitrella patens), green algae (Chlamydomonas reinhardtii), Arabidopsis, rapeseed, soybean, poplar, maize, and rice (Romanel et al., 2009; Peng and Weselake, 2013). However, B3 TFs have not yet to be identified in the important non-edible oilseed crop castor bean.
Members of the B3 superfamily play diverse roles in regulating plant growth and development. These regulatory roles are mediated through DNA binding by the B3 domain and additional domains, including AP2/EREBP (AP2) (Kagaya et al., 1999), auxin response factor (Aux_resp) (Ulmasov et al., 1999), auxin/indole-3-acetic acid domain (Aux/IAA) (Ulmasov et al., 1997), and zinc finger CW domain (zf-CW) (Suzuki et al., 2007). Members of the RAV family are characterized by a C-terminal B3 domain and an N-terminal AP2 domain, both of which bind to CAACA sequences (Kagaya et al., 1999). The RAV subfamily might act as a negative component in regulating developmental retardation of lateral root, leaves, flowers and early seeding (Hu et al., 2004; Feng et al., 2014). The highly divergent N-terminal B3 domain of ARF subfamily genes recognizes auxin response elements (AuxREs; TGTCTC) (Ulmasov et al., 1999). The ARF subfamily is functionally involved in regulating auxin-mediated physiological processes such as lateral root formation, gravitropism in both hypocotyl and root, gradual partitioning of lateral organs along the abaxial/adaxial axis, and flower development (Okushima et al., 2005a, 2005b; Pekker et al., 2005). Little is known about the DNA binding activity in the REM subfamily. However, the REM subfamily has been shown to participate in mediating the maintenance of the vernalization response for promoting flowering (Levy et al., 2002), and regulating the embryo sac differentiation (Matias-Hernandez et al., 2010). Interestingly, although B3 domains unique to different subfamilies share a common structural framework, these domains are able to bind to different DNA motifs (Yamasaki et al., 2004; Waltner et al., 2005). In addition, although the DNA binding specificity of ABI3/VP1, HSI, RAV and ARF has been characterized, few studies have comprehensively characterized B3 superfamily structure, expression and function in a given species.
One B3 subfamily that may play an important role in storage lipid biosynthesis is the ABI3/VP1 subfamily. The B3 domains of ABI3/VP1 members recognize the Sph/RY element (CATGCA) (Suzuki et al., 1997; Crowe et al., 2000; Guerriero et al., 2009; Guo et al., 2013). Members of the ABI3/VP1 subfamily, including LEC2 (LEAFY COTYLEDON 2), ABI3 and FUS3 (FUSCA 3), are specifically expressed during seed development and play a vital role in regulating embryogenesis, endosperm development and filling storage reservoirs in developing seeds (Swaminathan et al., 2008). Most studies on the ABI3/VP1 subfamily have focused on the regulation of embryogenesis and endosperm development in Arabidopsis and cacao (Keith et al., 1994; Stone et al., 2001, 2008; Tsuchiya et al., 2004; Braybrook et al., 2006; Zhang et al., 2014). However, several lines of evidence indicate that LEC2, ABI3 and FUS3 are involved in regulating storage material accumulation in developing seeds, including fatty acid biosynthesis, storage lipid assembly, and storage protein accumulation (Crowe et al., 2000; Braybrook et al., 2006; Baud et al., 2007; Stone et al., 2008; Andrianov et al., 2010; Yamamoto et al., 2010; Angeles-Núñez and Tiessen, 2011; Mönke et al., 2012; Zhang et al., 2014). Because storage material accumulation in developing seeds is often species-specific, little is known about how ABI3/VP1 members regulate storage lipid biosynthesis in different plants.
In this study, we performed a genome-wide analysis of the B3 superfamily in castor bean to better understand the regulatory role of the B3 superfamily genes in castor seed development and material accumulation. For this purpose, we identified 61 B3 TFs in castor bean and characterized their structure and gene expression. Expression analysis of B3 genes suggested that ABI3/VP1 subfamily members (RcLEC2, RcABI3 and RcFUS3) are seed-specific TFs in castor bean. In addition, yeast one-hybrid and transient expression assays indicated that RcLEC2 proteins are potential active regulators of seed lipid biosynthesis.
2. Materials and methods 2.1. Plant materials and growth conditionsCastor bean seeds (ZB306), provided by Zibo Academy of Agricultural Science, were sterilized and germinated in an incubator at 30 ℃. Seeds were collected at five different developmental stages (seed1−seed5) from 10 to 50 days after pollination. Roots and leaves were collected 14 days after germination. The pollen, inflorescence, ovules, capsule, endosperms, and embryos were separated from the developing castor bean, then immediately frozen in liquid nitrogen for storage at −80 ℃.
2.2. Identification and classification of B3 TFs from the castor bean genomeTo identify candidate B3 genes in the sequenced genome of castor bean (http://castorbean.jcvi.org/index.php), we performed a local BLAST search using Arabidopsis B3 protein sequences as queries (https://www.arabidopsis.org/). SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.sanger.ac.uk/) tools were then used to confirm putative B3 genes (Punta et al., 2011; Letunic et al., 2012). Multiple alignments of full-length amino acid sequences of the B3 TFs in castor bean and Arabidopsis were conducted with the MUSCLE tool (Edgar, 2004). Neighbor-joining (NJ) phylogenetic trees were generated using MEGA 5.0 (Tamura et al., 2011) with 1000 bootstrap replicates.
2.3. MEME motif and gene structural analysesConserved motifs in castor bean B3 TFs were identified by the online MEME analysis tool (http://meme.ebi.edu.au/meme/intro.html). The exon and intron structures of B3 genes were illustrated using Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/).
2.4. Transcriptome data analysisTo investigate the expression patterns of B3 genes, we used our castor bean RNA-seq database (https://woodyoilplants.iflora.cn/) to obtain FPKM values of B3 and lipid genes in different tissues, including root, stem, leaf, seedling, ovule, capsule, inflorescence, germinating seed (G_seed), endosperm, embryo, and seeds of five different development stages (seed1−seed5). Expression levels were normalized basing on FPKM values and heatmaps were generated in the R package. Hierarchical cluster analysis was performed based on the ‘complete’ clustering method.
2.5. Co-expression gene network analysisCo-expression analysis was conducted by repeated iteration of network topological parameters as a function of the Pearson correlation coefficient (PCC) threshold in order to select significant correlation values. The correlation matrix was visualized and analyzed by R package and Cytoscape (v.3.6.1) (http://www.cytoscape.org) for co-expression network of genes.
2.6. Transient expression assayFor the transient expression assay, the 548-bp promoter sequence of RcOleosin1 and 321-bp promoter sequence of RcOleosin2 were amplified by PCR. The products were cloned into the HindIII and SalI digested pRI101-LUC (luciferase) vector to construct firefly LUC reporter plasmids including pRcOleosin1:LUC and pRcOleosin2:LUC. The full-length coding regions of RcLEC2 and RcABI3 were amplified and then inserted into the plasmid PWXP 1.0 to generate the effecter constructs of 35S:RcLEC2 and 35S:RcABI3. Agrobacterium tumefaciens-mediated transient transformation (Liu et al., 2010) and detection of luminescence (Xiong et al., 1999) were performed as described previously respectively. To take a luminescence image, the treated leaves were sprayed with luciferin and the plate was left in a dark box for 5 min to eliminate interference from chlorophyll fluorescence. Photos were taken by an IVIS Lumina XR. Primers are listed in Table S6.
2.7. Yeast one-hybrid (Y1H) assayFor the Y1H assay, oligonucleotides were synthetized that contained four tandem copies of each Sph/RY element with flanking sequences of the RcOleosin2 promoter. The products were cloned into pAbAi plasmids digested with HindIII and SmaI. RcLEC2 was cloned into the plasmid pGADT7. Each pRcOleosin2-E-AbAi, pRcOleosin2-Em (Mutant)-AbAi, and p53-AbAi vectors were linearized with BstBI and then transformed into Y1HGold strain. Transformants were selected on SD/-Ura media. Subsequently, pGADT7-RcLEC2 was co-transformed into Y1HGold without digesting. Yeast was grown in SD–Ura–Leu medium and then spotted on SD–Ura–Leu medium in the presence or absence of 200 ng/ml AbA (Aureobasidin A). The plates were incubated for 4 d at 28 ℃. The pGADT7-RcLEC2/p53-AbAi and pGADT7-P53/p53-AbAi constructs were used as negative and active controls, respectively. The primers and flanking sequences used for plasmid construction are shown in Tables S6 and S7.
3. Results 3.1. Identification of genes encoding B3 proteins in castor beanA total of 61 putative B3 genes were identified in the castor bean, encoding proteins that ranged from 109 to 1119 amino acids in length (Table S1). Putative B3 genes were classified into five subfamilies: AFR (17 genes), RAV (7), HSI (3), ABI3/VP1 (3), and REM (31) (Table S1). Distinct B3 domain organization within different B3 subfamilies was observed (Fig. S1). B3 domain sequences identified from ABI3/VP1, HSI, RAV and ARF subfamilies were highly conserved, whereas B3 domain sequences of REM members varied (Fig. 1). Multiple sequence alignments identified 44 and 85 conserved amino acid residues in the ABI3/VP1 subfamily and HSI subfamily, respectively. Also, most residues were conserved in the RAV and ARF subfamilies. The amino acid residues were, however, highly variable in the REM subfamily.
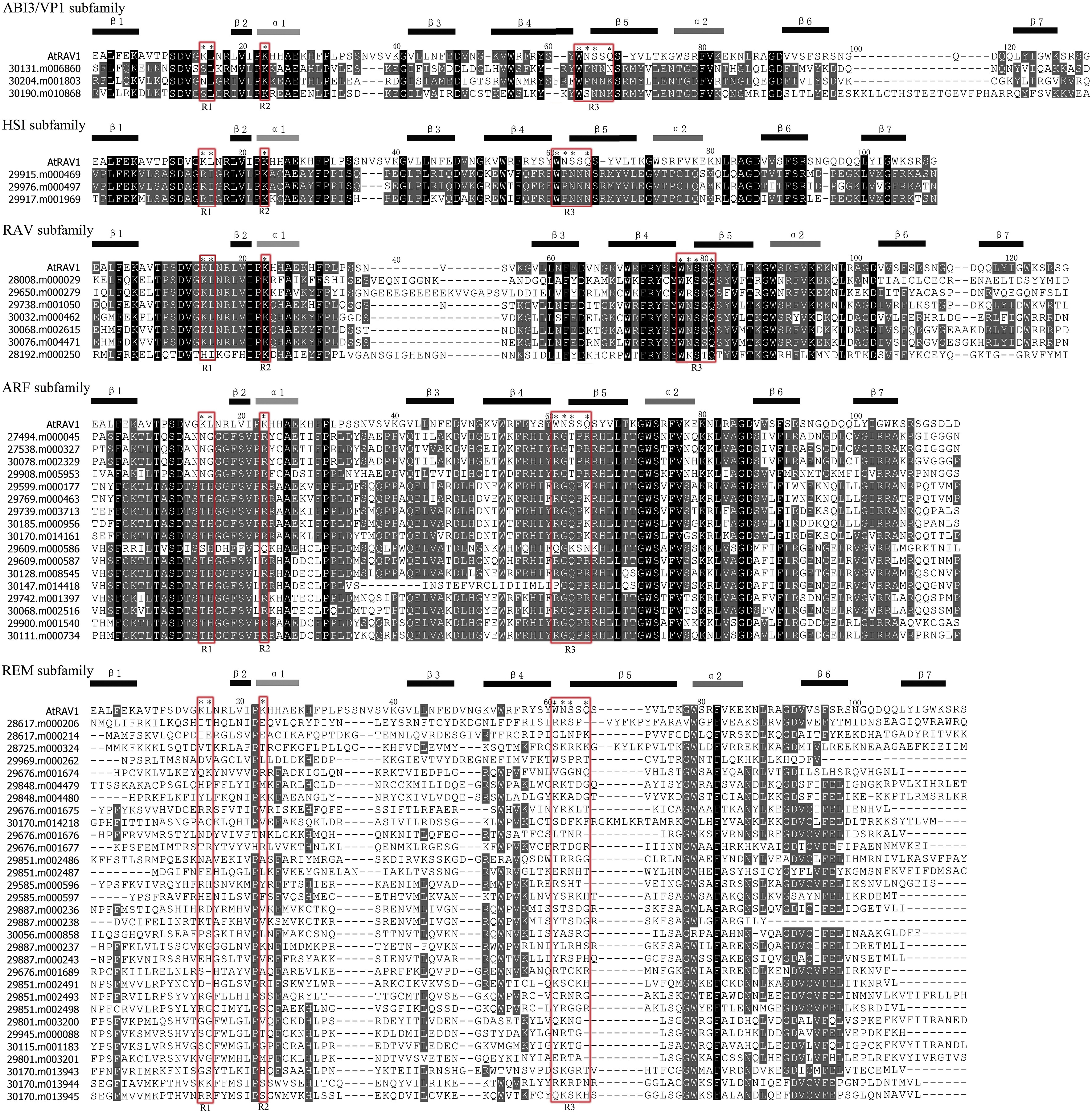
|
| Fig. 1 Alignment of the B3 domain sequences between AtRAV1 and castorbean B3 proteins. A total of 61 B3 domains were aligned using vector NTI soft. The structural location of α1, α2 and β1–β7 is indicated upon the alignment. Black stars represent the positions in the AtRAV1 sequence that are proposed to contact DNA (Yamasaki et al., 2004). The DNA contact regions are marked using red rectangles and designed as R1–R3. |
Previous research found that key DNA-contacting regions, designated as R1–R3, in the AtRAV1 of Arabidopsis (corresponding to K15, L16, K32, W78, N79, S80 and Q82) have been evolutionarily conserved and critical for proper function of the B3 domain (Yamasaki et al., 2004). In castor bean, the amino acid residues of R1, R2 and R3 regions were highly conserved within each subfamily (including ABI3/VP1, HSI, RAV, and ARF), whereas these residues were highly variable in the REM subfamily (Fig. 1). Particularly, we noted that the amino acid sequences were distinctly differentiated among different subfamilies, though they were conserved within each subfamily (except for the REM). For example, the amino acid residues of R3 region were W [N/K/R]SSQ within the RAV subfamily, whereas the corresponding residues of ABI3/VP1, HSI and ARF subfamilies were W [P/S]NN [K/N], WPNNN and RG [Q/T]P [R/K], respectively.
To examine the phylogenetic relationships of B3 proteins identified in castor bean, we generated phylogenetic trees based on the alignments of full-length protein sequences. When four REM members (29969.m000262, 28617.m000214, 28725.m000324, and 28617.m000206) were excluded from our analysis, B3 proteins of the same subfamily clustered into the same clades with well supported bootstrap values (Fig. 2). When we compared the evolutionary patterns of castor bean and Arabidopsis B3 proteins, we also found that ABI3/VP1, HSI, RAV, and ARF subfamily members were clustered in clades with members of the same subfamily (Fig. S2). Particularly, all major clades and subclades contained interspecific members. However, members of REM subfamily were diversely distributed in different clades of the phylogenetic tree and most clades contained intra-species members.

|
| Fig. 2 An unrooted phylogenetic tree of the B3 family in castorbean. The aminoacidsequences were aligned using MUSCLE tool and then a NJ phylogenetic tree was generated using MEGA5.0 with 1000 bootstrap replicates. Each subfamily of B3 members is indicated by an arc. |
To reveal specific properties for DNA-binding of B3 proteins in castor bean, putative motifs were analyzed by the program MEME. A total of 25 conserved motifs were detected in the 61 B3 proteins and named as motifs 1–25 (Table S2). The distribution of motifs in each B3 proteins is shown in Fig. 3. Some motifs are shared by several groups. For example, motif 2 is present in the ABI3/VP1, HSI, RAV and ARF subfamily; motif 12 is present in the ABI3/VP1 and HSI subfamily; in addition, motif 1 and motif 9 are shared by the RAV and ARF subfamily. As expected, most subfamily members clustered together in the phylogenetic tree shared one or more conserved motifs (Fig. 3). For example, all members of the ABI3/VP1 and HSI subfamilies shared motif 2 and motif 12; all members of the HSI subfamily shared motifs 12, 13 and 22; most members of the ARF subfamily shared motifs 1, 2, 4, 5, 6, 7, 9, 10, 11 and 25; virtually all members of the ARF subfamily also shared motifs 8, 16 and 18; all members of the REM subfamily contained motif 3; and all members of RAV subfamily contained motifs 1 and 9.
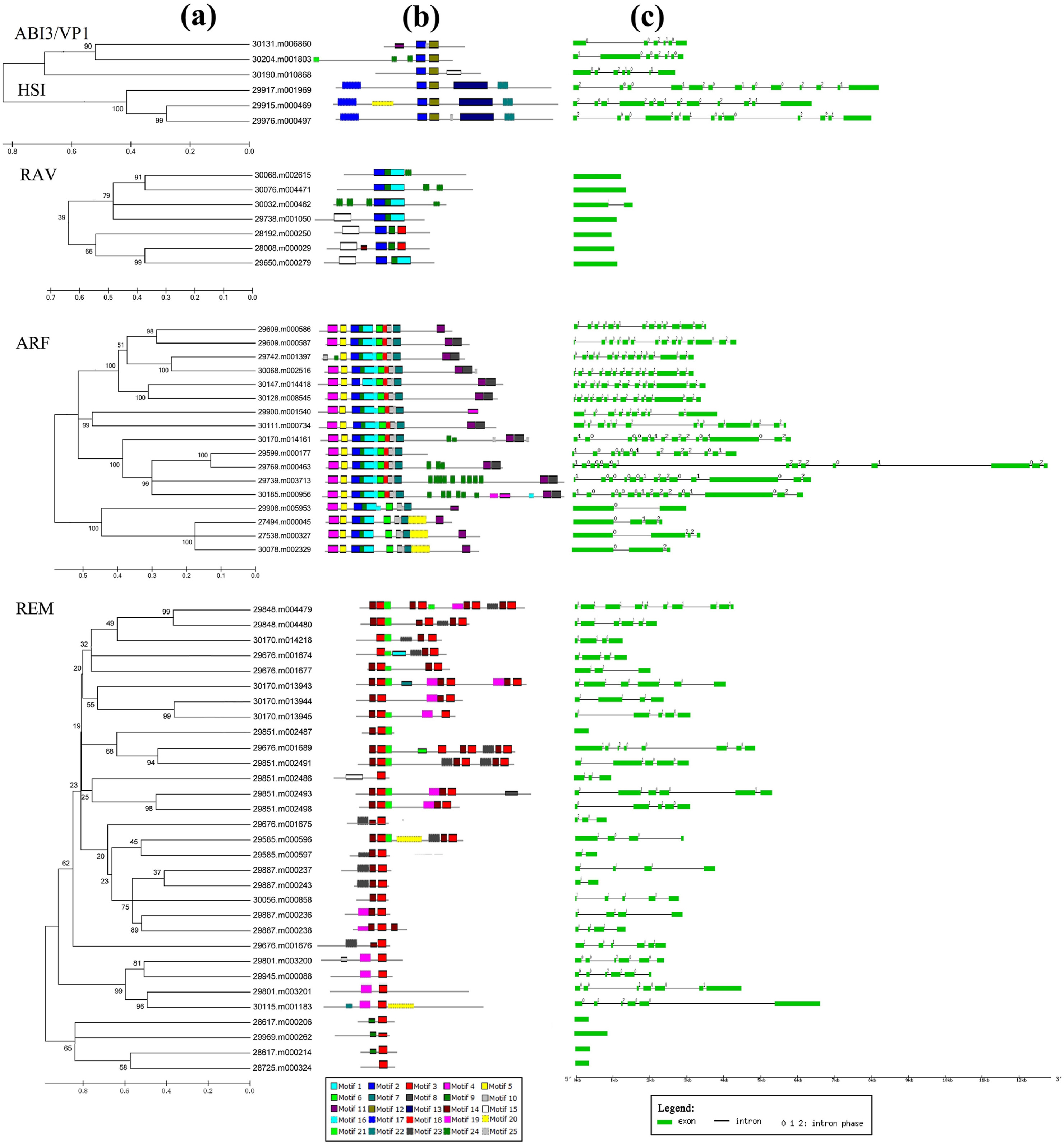
|
| Fig. 3 Structural features and the distribution of conserved motifs within each B3 subfamily in castorbean. (a) Phylogenetic clades identified within each B3 subfamily. (b) The distribution of conserved motifs within aminoacidsequences of each B3 protein. Each motif is represented by a number of colored boxes. (c) Exon/intron structures of castor bean B3 genes. The exons and introns are represented by boxes and lines, respectively. The number above the line indicates the splicing phases. |
Gene structural diversity may elucidate the emergence and evolution of multigene families. Thus, we analyzed the exon/intron organization of B3 genes in castor bean (Fig. 3). We found that the intron number of B3 genes ranged from 0 (11 genes) to 13 (8 genes). Excluding REM subfamily genes, which had diverse intron numbers (0–9), most genes within each B3 subfamily shared the same or similar intron patterns. For instance, six of seven members in the RAV subfamily are intron-less and just one member (30032.m000462) has one intron; three members in the ABI3/VP1 subfamily contain five or six introns; three members of the HSI subfamily include eleven or twelve introns. In addition, the phylogenetic tree of the ARF subfamily mainly consisted of two clades. One clade contained members with more than ten introns, whereas the other included members with less than four introns.
The pattern of exon/intron splicing phase was also considered to be informative for exploring the emergence and evolution of the B3 gene family. For genes containing introns, we detected the intron position pattern (Fig. 3). Although the number of introns varied, the intron positions were conserved in each B3 subfamily except REM, whose members showed variation in intron position. Three splicing phases were designated: phase 0, splicing occurs after the third nucleotide of the codon; phase 1, splicing occurs after the first nucleotide of the codon; and phase 2, splicing occurs after the second nucleotide. We found that most members in the ABI3/VP1 subfamily shared similar splicing phase patterns, and the pattern of splicing phase was also conserved in the HSI subfamily (Fig. 3). Similarly, most members in each clade of the ARF subfamily shared similar splicing phase patterns. However, the splicing phase patterns in the REM subfamily were diverse.
3.3. Tissue-specific expression profiles of B3 genes in castor beanA total of 48 B3 genes showed expression profiles in our libraries, whereas 13 additional B3 genes were not expressed in any tissue tested (Table S3). This lack of expression may be the result of functional redundancy of genes within the same subfamily. Most B3 genes displayed specific expression patterns across different tissues (Fig. 4). RAV subfamily genes (especially 28192.m000250 and 29738.m001050) were more highly expressed in reproductive tissues (ovule and inflorescence) and stem; HSI subfamily genes were expressed in all tissues. ARF subfamily members were expressed at lower levels in seeds, except 29742.m001397, but were highly expressed in leaf, stem, ovule, and inflorescence. Interestingly, the ARF subfamily gene 30078.m002329 was highly expressed in the early stages of seeds development (seed1 and seed2), but not the mid-late stages (seed3–seed5). Furthermore, the REM subfamily genes were more highly expressed in reproductive tissues (ovule, inflorescence, and embryo) and early seed stages (seed1 and seed2). Some genes, including 29676.m001674, 29848.m004479, 30170.m013944, 29887.m000238 and 29945.m000088, were more highly expressed in embryos and are probably related to embryonic development.
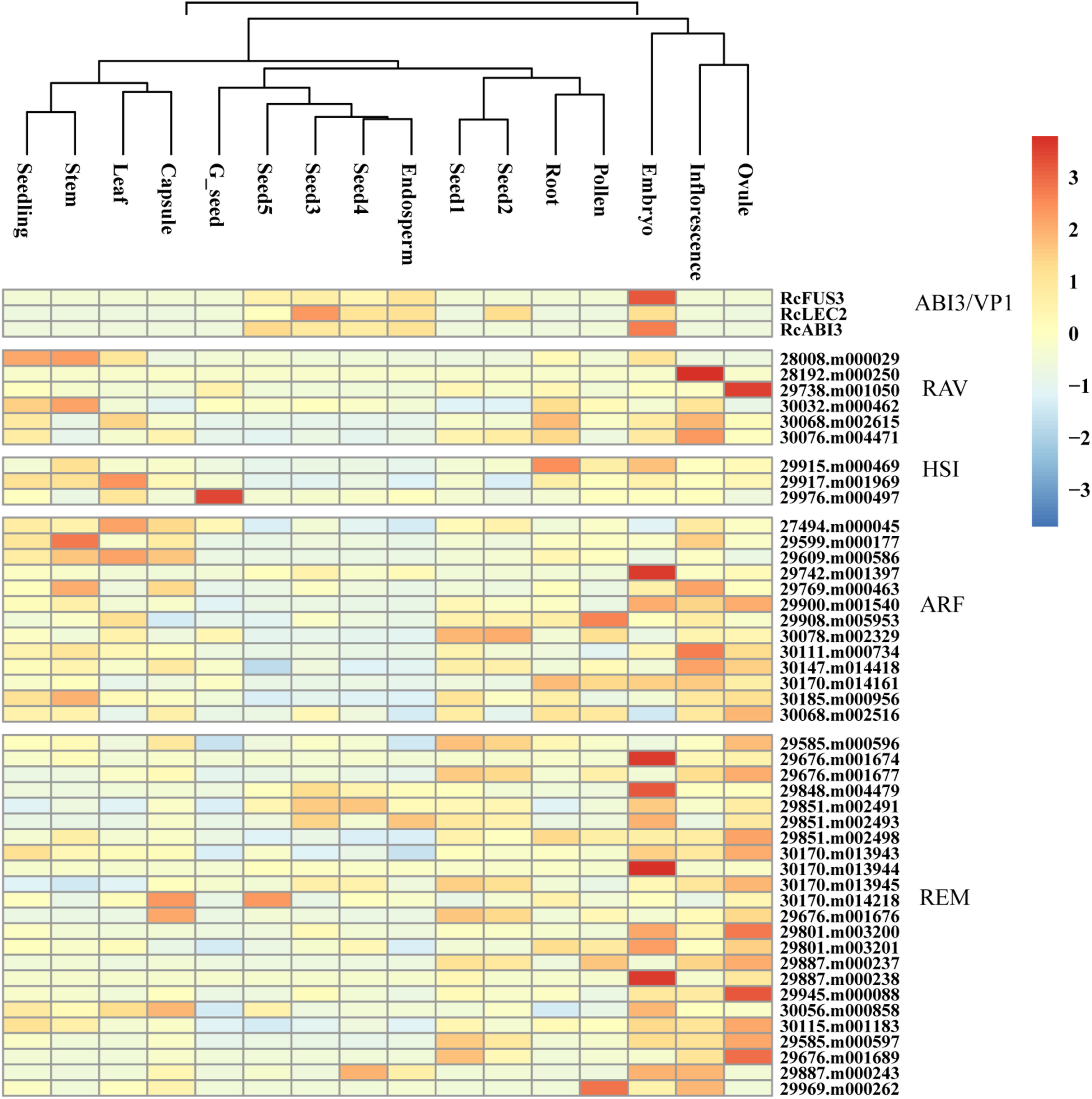
|
| Fig. 4 Expression profiles of B3 genes in different tissues. Blue and red boxes indicate the lower and higher transcriptional levels, respectively. The scale bar represents the relative expression level after normalization. Based on the ‘complete’ clustering method, hierarchical cluster analysis was performed. |
Notably, all members of the ABI3/VP1 subfamily, RcLEC2 (30190.m010868), RcABI3 (30204.m001803) and RcFUS3 (30131.m006860), were only highly expressed in seeds, especially in the embryo and endosperm (Fig. 4, Table S3). RcLEC2 was expressed in stages of early and middle seed development, whereas RcABI3 and RcFUS3 were expressed in the middle and late stages of seed development. Previous studies have revealed that AtLEC2, AtABI3 and AtFUS3 are often expressed specifically in developing seeds, participating in the regulation of seed oil accumulation (Brown et al., 2012). These results suggest that certain B3 genes, especially RcABI3/VP1 subfamily members, might play important roles in regulating different stages of seed development, including the endosperm, which is the tissue where stored material is accumulated during seed development.
3.4. Lipid gene co-expression network with RcABI3/VP1 subfamilyPrevious research reported that the AtABI3/VP1 subfamily regulates lipid genes by binding the cis-acting element Sph/RY (Andrianov et al., 2010; Brown et al., 2012). To study if and how ABI3/VP1 subfamily members regulate the transcriptional activity of oil synthesis genes, we examined the promoter regions of 71 oil synthesis genes (Chan et al., 2010) for Sph/RY elements in castor bean. We found that 26 oil genes were expressed in different tissues and contained at least one Sph/RY element in their 2-kb promoter regions, indicating that the expression of these genes may be regulated by RcABI3/VP1 subfamily (Fig. 5, Table S4). We found that 16 of these oil genes were expressed in all examined seed stages, including seed1 to seed5, embryo and endosperm. Interestingly, two core oil genes, RcOleosin1 (29917.m001992) and RcOleosin2 (30147.m014333), were extremely highly expressed at the stages during which seed oil is rapidly accumulated (seed3–seed5), as well as in the embryo and endosperm. This pattern of gene expression is similar to the expression patterns of the genes in the RcABI3/VP1 subfamily.
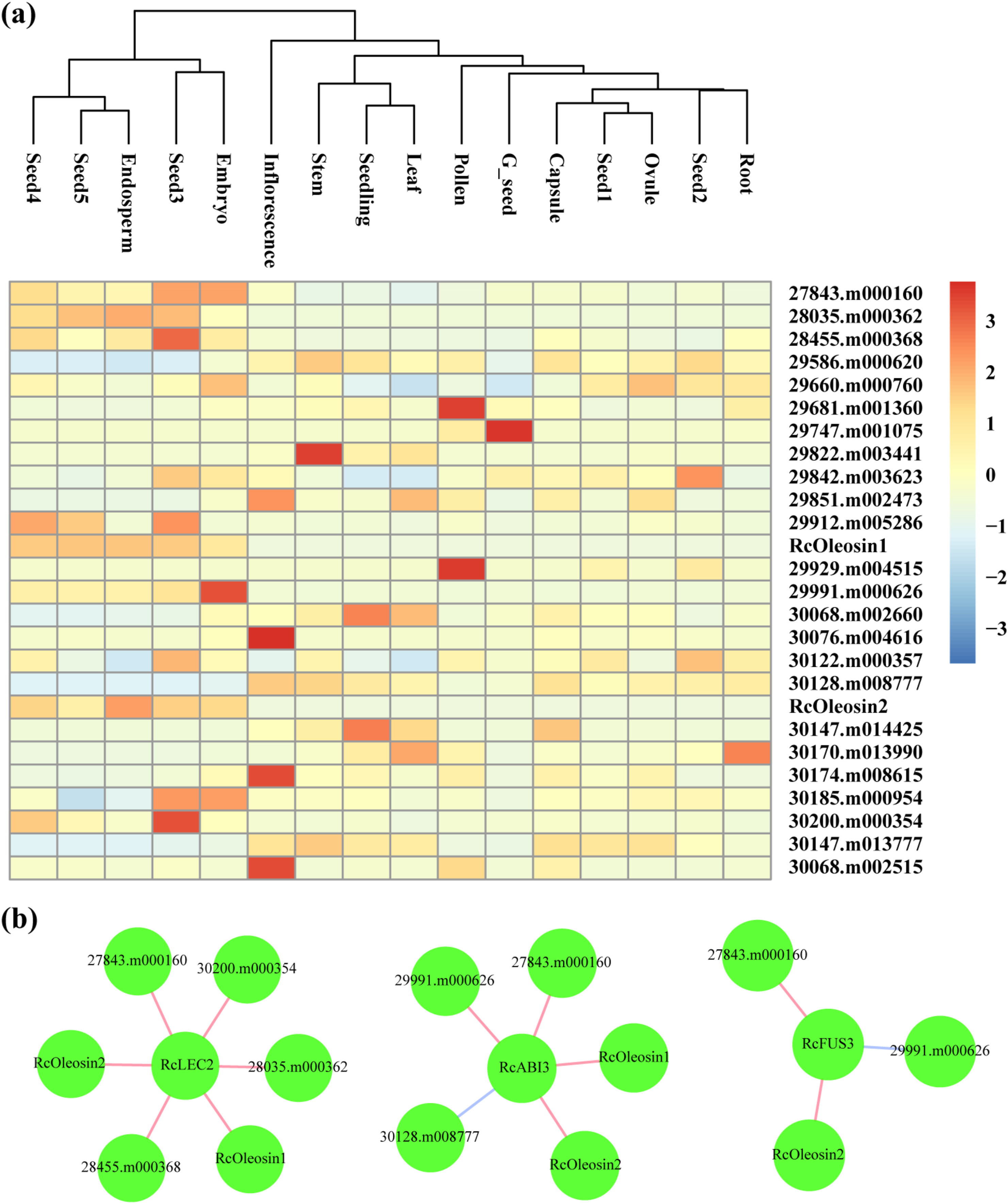
|
| Fig. 5 Expression profiles and co-expression network analysis of putative target lipid genes of RcABI3/VP1 subfamily. (a) Expression profiles of putative target lipid genes of RcABI3/VP1 subfamily among different tissues. Blue and red boxes indicate the lower and higher transcriptional levels, respectively. The scale bar represents the relative expression level after normalization. Based on the ‘complete’ clustering method, hierarchical cluster analysis was performed. (b) The co-expressed lipid genes were identified. Each gene is represented by one circle. Red and blue lines indicate positive and negative correlation among RcABI3/VP1 subfamily and putative target lipid genes, respectively. Absolute correlation coefficient was greater than 0.7. |
To further investigate transcriptional regulation in seed oil synthesis, we constructed a gene co-expression network of RcABI3/VP1 subfamily and 26 putative target lipid genes using the RNA-seq data from different tissues. We found that all members of the RcABI3/VP1 subfamily and 11 oil genes were included in the gene co-expression module (absolute correlation coefficient > 0.7) (Fig. 5, Table S5). The expression of three oil genes highly expressed in seeds (e.g., RcOleosin2) was positively correlated with RcFUS3 expression. The expression of four oil genes highly expressed in seeds (e.g., RcOleosin1 and RcOleosin2) was positively correlated with RcABI3 expression. In contrast, the expression of 30128.m008777, which is expressed at low levels during seed development, was negatively corelated with RcABI3 expression. Similarly, the expression of six oil genes highly expressed in seeds (e.g., RcOleosin1 and RcOleosin2) were also positively corelated with RcLEC2 expression. Among these oil genes, the expression of two genes, RcOleosin2 and 27843.m000160, was positively correlated with all members of the RcABI3/VP1 subfamily; expression of one gene, 29991.m000626, was positively correlated with RcFUS3 and RcABI3, and RcOleosin1 expression was positively correlated with RcLEC2 and RcABI3. These results indicate that members of the RcABI3/VP1 subfamily may regulate the transcription of seed oil genes in castor bean.
3.5. Transcription activity of RcOleosin2 regulated by RcLEC2Given the positively correlated expression patterns of members of the RcABI3/VP1 subfamily and oleosin genes during seed development, we used a tobacco transient expression assay system to determine whether RcLEC2 and RcABI3 activate expression of RcOleosin1 and RcOleosin2. Reporter plasmids were constructed by fusing Sph/RY elements and either the RcOleosin1 promoter sequence (548-bp) or the RcOleosin2 promoter sequence (321 bp) to the luciferase (LUC) reporter gene (Fig. 6, Table S6). Infiltration of tobacco leaves with negative control (i.e., Agrobacterium tumefaciens cells only with the reporter plasmids) produced low luminescence intensity (Fig. 6). Co-infiltration of these constructs with 35S:RcLEC2 produced a high luminescence intensity within 72 h in pRcOleosin2:LUC injected leaves, but not in pRcOleosin1:LUC injected leaves. Co-expression with 35S:RcLEC2 showed about 1.5-fold activation of pRcOleosin2:LUC (Fig. 6). These results suggest that RcLEC2 actively regulates the expression of RcOleosin2. However, when these constructs were co-infiltrated with 35S:RcABI3, no significant increase of luminescence intensity was detected (Fig. 6).
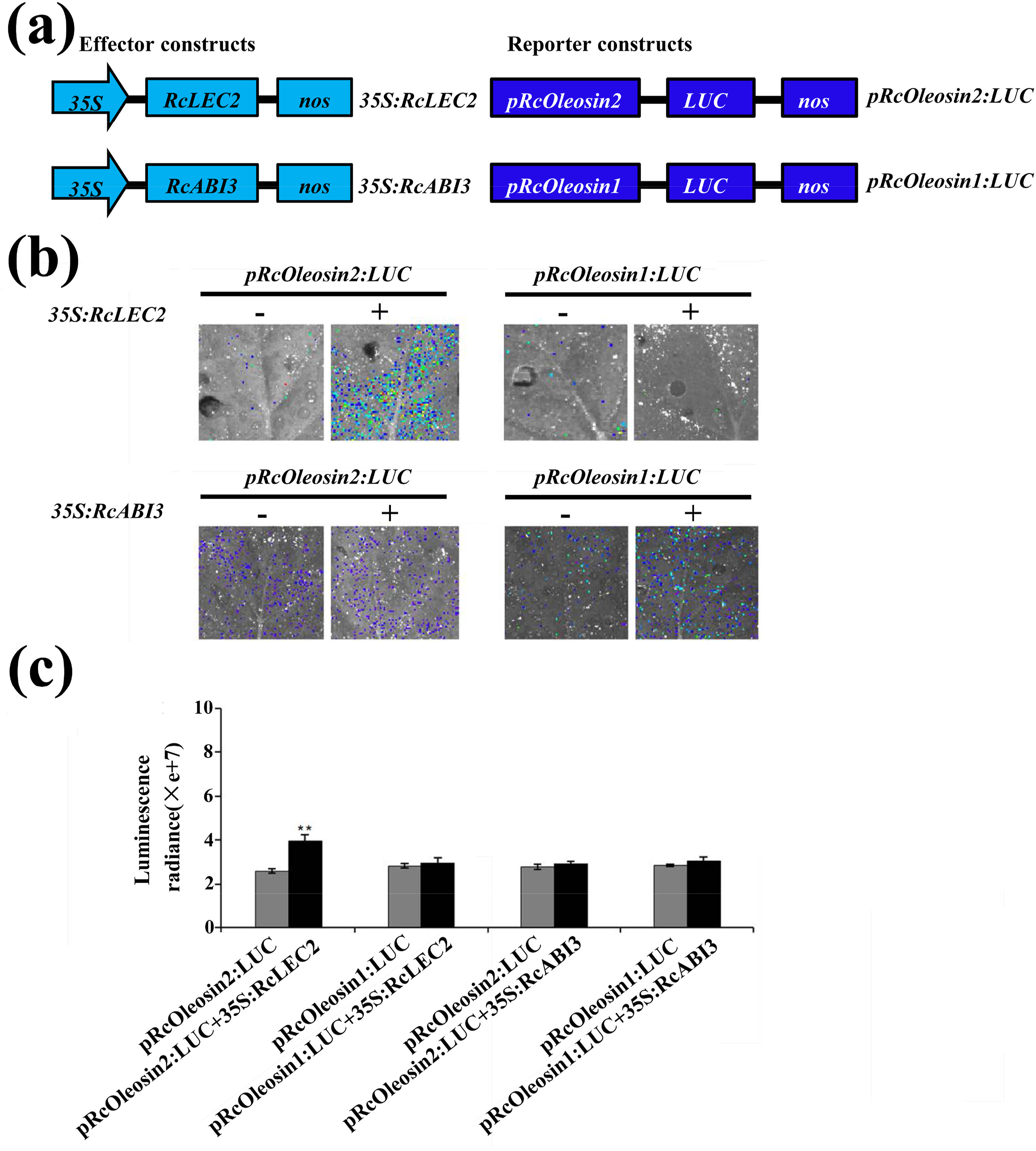
|
| Fig. 6 Interaction analysis of RcLEC2 and RcABI3 with putative target oil genes RcOleosin1 and RcOleosin2. (a) Schematic diagrams of effector and reporter constructs used in Transient Tobacco Assay. (b) RcLEC2 and RcABI3 regulated the expression of target genes by transient expression in tobacco leaves. (c) Quantitative analysis of luminescence intensity for each comparison in (b). For each column, bars indicate SD (n ≥ 3). Double asterisks denote a significant difference from the control groups (P < 0.01). Luminescence intensity was quantized by calculating the radiance of same-sized areas on each tobacco leaf. |
To further determine the interaction between the promoter of RcOleosin2 and RcLEC2 in yeast, four tandem copies of the Sph/RY element (designed as E) with flanking sequences of RcOleosin2 promoter were cloned into the pAbAi vector (Fig. 7, Table S7). When transformed together with pGADT7-RcLEC2, the yeast transformed with mutant Sph/RY sequences (designed as Em) and negative control p53-AbAi were suppressed by 200 ng/ml Aureobasidin A (AbA), whereas the yeast transformed with pRcOleosin2-E-AbAi and positive control pGADT7-P53/p53-AbAi both grew well in medium containing AbA (Fig. 7). These results indicate that RcLEC2 bound to the Sph/RY element of the RcOleosin2 promoter in yeast cells.
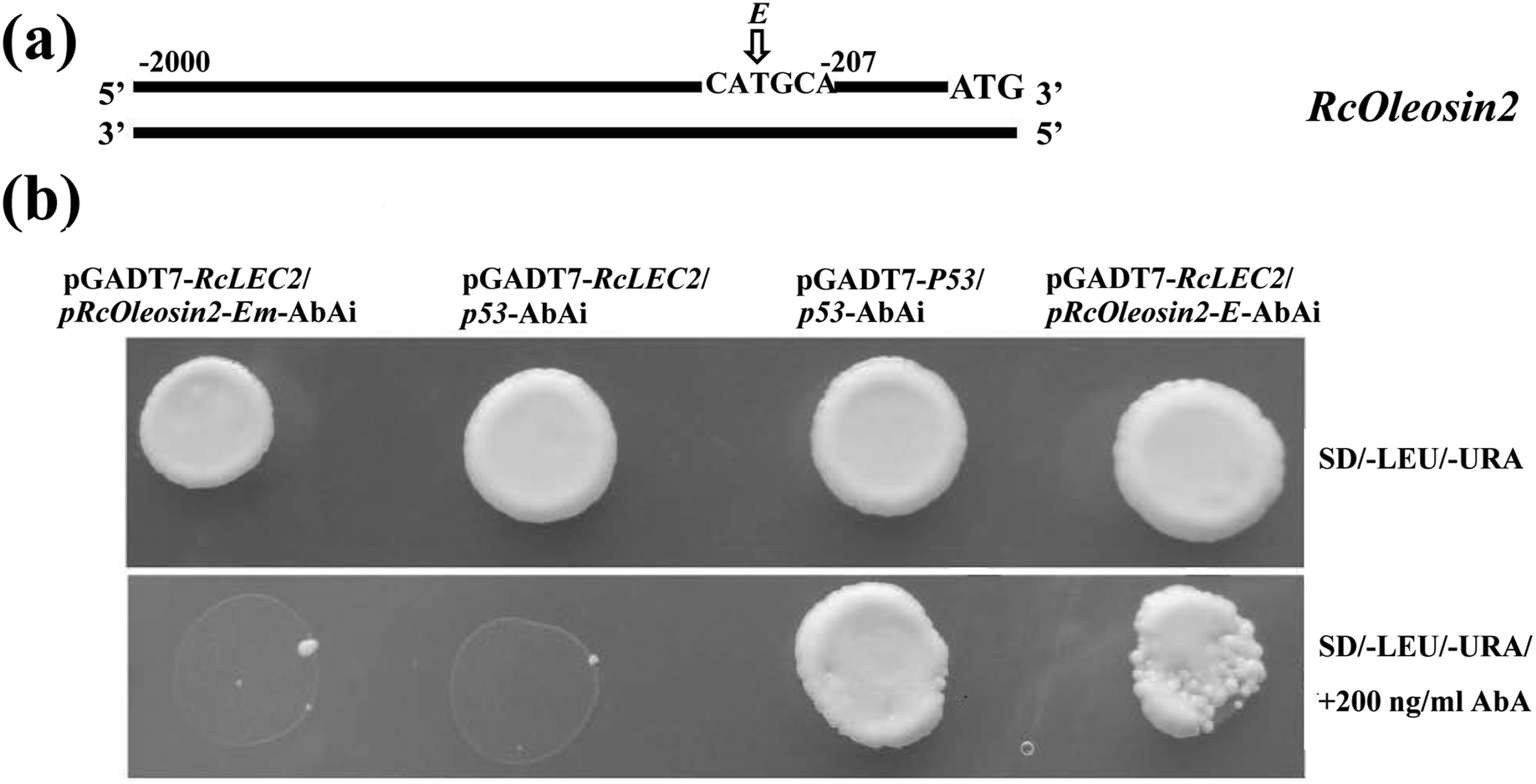
|
| Fig. 7 RcLEC2 bound to the promoter of RcOleosin2. (a) Diagram of RcOleosin2 promoterregion. The Sph/RY element of RcOleosin2 promoter designed as E. (b) RcLEC2 bound to the Sph/RY element of RcOleosin2 promoter. Four tandem copies of elements E and mutant Em with flanking sequences of RcOleosin2 promoter were cloned into pAbAi vector and these plasmids were co-transfected into yeast Y1HGold cells with pGADT7-RcLEC2. Detailed sequences are shown in . Yeast was grown in SD–Ura–Leu medium and then spotted on SD–Ura–Leu medium in the presence or absence of 200 ng/ml AbA. The pGADT7-RcLEC2/p53-AbAi and pGADT7-P53/p53-AbAi were used as negative and active control, respectively. Growth of transfected yeast cells on AbA medium indicates binding of RcLEC2 to the corresponding elements. |
The B3 family has been widely studied in diverse plants because of its extensive role in plant physiology (Swaminathan et al., 2008). The present study is, however, the first to report the structure, expression and function of members of the B3 superfamily in castor bean. In this study, we used the castor bean genome database to identify 61 putative B3 proteins (Table S1). These B3 proteins identified were unambiguously classified into five subfamilies, consistent with those of the B3 superfamily in other plants. We grouped the 61 proteins into five subfamilies based on structural features and sequence similarity. In addition, structural analysis of the B3 TFs in castor bean showed that excluding REM subfamily, which exhibited diverse structural features, members of each B3 subfamily shared conserved B3 domain sequences, motif distribution, and intron/exon structures (Figs. 1 and 3). Our categorization of B3 genes in castor bean is similar to that in Arabidopsis and rice (Swaminathan et al., 2008; Romanel et al., 2009). Specifically, there are a high number of REM subfamily genes, a small number of ABI3/VP1 and HSI subfamily genes, and the number of ARF and RAV subfamily members is in the middle of the range. Excluding four members of the REM subfamily, our phylogenetic analysis confirmed our categorization of B3 members based on structural features (Fig. 2). Phylogenetic analysis combining castor bean and Arabidopsis B3 proteins showed that for ABI3/VP1, HSI, RAV, and ARF subfamily, all major clades and subclades were clustered by interspecies members, indicating that the B3 genes belong to these four subfamilies are homologous between castor bean and Arabidopsis (Fig. S2). This is consistent with previous studies that found members of rice and Arabidopsis ABI3/VP1, HSI, RAV, and ARF subfamilies are closely related (Swaminathan et al., 2008).
Previous studies have shown that most members of the ABI3/VP1, HSI, RAV and ARF subfamilies are structurally conserved among angiosperm species (e.g., rice, poplar and Arabidopsis) (Romanel et al., 2009). Our research suggests that these subfamilies (ABI3/VP1, HSI, RAV, and ARF) might have an ancient origin in angiosperm evolution. We also found that members of the REM subfamily had divergent relationships and varied structurally in castor bean, which is consistent with findings in other angiosperms (e.g., rice, poplar and Arabidopsis) (Romanel et al., 2009), and indicates that REM genes have undergone divergent evolution. One remaining puzzle is how B3 domains from distinct subfamilies share a common structural framework yet bind to different DNA sites (Yamasaki et al., 2004; Waltner et al., 2005). Previous studies have indicated that different amino acids are conserved at DNA-contacting residues in distinct B3 subfamilies in Arabidopsis (Swaminathan et al., 2008). This may be in part responsible for the different DNA binding sites recognized by different B3 subfamilies. This finding also suggests that variation in the B3 domain may explain why members of the REM subfamily have fewer specific contacts with DNA. Alignment of B3 domain sequences of castor bean, with a special focus on those residues thought to contact the regulatory regions of the AtRAV1 protein, revealed that the amino acids in each B3 subfamily are conserved, except for members of the REM subfamily, in which residues that contact DNA varied greatly (Fig. 1). This finding supports and extends previous reports in Arabidopsis (Swaminathan et al., 2008). Specifically, we found that the sequences of all three DNA contact regions in each B3 subfamily (except for REM subfamily) are conserved between castor bean and Arabidopsis, which suggests that the DNA-binding specificities in different B3 subfamilies might be consistent in angiosperms, or at least between castor bean and Arabidopsis.
This study is the first to identify the complete array of motifs in B3 TFs in castor. In addition to the B3 domain, four additional domains (Fig. S1) coexist in B3 proteins, including the AP2/EREBP domain (Xu et al., 2013), the Aux_resp domain (Ulmasov et al., 1999; Guilfoyle and Hagen, 2007), the Aux/IAA domain (Guilfoyle and Hagen, 2007), and the zf-CW domain (Tsukagoshi et al., 2005; He et al., 2010), which have been shown to be present in other plants and involved in regulation of diverse physiological processes. As in Arabidopsis, these extra B3 domains might allow these TFs to have multiple functions in castor bean. Generally, conserved motifs within TFs are related to transcriptional activity, nuclear localization, and protein–protein interactions (Xu et al., 2013). As a result, most conserved motifs display a group-specific distribution pattern in each B3 subfamily (Fig. 3, Table S2). Taken together, the different gene structures and different conserved amino acids in DNA-binding regions among each B3 subfamily strongly indicate that functional divergence exists among each B3 subfamily. Although little is known about the functions of these motifs, conserved motifs identified in this study apply a frame to search the motif constitution in B3 family among species, which might help to understand the functional diversification of B3 TFs among species.
Castor bean is an important non-edible oilseed crop and its oil is considered a unique feed-stock ideal for biodiesel production (Ogunniyi, 2006; Scholz and Silva, 2008). Understanding the molecular mechanisms of oil accumulation in castor bean would improve genetic breeding and accelerate the development of high-oil producing castor bean cultivars. Previous research has shown that Arabidopsis LEC2 and ABI3 are involved in seed development and maturation (Swaminathan et al., 2008). Our expression analysis of castor bean showed that most B3 genes display tissue-specific expression patterns (Fig. 4, Table S3), indicating tissue-specific functions. For example, the three members of ABI/VP1 subfamily (RcLEC2, RcABI3 and RcFUS3) were specifically expressed in seed stages, including in the embryo and endosperm, which implies that ABI/VP1 genes may play roles during embryo and endosperm development. To explore the mechanisms of transcriptional regulation of the ABI/VP1 subfamily in castor bean, we screened RNA-seq data and found 26 oil genes (Fig. 5, Table S4) whose promoter regions contained at least one Sph/RY cis-acting element recognized by ABI/VP1 subfamily proteins (Andrianov et al., 2010). Expression profiles indicated that several oil genes were highly expressed in seed oil fast accumulation stages (seed3–seed5), especially two core genes, RcOleosin1 and RcOleosin2, suggesting the possible relationship between these genes and ABI/VP1 subfamily in castor bean.
Furthermore, our gene co-expression module revealed that RcOleosin1 and RcOleosin2 were highly positively associated with ABI/VP1 subfamily genes in castor bean (Fig. 5, Table S5). Notably, these two oleosin genes were both corelated with RcLEC2 and RcABI3. In Arabidopsis, ABI3 has been shown to play a role in seed maturation and regulation of oleosin gene expression (Crowe et al., 2000; Mönke et al., 2012). In addition, LEC2 has been reported to be extensively involved in oil deposition and accumulation (Braybrook et al., 2006; Stone et al., 2008; Angeles-Núñez and Tiessen, 2011; Kim et al., 2013), which strongly supports our hypothesis that ABI/VP1 subfamily genes are likely the activators of lipid biosynthesis in castor bean. Further examination indicated that RcLEC2 can directly target the promoter of RcOleosin2 but not RcOleosin1 through the Sph/RY element (Fig. 6, Fig. 7), which indicates that RcLEC2 might be a key active regulator of seed oil biosynthesis in castor bean. However, RcABI3 does not bind the promoters of either RcOleosin1 or RcOleosin2. These results suggest that the mechanisms by which different ABI3/VP1 subfamily members regulate seed oil accumulation may be distinct and require further study.
This study identified the domains, motifs, structures and tissue-specific expression patterns of 61 B3 TFs in the non-edible oilseed crop castor bean. In addition, we demonstrated that one member of the ABI3/VP1 subfamily, RcLEC2, might play important roles in seed development and oil accumulation through direct positive regulation of the transcription of the oil-body structural gene RcOleosin2. This study provides an important foundation and novel insights that will benefit further studies into the mechanisms by which B3 TFs regulate seed development and material accumulation in castor bean. However, elucidating the exact regulatory mechanisms of B3 TFs, including ABI3/VP1 subfamily, in these physiological processes should be more fully investigated in the future.
Author contributionsWBW and TA carried out sequence data analyses and experiments. YYZ, DW, BH and WX contributed to sample collection and assisted with data analyses. WBW and TA wrote the paper. AZL designed and managed the experiments, and organized the manuscript. All authors read and approved the final version of the manuscript.
Declaration of competing interestThe authors declare that they have no competing interests.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.06.008.
AcknowledgementsAuthors gave many thanks to Zibo Academy of Agricultural Sciences, Shandong, China for providing the seeds of castor bean var. ZB306 elite inbred line. This work was supported by National Natural Science Foundation of China (31661143002, 81760507, 31571709, 31771839, 31701123 and 31501034), Yunnan Applied Basic Research Projects (2016FA011, 2016FB060 and 2016FB040), the National R&D Infrastructure and Facility development Program of China “Fundamental Science Data Sharing Platform (DKA 2017-12-02-16) and the 13th Five-year informatization Plan of Chinese Academy of Sciences (No. XXH13506).
Akpan, U., Jimoh, A., Mohammed, A., 2006. Extraction, characterization and modification of castor seed oil. Leonardo J. Sci., 8: 43-52. |
Andrianov, V., Borisjuk, N., Pogrebnyak, N., et al., 2010. Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J., 8: 277-287. DOI:10.1111/j.1467-7652.2009.00458.x |
Angeles-Núñez, J.G., Tiessen, A., 2011. Mutation of the transcription factor LEAFY COTYLEDON2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol., 168: 1891-1900. DOI:10.1016/j.jplph.2011.05.003 |
Baud, S., Mendoza, M.S., To, A., et al., 2007. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J., 50: 825-838. DOI:10.1111/j.1365-313X.2007.03092.x |
Braybrook, S.A., Stone, S.L., Park, S., et al., 2006. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A., 103: 3468-3473. DOI:10.1073/pnas.0511331103 |
Brown, A.P., Kroon, J.T., Swarbreck, D., et al., 2012. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One, 7: e30100. DOI:10.1371/journal.pone.0030100 |
Chan, A.P., Crabtree, J., Zhao, Q., et al., 2010. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol., 28: 951-956. DOI:10.1038/nbt.1674 |
Crowe, A.J., Abenes, M., Plant, A., et al., 2000. The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci., 151: 171-181. DOI:10.1016/S0168-9452(99)00214-9 |
Edgar, R.C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res., 32: 1792-1797. DOI:10.1093/nar/gkh340 |
Feng, C.Z., Chen, Y., Wang, C., et al., 2014. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J., 80: 654-668. DOI:10.1111/tpj.12670 |
Franco-Zorrilla, J.M., Cubas, P., Jarillo, J.A., et al., 2002. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiol., 128: 418-427. DOI:10.1104/pp.010323 |
Giraudat, J., Hauge, B.M., Valon, C., et al., 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell, 4: 1251-1261. |
Guerriero, G., Martin, N., Golovko, A., et al., 2009. The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth. New Phytol., 184: 552-565. DOI:10.1111/j.1469-8137.2009.02977.x |
Guilfoyle, T.J., Hagen, G., 2007. Auxin response factors. Curr. Opin. Plant Biol., 10: 453-460. DOI:10.1016/j.pbi.2007.08.014 |
Guo, X., Hou, X., Fang, J., et al., 2013. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J., 75: 403-416. DOI:10.1111/tpj.12209 |
He, F., Umehara, T., Saito, K., et al., 2010. Structural insight into the zinc finger CW domain as a histone modification reader. Structure, 18: 1127-1139. DOI:10.1016/j.str.2010.06.012 |
Hu, Y.X., Wang, Y.H., Liu, X.F., et al., 2004. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res., 14: 8-15. DOI:10.1038/sj.cr.7290197 |
Kagaya, Y., Ohmiya, K., Hattori, T., 1999. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res., 27: 470-478. DOI:10.1093/nar/27.2.470 |
Keith, K., Kraml, M., Dengler, N.G., et al., 1994. fusca 3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell, 6: 589-600. DOI:10.2307/3869865 |
Kim, H.U., Jung, S.J., Lee, K.R., et al., 2013. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio, 4: 25-32. |
Letunic, I., Doerks, T., Bork, P., 2012. Smart 7: recent updates to the protein domain annotation resource. Nucleic Acids Res., 40: D302-D305. DOI:10.1093/nar/gkr931 |
Levy, Y.Y., Mesnage, S., Mylne, J.S., et al., 2002. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science, 297: 243-246. DOI:10.1126/science.1072147 |
Liu, L., Zhang, Y., Tang, S., et al., 2010. An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J., 61: 893-903. DOI:10.1111/j.1365-313X.2009.04109.x |
Matias-Hernandez, L., Battaglia, R., Galbiati, F., et al., 2010. VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell, 22: 1702-1715. DOI:10.1105/tpc.109.068627 |
McCarty, D.R., Carson, C.B., Stinard, P.S., et al., 1989. Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell, 1: 523-532. DOI:10.2307/3868973 |
Mönke, G., Seifert, M., Keilwagen, J., et al., 2012. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res., 40: 8240-8254. DOI:10.1093/nar/gks594 |
Ogunniyi, D.S., 2006. Castor oil: a vital industrial raw material. Bioresour. Technol., 97: 1086-1091. DOI:10.1016/j.biortech.2005.03.028 |
Okushima, Y., Mitina, I., Quach, H.L., et al., 2005. Auxin response factor 2 (ARF2): a pleiotropic developmental regulator. Plant J., 43: 29-46. DOI:10.1111/j.1365-313X.2005.02426.x |
Okushima, Y., Overvoorde, P.J., Arima, K., et al., 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell, 17: 444-463. DOI:10.1105/tpc.104.028316 |
Pekker, I., Alvarez, J.P., Eshed, Y., 2005. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell, 17: 2899-2910. DOI:10.1105/tpc.105.034876 |
Peng, F.Y., Weselake, R.J., 2013. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet., 126: 1305-1319. DOI:10.1007/s00122-013-2054-4 |
Punta, M., Coggill, P.C., Eberhardt, R.Y., et al., 2011. The Pfam protein families database. Nucleic Acids Res., 40: D290-D301. |
Qiu, L., Yang, C., Tian, B., et al., 2010. Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol., 10: 278. DOI:10.1186/1471-2229-10-278 |
Romanel, E.A., Schrago, C.G., Counago, R.M., et al., 2009. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One, 4: e5791. DOI:10.1371/journal.pone.0005791 |
Scholz, V., Silva, J.N.D., 2008. Prospects and risks of the use of castor oil as a fuel. Biomass Bioenerg., 32: 95-100. DOI:10.1016/j.biombioe.2007.08.004 |
Stone, S.L., Braybrook, S.A., Paula, S.L., et al., 2008. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A., 105: 3151-3156. DOI:10.1073/pnas.0712364105 |
Stone, S.L., Kwong, L.W., Yee, K.M., et al., 2001. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A., 98: 11806-11811. DOI:10.1073/pnas.201413498 |
Suzuki, M., Kao, C.Y., McCarty, D.R., 1997. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell, 9: 799-807. |
Suzuki, M., Wang, H.H., McCarty, D.R., 2007. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol., 143: 902-911. DOI:10.1104/pp.106.092320 |
Swaminathan, K., Peterson, K., Jack, T., 2008. The plant B3 superfamily. Trends Plant Sci., 13: 647-655. DOI:10.1016/j.tplants.2008.09.006 |
Tamura, K., Peterson, D., Peterson, N., et al., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28: 2731-2739. DOI:10.1093/molbev/msr121 |
Tsuchiya, Y., Nambara, E., Naito, S., et al., 2004. The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J., 37: 73-81. DOI:10.1046/j.1365-313X.2003.01939.x |
Tsukagoshi, H., Saijo, T., Shibata, D., et al., 2005. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol., 138: 675-685. DOI:10.1104/pp.104.057752 |
Ulmasov, T., Hagen, G., Guilfoyle, T.J., 1999. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U.S.A., 96: 5844-5849. DOI:10.1073/pnas.96.10.5844 |
Ulmasov, T., Murfett, J., Hagen, G., et al., 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell, 9: 1963-1971. |
Waltner, J.K., Peterson, F.C., Lytle, B.L., et al., 2005. Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Sci., 14: 2478-2483. DOI:10.1110/ps.051606305 |
Xiong, L., David, L., Stevenson, B., et al., 1999. High throughput screening of signal transduction mutants with luciferase imaging. Plant Mol. Biol. Rep., 17: 159-170. DOI:10.1023/A:1007519200505 |
Xu, W., Li, F., Ling, L., et al., 2013. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genom., 14: 785. DOI:10.1186/1471-2164-14-785 |
Yamamoto, A., Kagaya, Y., Usui, H., et al., 2010. Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol., 51: 2031-2046. DOI:10.1093/pcp/pcq162 |
Yamasaki, K., Kigawa, T., Inoue, M., et al., 2004. Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell, 16: 3448-3459. DOI:10.1105/tpc.104.026112 |
Zhang, Y., Clemens, A., Maximova, S.N., et al., 2014. The Theobroma cacao B3 domain transcription factor TcLEC2 plays a duel role in control of embryo development and maturation. BMC Plant Biol., 14: 106. DOI:10.1186/1471-2229-14-106 |



