Individuals or populations with similar phenotypes are generally classified as one species, which is the traditional taxonomic approach (Duminil and Di Michele, 2009). Populations with similar phenotypes but multiple origins would pose a challenge to this definition of a species. Nevertheless, sharp changes in the plant environment that occur over short distances (e.g., changes in soil type, moisture and temperature) may cause individual species to adapt to environmental changes by adaptive genetic divergence (Hoekstra and Coyne, 2007). Local adaptation across an environmental gradient, either with or without gene flow, may be the first step that ultimately leads to the origin of fully reproductively isolated forms (i.e., biological species concept) (Mayr, 1942; Abbott, 2017). Therefore, investigating how certain traits originate and evolve in response to environmental changes and whether adaptive genetic divergence is impaired by gene flow would provide new insights into the delimitation of species with complicated evolutionary histories.
Recently, Aquilegia L. (also known as columbine) has emerged as a model system for floral evolution due to its unusual floral morphology (Kramer, 2009; Kramer and Hodges, 2010). The genus Aquilegia represents a recent adaptive radiation, with a Eurasia origin followed by an expansion to North America over the last 1–3 million years (Fior et al., 2013). The radiation of Aquilegia species was driven by diversification in both pollinators and ecological habitats (Hodges and Arnold, 1995; Hodges, 1997; Hodges et al., 2003). Different lengths of nectar spur are associated with different pollinators in North America (Whittall and Hodges, 2007). Nevertheless, there is a spurless species, Aquilegia ecalcarata Maxim., that has a reduced nectar spur that makes it appear spurless. The extremely reduced spur (referred as the spurless status) and the loss of nectar in A. ecalcarata is unique in the genus and believed to be a derived character rather than ancestral state (Hodges and Arnold, 1995; Fior et al., 2013). A. ecalcarata together with Aquilegia yabeana Kitag., Aquilegia oxysepala Trautv. and C.A. Mey. var. kansuensis Brühl (treated as Aquilegia kansuensis by Erst et al., 2014) and Aquilegia rockii Munz. are clustered as a monophyletic clade (Fior et al., 2013). Interestingly, these four closely related species are all endemic to China and differ in spur length and degree of curvature (Xiao, 1979; Fu and Robinson, 2001). A. rockii has long but straight or slightly incurved spurs, while A. yabeana and A. kansuensis (=A. oxysepala var. kansuensis) both have long and hooked or coiled spurs. The petal blade is the same color as the sepals in A. yabeana and A. rockii, but different in A. kansuensis (Fig. S1). All three species have nectar tissue at the apex of their spurs. Given that A. ecalcarata harbors unique phenotypes in lacking both spurs and nectar tissue, we regard A. ecalcarata and its relatives (A. yabeana, A. kansuensis and A. rockii) as a good model for investigating floral evolution, species divergence and speciation at the infrageneric level. These four species also differ in habitat (Fig. S2). The different habitat makes the origin of A. ecalcarata more intriguing and may provide important information about how A. ecalcarata adapted to a new environment.
Although Fior et al. (2013) constructed the phylogeny of the four closely related species, only one individual for each species was sampled; thus, how the spurless status originated remains unknown. Our previous research indicated that Aquilegia ecalcarata shows significant genetic divergence among populations (Huang et al., 2018). Morphological studies have suggested that the floral traits (such as the length of sepals, spurs and carpels) differ significantly among 19 sampled populations of A. ecalcarata (Xue et al., 2020). Therefore, we propose that spur loss in Aquilegia, as well as in A. ecalcarata, had independent origins. However, researchers have shown that gene flow has been a common feature throughout Aquilegia (Filiault et al., 2018). Therefore, the possibility of hybridization between A. ecalcarata and its closest relatives may not be excluded. Further analyses, such as gene flow estimation, is required to further clarify how these evolutionary forces may have impacted the origin of A. ecalcarata. Moreover, continuous variations in both vegetative and floral traits have been observed in some populations among A. yabeana, A. kansuensis and A. rockii in the field. Whether introgression occurred in sympatric areas among the three species deserves more investigation. Here we use both nuclear and chloroplast gene fragments to determine whether hybridization affected the evolutionary origin of A. ecalcarata, and to characterize the genetic differentiation and gene flow among A. ecalcarata and its relatives. This study will enhance our understanding of how floral evolution, especially the diversity of nectar spurs, contributes to parallel speciation in closely related species.
2. Materials and methods 2.1. Taxon sampling and lociIn this study, we sampled a total of 634 individuals (Fig. 1 and Table 1). We sampled nineteen populations of Aquilegia ecalcarata, nine of A. kansuensis, six of A. rockii and eight of A. yabeana. Fifteen and five individuals were sampled from each population for nuclear gene fragment and chloroplast locus sequencing, respectively. For outgroups, we sampled four individuals each from Aquilegia canadensis L., A. chrysantha A. Gray, A. atrovinosa Popov ex Gamojun. and A. oxysepala.
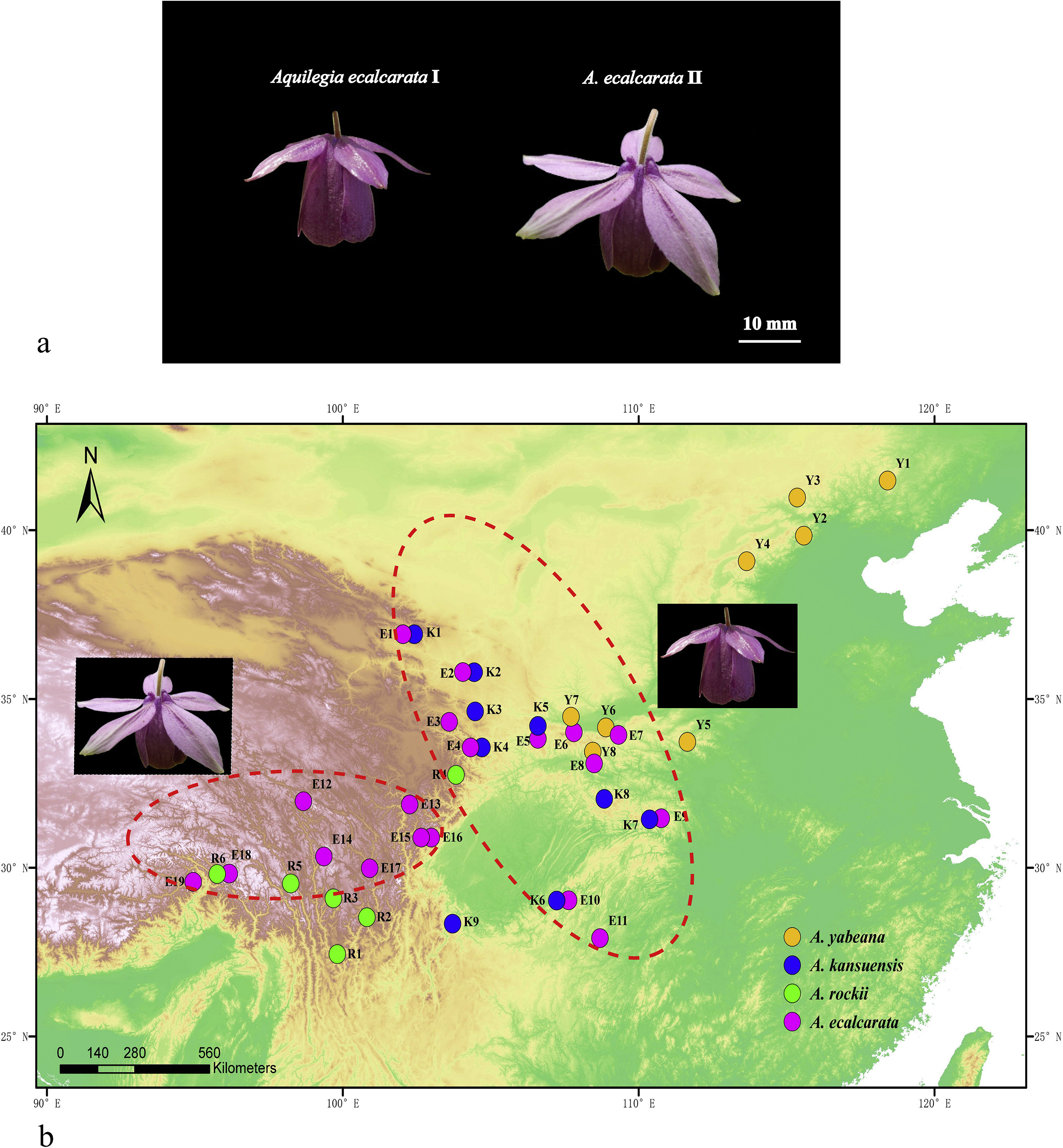
|
| Fig. 1 The phenotypic comparison between Aquilegia ecalcarata I and II (a) and the geographicaldistribution of 42 sampled populations (b), with red dash circles for A. ecalcarata I and II. |
| Taxon | Population name | Sample size | Locality | Latitude (N) | Longitude (E) | Altitude (m) |
| Aquilegia yabeana | Y1 | 15 | Ningcheng, Inner Mongolia | 41°27′ | 118°24′ | 1128 |
| Y2 | 15 | Mentougou, Beijing | 39°55′ | 115°26′ | 1550 | |
| Y3 | 15 | Chongli, Hebei | 40°58′ | 115°21′ | 1720 | |
| Y4 | 15 | Fanshi, Shanxi | 39°05′ | 113°38′ | 2125 | |
| Y5 | 15 | Luanchuan, Henan | 33°43′ | 111°38′ | 1789 | |
| Y6 | 15 | Huxian, Shaanxi | 34°09′ | 108°53′ | 1800 | |
| Y7 | 15 | Meixian, Shaanxi | 34°05′ | 107°42′ | 1182 | |
| Y8 | 15 | Ningshan, Shaanxi | 33°26′ | 108°26′ | 1582 | |
| A.kansuensis | K1 | 15 | Huzhu, Qinghai | 36°55′ | 102°24′ | 2517 |
| K2 | 15 | Yuzhong, Gansu | 35°47′ | 104°03′ | 2561 | |
| K3 | 15 | Zhangxian, Gansu | 34°38′ | 104°28′ | 2451 | |
| K4 | 15 | Zhouqu, Gansu | 33°33′ | 104°19′ | 2705 | |
| K5 | 15 | Fengxian, Shaanxi | 34°11′ | 106°35′ | 1578 | |
| K6 | 15 | Nanchuan, Chongqing | 29°01′ | 107°13′ | 1953 | |
| K7 | 15 | Xingshan, Hubei | 31°26′ | 110°21′ | 1608 | |
| K8 | 15 | Chengkou, Chongqing | 32°02′ | 108°50′ | 2245 | |
| K9 | 15 | Leibo, Sichuan | 28°20′ | 103°42′ | 1822 | |
| A.rockii | R1 | 15 | Shangarila, Yunan | 27°26′ | 099°48′ | 2934 |
| R2 | 15 | Muli, Sichuan | 28°31′ | 100°48′ | 3263 | |
| R3 | 15 | Xiangcheng, Sichuan | 29°05′ | 099°40′ | 3412 | |
| R4 | 15 | Songpan, Sichuan | 32°45′ | 103°49′ | 3199 | |
| R5 | 15 | Mangkang, Tibet | 29°32′ | 098°14′ | 3489 | |
| R6 | 15 | Bomi, Tibet | 29°48′ | 095°44′ | 3253 | |
| A.ecalcarata | E1 | 15 | Huzhu, Qinghai | 36°54′ | 102°24′ | 2583 |
| E2 | 15 | Yuzhong, Gansu | 35°47′ | 104°3′ | 2349 | |
| E3 | 15 | Zhuoni, Gansu | 34°19′ | 103°35′ | 2877 | |
| E4 | 15 | Zhouqu, Gansu | 33°33′ | 104°18′ | 2770 | |
| E5 | 15 | Fengxian, Shaanxi | 34°12′ | 106°35 | 1632 | |
| E6 | 15 | Meixian, Shaanxi | 34°00′ | 107°48′ | 2770 | |
| E7 | 15 | Huxian, Shaanxi | 33°49′ | 108°36′ | 2439 | |
| E8 | 15 | Ningshan, Shaanxi | 33°28′ | 108°29′ | 2167 | |
| E9 | 15 | Xingshan, Hubei | 31°27′ | 110°16′ | 2580 | |
| E10 | 15 | Nanchuan, Chongqing | 29°02′ | 107°11′ | 2121 | |
| E11 | 15 | Jiangkou, Guizhou | 27°54′ | 108°41′ | 2426 | |
| E12 | 15 | Dege, Sichuan | 31°57′ | 98°39′ | 3532 | |
| E13 | 15 | Barkam, Sichuan | 31°52′ | 102°37′ | 3193 | |
| E14 | 15 | Batang, Sichuan | 30°19′ | 099°21′ | 3470 | |
| E15 | 15 | Xiaojin, Sichuan | 30°53′ | 102°38′ | 3268 | |
| E16 | 15 | Wenchuan, Sichuan | 30°53′ | 102°59′ | 2646 | |
| E17 | 15 | Yajiang, Sichuan | 29°59′ | 100°54′ | 3545 | |
| E18 | 15 | Bomi, Tibet | 29°49′ | 095°42′ | 3211 | |
| E19 | 15 | Milin, Tibet | 29°35′ | 094°56′ | 3282 |
We used three chloroplast loci (rpl32-trnL, trnK-rps 16, rps16-trnQ), following previous research (Fior et al., 2013). For nuclear gene fragments, we developed markers based on the genome of Aquilegia coerulea E. James in the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal). In addition to six markers used in our previous work (Huang et al., 2018), we selected an additional nine gene fragments; these fifteen loci cover seven chromosomes. We screened genes of lengths from 2 kb to 5 kb that contained both exons and introns. Then we blasted the candidate genes against the database to ensure they only mapped to one genome position. Details are shown in Fig. S3 and Table S1. Total DNA was extracted from 1 g of silica gel dried leaf material using a CTAB protocol (Zhu et al., 2007).
2.2. Amplification, sequencing and sequence analysisPCR amplification followed a protocol described in our previous study (Zhang and Ge, 2007). Sequencing was done on an ABI3730XL automatic sequencer (Applied Biosystems Corp.). If dual peaks were found, PCR fragments were cloned into pGEM T-easy vectors (Promega Corp.) with a Pharmacia purification kit (Amersham Pharmacia Biotech) and three cloned DNA fragments were sequenced for each individual. To correct for errors in cloned fragments, we identified individuals in the alignments that contained singletons and then re-sequenced at least four clones after a second round of PCR. All sequences have been deposited in GenBank, with the accession numbers KY582911- KY582935, MG710812-MG711310, MH238358-MH238411, MH720548-MH720938, MH766660-MH766870, and MH788640-MH788921. Sequences were assembled with ContigExpress (Informax Inc., North Bethesda, MD), aligned with Clustal X1.83 (Thompson et al., 1997) and further refined with BioEdit 7.0.9.0 (Hall, 1999).
2.3. Genetic diversity and neutrality testFor each locus, we calculated the number of segregating sites (S), number of haplotypes (h), pairwise differences π (Nei, 1987) and Watterson's θ (Watterson, 1975). We also estimated the minimum number of recombination events (Rm) with the four-gamete test (Hudson and Kaplan, 1985). Tajima's D (Tajima, 1989) and D∗ and F∗ (Fu and Li, 1993) were calculated to test the neutrality of each locus. We obtained the associated one-tailed test p-value for Tajima's D and Fu and Li's D∗ and F∗ by computing 1000 coalescent simulations, accounting for estimates of the recombination per gene. The multilocus HKA tests (Hudson et al., 1987) across loci were conducted at the species level using Aquilegia canadensis, A. chrysantha, A. atrovinosa and A. oxysepala as outgroups. All analyses were performed in DnaSP 5.1 (Librado and Rozas, 2009).
2.4. Phylogenetic analysisThe genealogical trees of all alleles were constructed by Mega 7 (Kumar et al., 2016), using the Neighbor-Joining (NJ) method with Kimura's 2-parameter distances (Kimura, 1980). Maximum Parsimony (MP) and Maximum Likelihood (ML) methods were also performed and a heuristic search with tree bisection–reconnection, ACCTRAN and 1000 random taxon-addition replicates was implemented in these analyses. The optimal model of sequence evolution for each data set was determined using Akaike's information criterion (AIC) implemented in MODELEST 3.7 (Posada and Crandall, 2001). We conducted bootstrap analysis to assess the confidence of internal nodes with 1000 replicates. Bayesian posterior probability was calculated by MrBayes 3.2 (Huelsenbeck and Ronquist, 2001). The incongruence length difference (ILD) test in PAUP 4.0 was used to evaluate whether it is appropriate to combine chloroplast and nuclear data to construct phylogenetic trees (Farris et al., 1994).
2.5. Divergence time estimationBEAST 2.3.2 was used to estimate the divergence time under a strict molecular clock model (Bouckaert et al., 2014). We set the nucleotide substitution model as GTR + I + G, and the chain-length of Markov chain Monte Carlo (MCMC) and sampling frequency to 2,000,000 and 2000, respectively. Tree prior was specified as a Yule process. Previous work indicated that Aquilegia diverged from other clades of Ranunculaceae 4.76 million years ago (Ma); the outgroups used in this study diverged from genus Aquilegia 3.88 Ma; and the four closely related species (A. ecalcarata, A. kansuensis, A. rockii and A. yabeana) diverged from the outgroups 2.79 Ma (Fior et al., 2013). These three time-scales were set as time constraints. The convergence was assessed by effective sample sizes (ESS) using Tracer v.1.5 (http://tree.bio.ed.ac.uk/software/tracer/). Finally, the divergence times were estimated by TreeAnnotator v.1.6.1 with half of the trees treated as burn-in and the divergence times visualized using FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
2.6. Population genetic structureThe overall distribution of nucleotide diversity among populations was investigated using an Analysis of Molecular Variance (AMOVA) in Arlequin 3.01. Sequence variation was hierarchically partitioned between two species, between populations, within species, and within populations. The significance of all estimated fixation indices was tested using 10,000 permutations as described in Excoffier et al. (1992). Pairwise FST was used to measure population differentiation within and between species. We performed a Bayesian clustering analysis in STRUCTURE 2.2.3 (Pritchard et al., 2000) and principal coordinate analysis (PCoA) using DARwin (Perrier and Jacquemond-Collet, 2006). For the former analysis, the program was run with SNP markers for K-values from 2 to 10, with 50,000 burn-in iterations followed by 300,000 MCMC iterations for accurate parameter estimates. To verify the consistency of the results, we performed 10 independent runs for each k using an admixture model with correlated allele frequencies. Estimation of the uppermost hierarchical level of the genetic structure was accomplished using a statistic ΔK following the procedure described in previous methods (Evanno et al., 2005).
2.7. Fitting an isolation-with-migration modelIMa 2 is based on an isolation-with-migration model and estimates effective population sizes (present and ancestral), splitting times, and population migration rates using MCMC simulations (Hey, 2010). As the isolation-with-migration model assumes no recombination within markers (Hey and Nielsen, 2004), we conducted recombination rate tests of fifteen nuclear loci using IMgc (http://hammerlab.biosci.arizona.edu/imgc_online.html) (Woerner et al., 2007). By discarding the recombination sections in each locus, the nonrecombining sections of the fifteen gene fragments (in total 8339 bp) were used for IMa2 calculation. After a burn-in of 107 generations and 2,000,000 steps, 20,000 genealogies were saved. The analysis was done with four independent runs with different random seeds. Convergence was assessed based on ESS >50, stable parameter trend plots, and similar parameter estimates from the first and the second half of the runs. We estimated the mutation rate of each locus based on a modified method (Tenaillon et al., 2004). The geometric average mutation rate of the fifteen gene fragments (2 × 10−5) was used to convert the effective population size (θ) to effective number of individuals (Ne). In “L mode” of IMa2, marginal posterior probability density estimates and LLR tests were used for assessing whether migration rates were significantly different from zero.
2.8. Re-examination of morphological dataXue et al. (2020) reported a detailed morphological investigation (i.e., 41 vegetative and floral traits) on the 42 populations of Aquilegia ecalcarata. Here we re-examined these data and compared what we refer to as A. ecalcarata I and II to demonstrate their morphological divergence. Statistical methods followed descriptions in Xue et al. (2020).
3. Results 3.1. Genetic diversity and neutrality testThe concatenated lengths of the fifteen nuclear gene fragments were 11,083 bp and the aligned sequence lengths ranged from 620 to 870 bp (739 bp on average). A total of 1880 segregating sites were detected (524 in Aquilegia ecalcarata, 297 in A. kansuensis, 259 in A. rockii and 520 in A. yabeana), and a total of 363 haplotypes were found across all individuals (Tables S2 and S3). For the chloroplast loci (rpl32-trnL, trnK-rps 16, rps16-trnQ), the concatenated lengths were 1809 bp. We detected a total of 41 segregating sites and 37 haplotypes (Hd = 0.946) in all 210 individuals.
We calculated πsil and θsil on combined nuclear sequence data to evaluate the level of genetic diversity among the four species. At the species level, A. yabeana possessed the highest genetic diversity (πsil 0.0114 and θsil 0.0141), whereas A. kansuensis had the lowest (πsil 0.0070 and θsil 0.0078). A. ecalcarata (πsil 0.0067/θsil 0.0135) and A. rockii (πsil 0.0074/θsil 0.0073) had a genetic diversity at intermediate levels. Notably, A. ecalcarata conserved more segregating sites (θsil 0.0135) than either A. kansuensis (θsil 0.0078) or A. rockii (θsil 0.0073).
We performed three neutrality tests (Tajima's D, Fu and Li's D, Fu and Li's F) in each species: the majority of the results showed non-significant values in each test, except for certain species in which significant values were shown for Lap 3 (A. ecalcarata), Cpt (A. ecalcarata), Bst 10 (A. rockii), Cer (A. ecalcarata and A. yabeana). We then conducted a multilocus HKA test to ensure all loci were neutral. The χ2 value of A. ecalcarata/outgroups was 0.112 (p = 0.738), A. kansuensis/outgroups was χ2 = 0.169 (p = 0.681), A. rockii/outgroups was χ2 = 0.047 (p = 0.829), and A. yabeana/outgroups was χ2 = 0.005 (p = 0.945). No significant values were found by the multilocus HKA test, implying that all nuclear sequence data were evolved neutrally and are appropriate for analyses.
3.2. Phylogeny with nuclear and chloroplast dataNeighbor-Joining, Maximum Parsimony, Maximum Likelihood and Bayesian methods were used to construct the genealogies of 42 populations of four species. Phylogenetic trees using these four methods with chloroplast sequence data showed similar topologies (Fig. 2a). The four outgroups were positioned as a sister clade; the ingroup populations were split into two clades, each with high bootstrap values (100) and Bayesian posterior probabilities. Interestingly, Aquilegia ecalcarata could be found in both clades and clustered with different species. Of the nineteen A. ecalcarata populations, seven (E5–E11) clustered with all A. yabeana populations and three A. kansuensis populations, whereas the other twelve clustered with all A. rockii populations and six A. kansuensis populations.
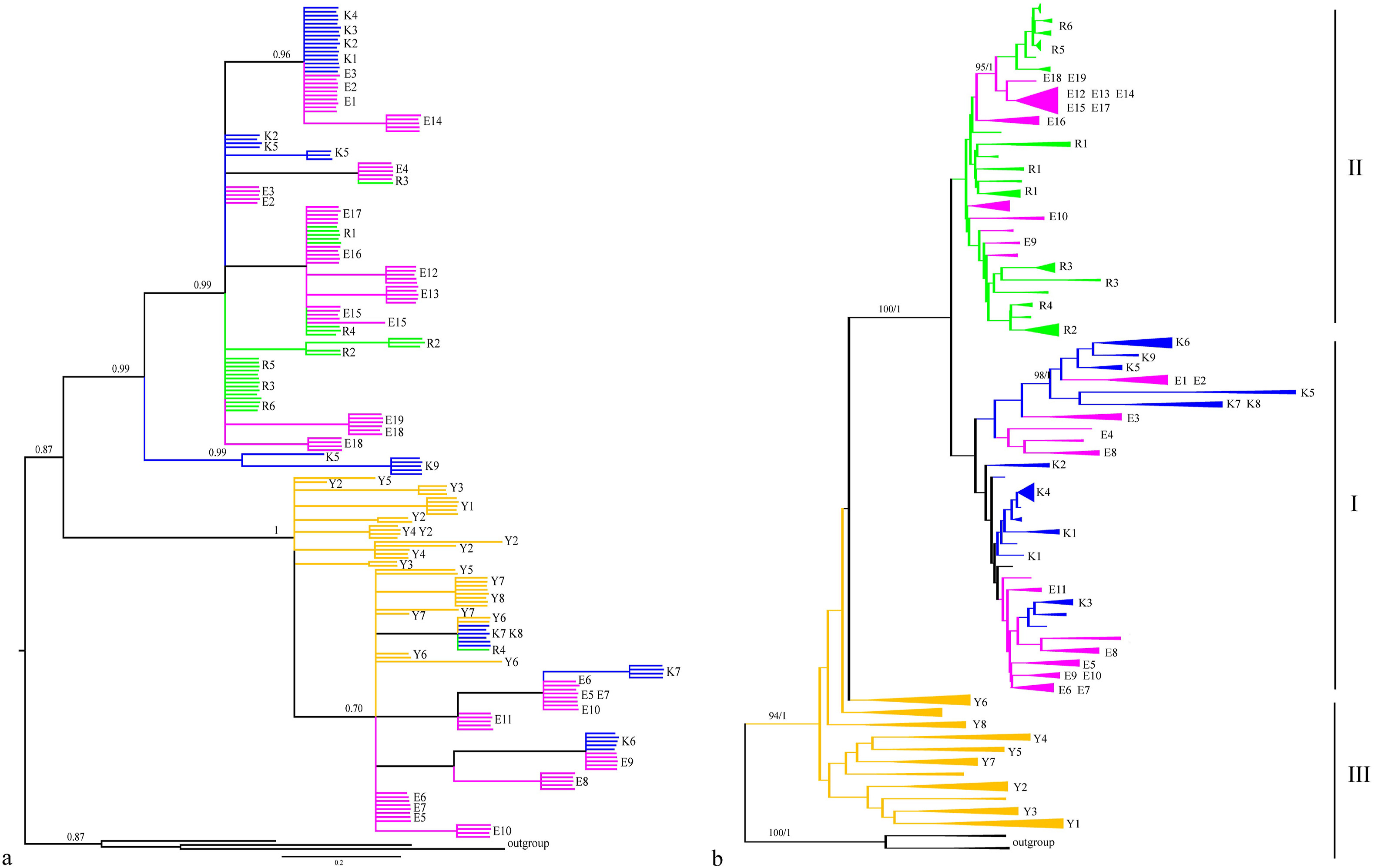
|
| Fig. 2 Phylogenetic trees of all 42 populations based on three combined chloroplast gene fragments (a); all populations except partial individuals from K9, R2, R4, R5 based on the combined nuclear gene fragments (b); Numbers along branches represent bootstrap support (1000 replicates) and Bayesian posterior probability. Aquilegia yabeana (orange), A. kansuensis (blue), A. rockii (green) and A. ecalcarata (pink). |
For fifteen nuclear gene fragments, we carried out an ILD test to decide whether we could combine all loci (p = 0.07), then we constructed phylogenetic trees with the three spurred species (A. yabeana, A. kansuensis and A. rockii) (Fig. S4). When we excluded A. ecalcarata, different methods generated phylogenetic trees with similar topologies, in which each of the three spurred species were monophyletic. Specifically, A. yabeana, A. kansuensis and A. rockii split into three clades with all outgroups falling out as sister. Although some individuals from population K9 (A. kansuensis) were positioned in the clade of A. rockii and other individuals from populations R2, R4 and R5 (A. rockii) fell into the clade of A. kansuensis, the majority of A. kansuensis and A. rockii populations remained monophyletic. One possible explanation for these exceptions could be introgression, as we found that some populations, such as R4, also fell into different clades in the chloroplast sequence NJ tree. Hence, we discarded these exceptions in later phylogenetic analyses.
We then analyzed the phylogeny of these population when the nineteen populations of A. ecalcarata were included. The phylogeny based on nuclear gene fragments differed from that based on chloroplast loci (Fig. 2b), although all populations split into three clades. The first diverging clade (named III, with a bootstrap value 94 and Bayesian posterior probability of 1) was composed only of A. yabeana, which is in contrast to our findings in the NJ tree based on chloroplast sequence data. The monophyly of A. yabeana suggested that A. yabeana is the ancestral species of the four taxa; furthermore, phylogenetic analysis based on nuclear gene fragments indicate that A. yabeana may not have contributed to the origin of A. ecalcarata. The nineteen populations of A. ecalcarata fell into two groups, with eleven populations clustered with A. kansuensis (named clade I), while eight populations clustered with A. rockii (named clade II). The branch lengths of clade I and clade II were relatively short with bootstrap values of only 56 and 63, respectively. Notably, different individuals from two populations of A. ecalcarata (E9 and E10) could be found in both clade I and clade II, while those from the remaining populations clustered into only one clade.
We also constructed gene trees based on each of the fifteen nuclear gene fragments (Fig. S5). Gene trees of six gene fragments (Hsh, Cta1, lap 3, Ger, Vap 4, and Cer) had different topologies compared to the remaining nine. A. ecalcarata did not split into two branches on those six loci. Two populations (E9 and E10) fell into different branches among different gene trees. The unstable phylogenetic positions of E9 and E10 indicates that they could have more complicated evolutionary histories.
Furthermore, we found that the populations of A. ecalcarata in the two clades varied slightly from the chloroplast gene tree to the nuclear gene tree. Specifically, the majority of clade I and II (K1–K5, K9, R1–R3, R5–R6, E1–E4 and E12–19) from nuclear gene tree belonged to the same clade as the chloroplast gene tree, whereas the remaining part of clade I and II (K6–K8, R4 and E5–E11) belonged to the other clade of the chloroplast gene tree. It was concerning that E5–E11 were clustered with A. yabeana in the chloroplast data. Four out of the seven of these populations (E5–E11) are sympatrically distributed with A. yabeana, which may have provided opportunity for gene introgression between population pairs.
We used an ILD test to decide whether we could construct phylogenetic trees with combined chloroplast and nuclear loci. The results showed the two data sets were not suitable for combination (p = 0.01).
The divergence time between outgroups and Aquilegia populations was about 2.86–3.00 million years (Myr); the divergence time of A. yabeana (clade III) from the other three species was about 2.57–2.64 Myr; the divergence time between clade I and II was 1.69–1.94 Myr; A. ecalcarata in clade I diverged from its closest relative about 0.71–0.92 Ma, whereas A. ecalcarata in clade II diverged from its closest relative about 0.73–0.85 Ma.
3.3. Structure and PCoA analysisWe found that nineteen populations of Aquilegia ecalcarata were split into different genetic groups and two populations (E9 and E10) appeared to be the products of hybridization (Fig. 3a). When K = 2, all individuals of A. ecalcarata had already been separated into different groups (in Fig. 3a, some were purple and the others were grey) with A. kansuensis or A. rockii, while A. yabeana and A. rockii still shared the same genetic component (grey), which indicated that different groups of A. ecalcarata diverged much earlier than A. rockii from A. yabeana. When K = 3, A. rockii and a proportion of A. ecalcarata were separated from A. yabeana, corresponding to clades I, II, III in the nuclear gene fragments tree. Notably, all A. ecalcarata still shared the same color with their closest relatives. When K = 4, the majority of A. kansuensis (K1–K6) and two populations of A. ecalcarata (E5 and E6) diverged from the others, implying that seven populations of A. ecalcarata (E1–E4, E7–E8, E11) were genetically differentiated from A. kansuensis. When K = 5, three populations of A. kansuensis (K7–K9) diverged from the rest six populations (K1–K6), indicating a substructure within the species. When K = 6, five populations of A. ecalcarata (E13–E16, E19) were separated into distinct genetic components compared with six populations of A. rockii (R1–R6) and K = 7 simply repeated the pattern of K = 6. Interestingly, E5 and E6 of A. ecalcarata were never separated from A. kansuensis (K1–K6) even when K = 7. In addition, E9 and E10 had two main different genetic components. Notably, partial individuals from K9, R2, R4 and R5, which showed different topology on the NJ tree (Fig. S4) were shown to have different genetic components when K = 6 and 7. K9 might have experienced gene exchange with A. rockii, while R2, R4 and R5 might have experienced gene flow from A. kansuensis. In Fig. 3b, we observed a significant increase of LnP(D) with K from 1 to 6 and a mild increase with K from 6 to 8. In Fig. 3c, ΔK showed peaks at K = 3 and K = 5. The two peaks implied that it would be more proper to divide 42 populations into three groups and there were substructures within two groups. Overall, STRUCTURE results were in accordance with the phylogeny based on nuclear data. The geographical distribution when K = 3 (Fig. S6) showed that the three colors had a distinct geographical pattern.
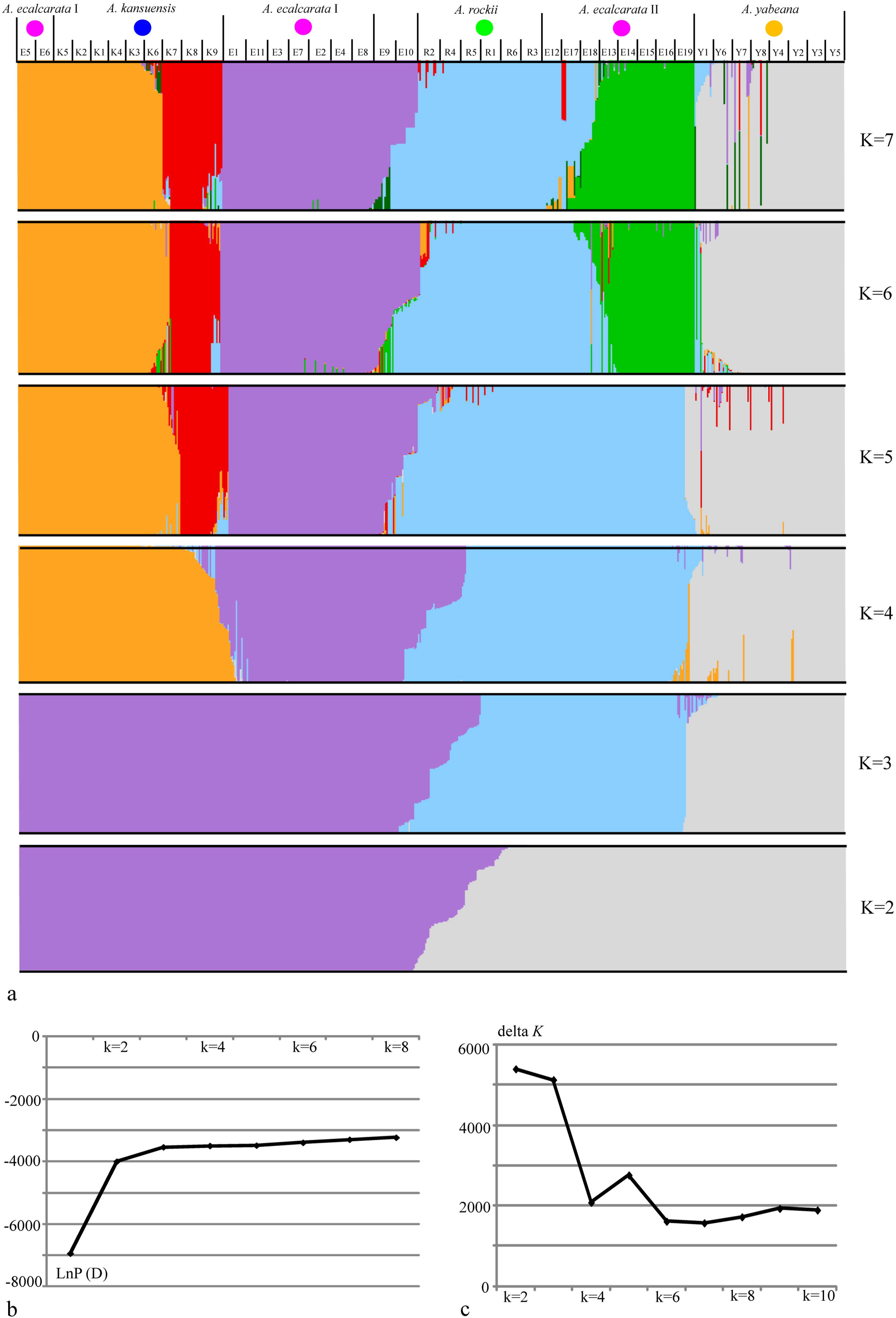
|
| Fig. 3 Structure analysis of 42 populations with combined nuclear data. Results of structure analysis at K = 2–7, (a); the maximum LnP(D) at different K (b); delta K at different K (c). |
PCoA results confirmed the species divergence revealed by STRUCTURE (Fig. S7). We observed three groups of scattered dots among 42 populations. On the PC1 and PC2 plot, the clades I and II were clustered separately, A. ecalcarata within each clade was surrounded by A. kansuensis or A. rockii. The three clades diverged greatly on PC1 axis and strikingly, PC1 accounts for 27.25% in total divergence. By contrast, PC2 and PC3 accounted for 11.18%, 6.84%, respectively. Those populations that showed evidence of introgression (i.e., E9, E10, K9, R2, R4 and R5) were located in the middle of clades I and II on the PCoA plot. All the above results suggest that A. ecalcarata had multiple origins of. A. ecalcarata in clades I diverged from its closest relative A. kansuensis when K = 4, while A. ecalcarata in clades II diverged from A. rockii when K = 6. Thus we suggested that the nine populations (E1–E8, E11) from clade I were addressed as A. ecalcarata I and the eight populations (E12–E19) from clade II were A. ecalcarata II. E9 and E10 showed evidence for hybridization between A. ecalcarata I and A. rockii.
3.4. Divergence among and within speciesThe pairwise FST values between and within species are shown in Tables 2 and S4. The minimum FST was between Aquilegia ecalcarata I and A. kansuensis (0.080); while the maximum FST was between A. ecalcarata II and A. yabeana (0.406). The genetic differentiation between A. ecalcarata I and II was 0.308, higher than 0.211 (A. kansuensis/A. rockii), 0.080 (A. ecalcarata I/A. kansuensis) or 0.127 (A. ecalcarata II/A. rockii) (see Fig. S8). If A. ecalcarata I or II resulted from hybridization with either A. kansuensis or A. rockii, the genetic divergence between A. ecalcarata I and II would not be very high and vice versa. Within species, the minimum FST was A. ecalcarata I (0.349), the maximum was A. rockii (0.610); A. yabeana and A. ecalcarata II were at intermediate levels (0.403 and 0.472). In addition, AMOVA showed that variations were mostly enriched among populations within species (up to 49.24% based on the combined nuclear gene fragments) (Table S5), which implies strong population structure in these closely related species. Mantel test results are shown in Table 3; the p-values of A. yabeana and A. ecalcarata II were significant, whereas those of A. kansuensis and A. rockii were not.
| A.ecalcarata II | A.kansuensis | A.rockii | A.yabeana | |
| Aquilegia ecalcarata I | 0.308 | 0.080 | 0.239 | 0.398 |
| A.ecalcarata II | 0.284 | 0.127 | 0.406 | |
| A.kansuensis | 0.211 | 0.404 | ||
| A.rockii | 0.381 |
| r | p-value | |
| Aquilegia ecalcarata I | 0.2827 | 0.163 |
| A.ecalcarata II | 0.4224 | 0.020 |
| A.kansuensis | 0.2149 | 0.214 |
| A.rockii | −0.6138 | 0.104 |
| A.yabeana | 0.5224 | 0.008 |
To test whether Aquilegia ecalcarata I underwent hybridization between A. ecalcarata II and A. kansuensis, or A. ecalcarata II underwent hybridization between A. ecalcarata I and A. rockii, we estimated the migration rate of the above species (Fig. S9; Table S6). A. ecalcarata populations E9 and E10 were excluded from IMa analysis because they were not ascribed as either A. ecalcarata I or II. The hypothesized hybridizations between A. ecalcarata I and A. rockii, or A. ecalcarata II and A. kansuensis are not supported by our gene flow results. For instance, the 2N1M1 between A. ecalcarata I and A. kansuensis was 0.177 (95% HPD 0.029–4.993) and 2N2M2 was significant 0.494 (95% HPD 0.207–4.841). Because A. kansuensis was assumed to be the closest relative of A. ecalcarata I, the gene flow from A. kansuensis to A. ecalcarata I was relatively high. In contrast, gene flow was only 0.064 (95% HPD 0.018–4.783) from A. ecalcarata I to A. rockii, and in the reverse direction, 0.091 (95% HPD 0.013–4.636). Thus, no significant introgressions were detected between A. ecalcarata I and A. rockii. Similarly, the gene flow between A. ecalcarata II and its closest relatives A. rockii was 2N2M2 0.306 (95% HPD 0.102–4.762), which was also significantly higher than that between A. ecalcarata II and A. kansuensis 2N1M1 0.106 (95% HPD 0.029–4.972) and 2N2M2 0.026 (95% HPD 0.008–4.993). The gene flow between A. ecalcarata I and A. ecalcarata II was calculated at a rate of 2N1M1 0 (95% HPD 0–4.993) and 2N2M2 was 0.172 (95% HPD 0.023–4.710). Interestingly, our analysis indicated that there was a low level of bidirectional gene flow between A. kansuensis and A. rockii, at a rate of 2N1M1 0.241 (0–4.552) and 2N2M2 0.192 (0–5.224). This low level of gene flow may explain the different topologies of partial individuals from populations K9, R2, R4 and R5. In addition, the gene flow between A. ecalcarata I and A. ecalcarata II for each gene fragment (Table S7) indicated that the majority of gene fragments showed very low level of gene flow, except Hsh, Cta1 and Exp.
3.6. Re-examination of morphological data between Aquilegia ecalcarata I and IIWe found that 13 out of 22 floral traits showed significant differentiation between Aquilegia ecalcarata I and II (Fig. S10). Notably, A. ecalcarata II had larger flower size (both the length and width of petal, sepal, carpel) and longer spurs than A. ecalcarata I, whereas A. ecalcarata I had more fertile stamens than A. ecalcarata II (Fig. 1a). More importantly, the geographical distribution of A. ecalcarata I and II also differed: A. ecalcarata I is mainly distributed in Gansu, Shaanxi, and Hubei provinces, whereas A. ecalcarata II is mostly distributed in Sichuan and Tibet provinces (Fig. 1b). In addition, we carried out Pearson correlation coefficients between the lengths of spurs and altitude in 42 populations (Fig. S11), the result was significant (r = −0.432, p = 0.009), suggesting the higher the population located, the shorter the spur length was.
4. Discussion 4.1. The distinct genetic divergence between Aquilegia ecalcarata I and II and the dispute over a single or multiple originsSample size and the number of molecular markers could affect the evaluation and interpretation of the genetic divergence or phylogenetic analysis in evolution studies. For instance, the origin of Oryza sativa (Asian cultivated rice) has been under debate for many years (Londo et al., 2006; Huang et al., 2012; Wang et al., 2018). One study sampled hundreds of rice individuals and used a few nuclear gene fragments, revealing that O. sativa was domesticated from its ancestor independently at different areas (Londo et al., 2006), whereas another study provided solid evidence of single origin of O. sativa based on whole-genome resequencing of more than 1000 individuals (Huang et al., 2012). Interestingly, the most recent research, which re-sequenced 3000 accessions of cultivated rice, supports the multiple-origin hypothesis of O. sativa (Wang et al., 2018). The debate over single- or multiple-origins of some species has been an ongoing topic in evolutionary biology, and studies commonly support opposite hypotheses. Given that Aquilegia is widely known for its ability to hybridize readily and easily (Hodges and Arnold, 1995), a hypothesis of a single origin followed by hybridization hypothesis would be a plausible explanation for the different types of A. ecalcarata. In fact, the Aquilegia genome project revealed that gene flow had been a common feature throughout the genus (Filiault et al., 2018). Nevertheless, evidence from the present study does not support the single-origin hypothesis. First, IMa 2 results indicate no strong gene flow between A. ecalcarata I and A. rockii, or A. ecalcarata II and A. kansuensis. Second, STRUCTURE analysis showed that both A. ecalcarata I and II have very pure genetic components. If hybridization occurred, A. ecalcarata I or II should have mixed genetic components similar to the A. ecalcarata populations E9 and E10. Besides, the fact that A. ecalcarata I or II diverged from its closest relatives at different times, the former emerged when K = 4 while the latter K = 6, this could be at least the evidence to against the single origin hypothesis. Third, chloroplast gene fragment trees showed that all A. ecalcarata are separated into different clades. If the single-origin hypothesis were true, A. ecalcarata I and II should be in the same clade since they would share the same maternal DNA. Fourth, the nucleotide polymorphism of all nineteen A. ecalcarata populations was much higher (θsil 0.0135) than that of A. ecalcarata I (θsil 0.0100) or A. ecalcarata II (θsil 0.0098). This indicated that more segregating sites emerged when we mixed two types of A. ecalcarata for calculation. Hybridization might also reduce the FST between A. ecalcarata I and II, but the real FST estimated in this study was even higher than that between A. kansuensis and A. rockii. Fifth, A. ecalcarata I and II have quite different distributions. The former is mainly distributed in the northwestern part of the entire distribution area (Shaanxi, Gansu, Qinghai, and Hubei provinces), whereas the latter is distributed in the southern part of distribution area (Sichuan, Tibety provinces). Finally, the genetic and phenotypic divergence between A. ecalcarata I and II are consistent. Morphological study on the same 42 populations divided A. ecalcarata into clusters referred to as E and F (Xue et al., 2020), with cluster E corresponding to A. ecalcarata II and cluster F corresponding to A. ecalcarata I (except E9 and E10) in this study.
Moreover, populations E9 and E10 appear to be an exception in that they showed signs of hybridization between A. ecalcarata I and A. rockii in the results of STRUCTURE. We inferred that A. ecalcarata I diverged and expanded into a new area where A. rockii was distributed. The possibility that A. ecalcarata populations E9 and E10 were the outcome of introgression suggests that A. ecalcarata might have a more complex evolutionary history and require further study based on more populations and molecular locus samplings. In addition, the phenotypic comparison indicates that divergence within A. kansuensis or between A. kansuensis and A. rockii are distinct; thus, spur loss is not the only phenotypic difference that should be considered when comparing A. ecalcarata with its closest relatives. Flower size, the extent of spur curvature and the color of dehiscent anthers are also major transitions. Taken together, evidence indicates that the genetic differentiation between A. ecalcarata I and II may be best explained by the multiple-origin hypothesis. However, to fully rule out the single-origin hypothesis and disentangle the complex evolutionary history A. ecalcarata and its relatives, whole-genome re-sequencing is required.
4.2. The complicated evolutionary history of an important trait and its challenges for species delimitationMorphological markers are the main characters used for plant species delimitation, as phenotypic similarity has been the criterion used historically by taxonomists to group individuals into species (Duminil and Di Michele, 2009). However, species diversification and morphological evolution are not always correlated. Consequently, morphological markers may fail to discriminate between morphologically similar species when complicated evolutionary histories have occurred. Researchers have reported independent origins of a series of traits from Oryza rufipogon Griff. to Oryza nivara Sharma et (Cai et al., 2019), but have not ascribed different populations of O. nivara to different species since the phenotypes are consistent within O. nivara. In this study, we recommend ascribing Aquilegia ecalcarata I and II to different species due to genetic, morphological and biogeographical differentiation. Moreover, A. ecalcarata contains a variety named A. ecalcarata form. semicalcarata. The longer spur length of A. ecalcarata II conforms to the description of A. ecalcarata form. semicalcarata (spur length >2 mm; Flora of China). In addition, the type specimen of A. ecalcarata form. semicalcarata was collected in Sichuan province, where A. ecalcarata II is distributed. Therefore, we suggest upgrading A. ecalcarata form. semicalcarata to Aquilegia semicalcarata (Schipcz) Huang et Ren. Thus, A. ecalcarata I can be referred to A. ecalcarata, whereas A. ecalcarata II can be referred to as A. semicalcarata. Furthermore, traditional taxonomy usually focuses on macroscopic changes between closely related species, such as spur loss, but neglects subtle changes in quantitative traits such as flower size. If pivotal traits show similarity but other phenotypes are differentiated, these differences should be evaluated. Our findings provide a good example of how to delimitate species that have independent evolutionary histories.
4.3. Implications for the adaptation of Aquilegia ecalcarata to new environmentAquilegia species diversity is assumed to be an example of ecological speciation, rather than being driven by the development of intrinsic barriers to gene flow (Filiault et al., 2018). In the present study, we note that both A. ecalcarata I and II grow in the stony environments along the slopes or peaks of mountains or by riversides. In contrast, A. kansuensis and A. rockii mostly grow in forest margins and riverside or under forest environments. These habitat differences suggest that A. ecalcarata I and II adapted to a new environment. Previous studies showed that natural selection by different pollinators, geographic isolation, and habitat shifts may all play important roles in diversification within Aquilegia (Hodges, 1997; Hodges et al., 2003; Whittall and Hodges, 2007; Bastida et al., 2010). Future work on pollination ecology and the investigation of ecological factors in stony habitats are required to help a better understanding of the driving forces of the adaptation in A. ecalcarata I and II. Because the spurless state is a derived rather than ancestral state in Aquilegia (Hodges and Arnold, 1995), we are still uncertain whether the loss of spurs is adaptive. Recently, Ballerini et al. (2020) identified a gene, POPOVICH (POP), crucial for nectar spur development in Aquilegia. POP plays a central role in regulating cell proliferation in the Aquilegia petal during the early phase of spur development. Neutrality tests on this gene would help us understand whether it was under positive selection. A. ecalcarata I and II might have independently fixed an ancestral spur loss allele, or have converged on a spur loss phenotype via completely different genetic means. Whether A. ecalcarata I and II share the alleles controlling the spurless status is yet to be explored.
5. ConclusionsIn this study, evidence indicates that the multiple-origin hypothesis might be more plausible for interpreting the genetic differentiation between Aquilegia ecalcarata I and II. However, whether spur loss has occurred independently within two distinct lineages of A. ecalcarata requires more evidence from whole genome re-sequencing data. Habitat differences relative to their closest relatives indicate that A. ecalcarata I and II may undergo habitat shift when they adapted to new environment. This study provides new insights into the relationship of floral diversification and species divergence.
Author contributionsY. Ren designed the study, L. Huang and F-D. Geng conducted the experiments and performed the analyses. J–J. Fan, X–Y. Zhang and C. Xue joined the sample collection, DNA extraction and phenotype measurement. J-Q. Kang and X–H. Zhang participated in data discussion. L. Huang and Y. Ren wrote the manuscript.
Declaration of competing interestThe authors declare that they have no conflict of interest.
Appendix ASupplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.06.006.
AcknowledgementsWe thank Li Sun, Xiao-peng Chang for sample collection and other members of Ren's group for technical assistance. We are grateful to Professor Hong-zhi Kong and Professor Song Ge (Institute of Botany, Chinese Academy of Science) for their valuable advice. We also thank professor Elena Kramer (Harvard University) for polishing this manuscript. This work was supported by the National Natural Science Foundation of China (31700180 and 31330007), and the fundamental Research Funds for the Central Universities of Shaanxi Normal University (GK202002011).
Abbott, R.J., 2017. Plant speciation across environmental gradients and the occurrence and nature of hybrid zones. J. Syst. Evol., 55: 238-258. DOI:10.1111/jse.12267 |
Ballerini, E.S., Min, Y., Edwards, M.B., et al., 2020. POPOVICH, encoding a C2H2 zinc-finger transcription factor plays a central role in the development of a key innovation floral nectar spurs in Aquilegia. Proc. Natl. Acad. Sci. U.S.A., 117: 22552-22560. DOI:10.1073/pnas.2006912117 |
Bastida, J.M., Alcantara, J.M., Rey, P.J., et al., 2010. Extended phylogeny of Aquilegia: the biogeographical and ecological patterns of two simultaneous but contrasting radiations. Plant Syst. Evol., 284: 171-185. DOI:10.1007/s00606-009-0243-z |
Bouckaert, R., Heled, J., Kühnert, D., et al., 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol., 10: e1003537. DOI:10.1371/journal.pcbi.1003537 |
Cai, Z., Zhou, L., Ren, N.N., et al., 2019. Parallel speciation of wild rice associated with habitat shifts. Mol. Biol. Evol., 36: 875-889. DOI:10.1093/molbev/msz029 |
Duminil, J., Di Michele, M., 2009. Plant species delimitation: a comparison of morphological and molecular markers. Plant Biosyst., 143: 528-542. DOI:10.1080/11263500902722964 |
Erst, A.S., Shaulo, D.N., Luferov, A.N., et al., 2014. On the taxonomical status of Aquilegia kansuensis (Ranunculaceae). Turczaninowia, 17: 24-25. DOI:10.14258/turczaninowia.17.4.4 |
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Excoffier, L., Smouse, P.E., Quattro, J.M., 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479-491. DOI:10.1093/genetics/131.2.479 |
Farris, J.S., Kallersjo, M., Kluge, A.G., et al., 1994. Testing significance of incon -gruence. Cladistics, 10: 315-319. DOI:10.1111/j.1096-0031.1994.tb00181.x |
Filiault, D., Ballerini, E.S., Mandakova, T., et al., 2018. The Aquilegia genome: adaptive radiation and an extraordinarily polymorphic chromosome with a unique history. eLife, 7: e36426. DOI:10.7554/eLife.36426 |
Fior, S., Li, M., Oxelman, B., et al., 2013. Spatiotemporal reconstruction of the Aquilegia rapid radiation through next-generation sequencing of rapidly evolving cpDNA regions. New Phytol., 198: 579-592. DOI:10.1111/nph.12163 |
Fu, D., Robinson, O.R. 2001. Aquilegia L. In: Wu ZY, Raven PH, eds. Flora of China 6. Beijing: Science Press, 278-281
|
Fu, Y.X., Li, W.H., 1993. Statistical tests of neutrality of mutations. Genetics, 133: 693-709. DOI:10.1093/genetics/133.3.693 |
Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser., 41: 95-98. |
Hey, J., 2010. Isolation with migration models for more than two populations. Mol. Biol. Evol., 27: 905-920. DOI:10.1093/molbev/msp296 |
Hey, J., Nielsen, R., 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics, 167: 747-760. DOI:10.1534/genetics.103.024182 |
Hodges, S.A., 1997. Floral nectar spurs and diversification. Int. J. Plant Sci., 158: 81-88. DOI:10.1086/297508 |
Hodges, S.A., Arnold, M.L., 1995. Spurring plant diversification: are floral nectar spurs a key innovation?. Proc. Roy. Soc. B: Biol. Sci., 262: 343-348. DOI:10.1098/rspb.1995.0215 |
Hodges, S.A., Fulton, M., Yang, J.Y., et al., 2003. Verne grant and evolutionary studies of Aquilegia. New Phytol., 161: 113-120. |
Hoekstra, H.E., Coyne, J.A., 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution, 61: 995-1016. DOI:10.1111/j.1558-5646.2007.00105.x |
Huang, L., Geng, F.D., Fan, J.J., et al., 2018. Genetic diversity and evolutionary history of four closely related Aquilegia species revealed by ten nuclear gene fragments. J. Syst. Evol., 56: 129-138. DOI:10.1111/jse.12298 |
Huang, X., Kurata, N., Wei, X., et al., 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature, 490: 497-501. DOI:10.1038/nature11532 |
Hudson, R.R., Kaplan, N.L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics, 111: 147-164. DOI:10.1093/genetics/111.1.147 |
Hudson, R.R., Kreitman, M., Aguade, M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics, 116: 153-159. DOI:10.1093/genetics/116.1.153 |
Huelsenbeck, J.P., Ronquist, F., 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17: 754-755. DOI:10.1093/bioinformatics/17.8.754 |
Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16: 111-134. DOI:10.1007/BF01731581 |
Kramer, E.M., 2009. Aquilegia: a new model for plant development, ecology, and evolution. Annu. Rev. Plant Biol., 60: 261-277. DOI:10.1146/annurev.arplant.043008.092051 |
Kramer, E.M., Hodges, S.A., 2010. Aquilegia as a model system for the evolution and ecology of petals. Phil. Trans. Biol. Sci., 365: 477-490. DOI:10.1098/rstb.2009.0230 |
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Librado, P., Rozas, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Londo, J.P., Chiang, Y.C., Hung, K.H., et al., 2006. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. U.S.A., 103: 9578-9583. DOI:10.1073/pnas.0603152103 |
Mayr, E., 1942. Systematics and the origin of species. Columbia University Press, New York
|
Nei, M. 1987. Molecular Evolutionary Genetics. New York: Columbia University Press
|
Perrier, X., Jacquemond-Collet, J.P. 2006. Darwin software. http://darwin.cirad.fr/darwin/
|
Posada, D., Crandall, K.A., 2001. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol., 16: 37-45. DOI:10.1016/S0169-5347(00)02026-7 |
Pritchard, J.K., Stephens, M., Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1093/genetics/155.2.945 |
Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123: 585-595. DOI:10.1093/genetics/123.3.585 |
Tenaillon, M.I., U'Ren, J., Tenaillon, O., Gaut, B.S., 2004. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol., 21: 1214-1225. DOI:10.1093/molbev/msh102 |
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G., 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 24: 4876-4882. |
Wang, W.S., Mauleon, R., Hu, Z.Q., et al., 2018. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature, 557: 43-49. DOI:10.1038/s41586-018-0063-9 |
Watterson, G.A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol., 7: 256-276. DOI:10.1016/0040-5809(75)90020-9 |
Whittall, J.B., Hodges, S.A., 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature, 447: 706-712. DOI:10.1038/nature05857 |
Woerner, A.E., Cox, M.P., Hammer, M.F., 2007. Recombination-filtered genomic datasets by information maximization. Bioinformatics, 23: 1851-1853. DOI:10.1093/bioinformatics/btm253 |
Xiao, P.K. 1979. Aquilegia. In: Delectis Florae Reipublicae Popularis Sinicae Agendae Academiae Sinicae Edita, Flora Reipublicae Popularis Sinicae 27. Beijing: Science Press, 490-502
|
Xue, C., Geng, F.D., Zhang, X.Y., et al., 2020. Pattern of variation in the morphological characteristics of Aquilegia ecalcarata and its closely related species. J. Syst. Evol., 58: 221-233. DOI:10.1111/jse.12494 |
Zhang, L.B., Ge, S., 2007. Multilocus analysis of nucleotide variation and speciation in null and its close relatives. Mol. Biol. Evol., 24: 769-783. |
Zhu, Q.H., Zheng, X.M., Luo, J.C., 2007. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol. Biol. Evol., 24: 875-888. DOI:10.1093/molbev/msm005 |



