b. College of Life Sciences, Shaanxi Normal University, Xi'an, 710062, China;
c. Eastern China Conservation Centre for Wild Endangered Plant Resources, Shanghai Chenshan Botanical Garden, Shanghai, 201602, China;
d. Key Laboratory of Environment Change and Resources Use in Beibu Gulf, Ministry of Education, Nanning Normal University, Nanning, 530001, China;
e. Department of Biology, Oberlin College, Oberlin, OH, 44074, USA;
f. State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, 100093, China;
g. Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization, the National Orchid Conservation Center of China and the Orchid Conservation & Research Center of Shenzhen, 518114, Shenzhen, China;
h. Life Science and Technology College, Harbin Normal University, Harbin, 150025, China
Staggering diversity of genome sizes has been found among eukaryotes, with available data for over 15,000 species showing that sizes vary c. 64,000-fold (Gregory, 2005). At present, the smallest known genome among eukaryotes is of the microsporidian, Encephalitozoon intestinalis (A. Cali, D.P. Kotler & J.M. Orenstein) R.A. Hartskeerl, T. Van Gool, A.R.J. Schuitema & E.S. Didier, which has only 0.0023 Gb of DNA (Biderre et al., 1999), while the largest reported genomes are found in the vascular plant species Paris japonica (Franch. & Sav.) Franch., Tmesipteris obliqua Chinnock, and Psilotum nudum (L.) P. Beauv. ‘gasa’, with 148.8 Gb (Pellicer et al., 2010), 147.3 Gb (Hidalgo et al., 2017a), and 142.4 Gb (Obermayer et al., 2002), respectively. Such enormous variation with lack of apparent correlation to organismal complexity has long attracted the attention of biologists, including Thomas (1971), who coined the phrase ‘the C-value paradox’ or ‘the C-value enigma’ (Gregory, 2001). Although the C-value paradox is widely accepted (Slijepcevic, 2018), the mechanisms underlying genome complexity, particularly in plants, remains poorly understood.
Lycophytes, one of two major lineages of extant vascular plants (the other major lineage consists of “ferns and seed plants”, called euphyllophytes in Banks et al., 2011), have genome sizes that range from extremely small (1C = 81.45 Mb in Selaginella selaginoides (L.) Link; Baniaga et al., 2016) to relatively large (1C = 11.7 Gb in Isoëtes lacustris L.; Hanson and Leitch, 2002), with an average of 1.66 Gb (Leitch and Leitch, 2013). Recent research shows that this extreme variation in genome size in lycophytes may be due to multiple paleo-polyploidization events, such as in Selaginella moellendorffii Hieron, a model lycophyte, in which two paleo-polyploidization events were detected by Wang et al. (2020). Even more paleo-polyploidization events may predate the divergence of lycophytes from the euphyllophytes, and, thus, help explain the successful establishment of land plants as well as large genome sizes in ferns, which represent early diverging euphyllophytes.
Ferns, which are sister to all seed plants, tend to have much larger genomes than other land plant lineages, including angiosperms (Klekowski and Baker, 1966; Barker and Wolf, 2010; Leitch and Leitch, 2012; Henry et al., 2014). In general, the genomes of ferns are underexplored compared to other eukaryotes, and some may have genome sizes comparable to the present record holder for the largest genome, Paris japonica (Hidalgo et al., 2017a; 2017b). Previous research noted that genome sizes in ferns may be correlated with many factors, such as chromosome number (Clark et al., 2016), spore size (Dyer et al., 2013; Henry et al., 2014), ecological traits, especially habitat types (Kang et al., 2014; Henry et al., 2014) and environmental change (Wakamiya et al., 1993; Nakazato et al., 2008a; Kang et al., 2014; Henry et al., 2014), and polyploidization (Haufler, 2014; Henry et al., 2014). Until recently, polyploidization has been assumed to be one of the most important mechanisms underlying large genome sizes in ferns; consequently, large genomes are predicted to contain a large number of chromosomes (Dolezel et al., 2003). Broadly, ferns do appear to have high chromosome numbers (Leitch and Leitch, 2012). However, genomic data increasingly show that large genomes contain many repeat sequences and transposons that help account for their large sizes. Thus, there may be no direct relationship between genome size and chromosome number (Rabinowicz et al., 2005; Hawkins et al., 2008). Two plausible mechanisms of genome size evolution ---polyploidy and transposon proliferation --- are not mutually exclusive. During the long evolutionary history of land plants, whole-genome duplications (WGDs) are thought to have occurred repeatedly, especially in ferns, but often with subsequent losses of chromosomes, while repeat sequences and transposons have also been incorporated and removed (One Thousand Plant Transcriptomes Initiative, 2019). Moreover, in ferns, lineage-specific average chromosome length may constrain genome size evolution (Liu et al., 2019). Thus, the evolutionary history of large genomes in ferns is likely to be complex, with several mechanisms at work simultaneously (Fujiwara et al., 2021; Szövényi et al., 2021).
In this study, we investigated how ferns have come to possess exceptionally high chromosome numbers and large genome sizes. To accomplish this, we measured the genome sizes of 255 samples (belonging to 27 family, 72 genera, and 240 species) of ferns and lycophytes using flow cytometry and calculated the phylogenetic signal of 2C values as well as the relationship between genome size and chromosome number. Additionally, we analyzed the correlations between genome size and spore size, which has often been used as a proxy for ploidy levels in ferns (Moran, 1982; Barrington et al., 1986; Beck et al., 2010). To better understand reciprocal effects of genome size evolution and shifts in habitat type, we assessed correlations between habitat types of species and their genome sizes, and we performed ancestral character reconstruction for different habitat types.
Our results provide insights into the factors that contribute to the large genomes of ferns and lycophytes. Additionally, our work greatly expands the knowledge of genome size in fern species with 228 reports that are new to science. Overall, our goals were to investigate the relationships of extant ferns and lycophytes to their habitat types, evolutionary history, and genome size using big data. Therefore, our large dataset may serve as a basic framework for subsequent applications such as genome sequencing, which requires a basic knowledge of genome size, and downstream approaches to inferring fern and lycophyte evolution.
2. Materials and methods 2.1. SamplingIn total, we collected genomic size data comprising 385 samples representing 355 species and several discrete subspecies and varieties of the Polypodiopsida (hereafter “ferns”; see PPG I from the Pteridophyte Phylogeny Group, 2016) and the Lycopodiopsida (hereafter “lycophytes”, including Lycopodiales, Isoëtales and Selaginellales). We used flow cytometry to quantify genome size of 240 species (255 samples), of which, 228 species (242 samples) were newly measured and validated. These 240 species belong to nine orders, 27 families, 72 genera. The vast majority of these species (233) were ferns, with the remaining seven species comprising two orders, two families, and three genera of lycophytes. Genome data from 123 species (130 samples) were downloaded from the Pteridophyte DNA C-values database (Bennett and Leitch, 2012) and recent publications. Among the 355 species (385 samples), 25 species (33 samples) belong to three families and five genera of lycophytes, and the rest are ferns.
To infer the taxonomic coverage of our sampling within lycophyte and fern orders, we used the estimates of species diversity for the orders in PPG I (The Pteridophyte Phylogeny Group, 2016), and, throughout, we followed the taxonomy in the Flora of China (http://www.efloras.org/flora_page.aspx?flora_id=2) for the classification of species within genera.
For all sampled species, including those newly sampled, we obtained chromosome numbers (2n), base number of chromosomes (x), and ploidy levels (Supplemental File 1) from the Missouri Botanical Garden's Index to Plant Chromosome Numbers (http://legacy.tropicos.org/project/ipcn).
2.2. Estimation by flow cytometryTo determine genome size by flow cytometry, we collected young, fresh leaves, bagged, sealed, and placed them in a low-temperature, dark ice box for express delivery. Upon arrival to the laboratory, a portion of the fresh material was immediately excised and the remaining tissue in silica gel for further experiments. All fresh leaf tissue was stored at 4 ℃ for a maximum of three days until use. Additionally, we collected all introduction records to include with voucher specimens representing the sampled individuals (Supplemental File 1 and Table 1), and we deposited the vouchers in the herbarium at Shanghai Chenshan Botanical Garden (CSH).
| Sampled taxonomic orders | Total number of extant species | Number sampled | Coverage (%) | Mean 2C (pg) | Min 2C (pg) | Max 2C (pg) | x-fold-2C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All | 11,916 | 255 | 2.1 | 14.56 | 0.64 | 103.72 | 162.06 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lycopodiales | 388 | 7 | 1.8 | 23.69 | 14.14 | 31.68 | 2.24 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Selaginellales | 700 | 1 | 0.1 | 19.08 | 19.08 | 19.08 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Equisetales | 15 | 1 | 6.7 | 33.32 | 33.32 | 33.32 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Psilotales | 17 | 1 | 5.9 | 56.15 | 56.15 | 56.15 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ophioglossales | 112 | 1 | 0.9 | 54.98 | 54.98 | 54.98 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Marattiales | 111 | 2 | 1.8 | 10.67 | 6.45 | 14.88 | 2.31 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Osmundales | 18 | 1 | 5.6 | 25.29 | 25.29 | 25.29 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Schizaeales | 190 | 3 | 1.6 | 21.06 | 12.03 | 28.26 | 2.35 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Salviniales | 82 | 2 | 2.4 | 1.70 | 0.95 | 2.44 | 2.57 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cyatheales | 713 | 3 | 0.4 | 11.22 | 9.22 | 13.39 | 1.45 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Polypodiales | 8,714 | 233 | 2.7 | 13.90 | 0.64 | 103.72 | 162.06 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Total number based on the Pteridophyte Phylogeny Group (2016) and 1 pg = 0.978 Gb. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
We followed previously published (Bennett and Leitch, 2001). protocols for flow cytometry. For accuracy, we compared the results of each sample in each experimental run to one of three reference standards: Pisum sativum L. ‘Bakana’, Vicia faba L., or Oryza sativa L. subsp. japonica S. Kato ‘Nipponbare’. For most samples, we found that O. sativa subsp. japonica ‘Nipponbare’ (2C = 0.795 pg, 1 pg = 0.978 Gb) represented a suitable standard (Bai et al., 2012). However, if the sample and O. sativa exhibited overlapping absorption peaks, we used either P. sativum ‘Bakana’ or V. faba for secondary determination.
Prior to flow cytometry, we applied either Otto (1990) or general purpose (GP) isolation buffers (Loureiro et al., 2007) to the fresh tissue according to Dolezel and Bartos (2005) as shown in Supplementary File 2. Due to the large number of samples processed in our study, we prepared all samples initially using GP buffer. If we detected too much fragmentation and overlap in images of a sample, we applied the Otto buffer in subsequent flow cytometry analyses for that species.
Following preparation of the nuclear suspension in buffer, we subjected each sample to a shaker for 5s before testing the reduced cohesion among cell nuclei. We analyzed the samples in a MILLIPORE® Guava PCA flow cytometer with the PM1 channel voltage set to 350 V and the sample collection parameter set to 20,000.
2.3. Statistical analysis 2.3.1. Relationship between genome size and palynological traitsWe calculated the relationship between genome sizes of the sampled ferns and lycophytes, their spore size, and other palynological traits, chromosomal traits (i.e., chromosome number, base number of chromosomes, and ploidy level), and habitat types. Correlations were plotted in R 3.5.1 (R Core Team, 2018). We estimated spore size using polar axis length (P), equatorial axis width (E), P/E ratio, and the area of the spore profile in equatorial view (Zhang et al., 1976, 2002, 2012; Tryon and Lugardon, 1991; Liu, 1997; Wang et al., 2001, 2006a, 2006b, 2015; Dai et al., 2002, 2005a, 2005b, 2005c; Lu et al., 2007; Jiang et al., 2010; Li et al., 2010; Xu et al., 2012; Yang et al., 2013; Ji et al., 2014; Shi et al., 2017). Initially, we measured P and E, which we used to obtain the P/E ratio, and we determined the area of the spore profile (hereafter spore area) using the formula πPE. We evaluated correlations between 2C values and (1) the four components of spore size and (2) the three chromosomal traits using Pearson's correlation coefficient. All data for spore size and genome size measurements are given in Supplemental File 1.
2.3.2. Phylogenetic framework and signalWe assembled a phylogenetic tree topology at the genus-level for the 96 genera of ferns and lycophytes sampled in this study. We based the topology primarily on Lehtonen et al. (2017) as well as more recently published molecular phylogenetic and taxonomic studies. We used Phylocom v.4.2 (Webb et al., 2008) to generate a time calibrated phylogeny with ages of important nodes constrained based on Lehtonen et al. (2017) and by applying bladj in Phylocom to distribute other node ages between the calibrated nodes.
2.3.3. Analyses of genome size and ecological traitsWe analyzed the relationships between 2C values and their habitat types using ANOVA and multinomial logistic regression with a Wald test for significance. We collected the habitat type information for each species primarily from the floras available via eFloras.org (http://www.efloras.org/index.aspx). We coded species to four habitat types, “epiphyte, hygrophyte, petrophyte and terrestrial”, according to their five life-forms: climber, epiphyte, hygrophyte, petrophyte and terrestrial plants (here all climbers were coded into the “terrestrial” habitat type for analyses; see Supplemental File 1).
2.3.4. Ancestral character reconstruction for habitat typesWe reconstructed the evolutionary history of habitat types of 96 genera across the phylogenetic tree. The polymorphic habitat type data were fitted with the “ER” model (equal rates for all permitted transitions) using the function “fitpolyMk” of the “phytools” package (Revell, 2012) in R 3.6.1. The function fitpolyMk fits an Mk model to data for a discrete character with intraspecific polymorphism. Under this model, it is assumed that transitions between states must occur through a polymorphic condition, whereas transitions cannot occur directly between two incompatible polymorphic conditions. Conditioned on the transition matrix of the fitted model, we inferred ancestral states by applying the maximum likelihood stochastic mapping approach implemented in the “make.simmap” function of phytools with 1000 replicates.
2.3.5. Correlations between changes in genome size and shifts in habitat typeBased on similar concepts in prior studies (e.g., Zanne et al., 2013), we sought to determine whether changes in genome size occur before, during, or after changes in habitat type. Specifically, we tested three hypotheses: Hypothesis 1 (H1) posits that shifts in habitat type predate changes in genome size; Hypothesis 2 (H2) posits that shifts in habitat type accompany changes in genome size; hypothesis 3 (H3) postulated that shifts in habitat type occur after changes in genome size. To test these hypotheses, we performed three correlation analyses using Pearson's r in R 3.6.1 (R Core Team, 2020). We used correlation analysis rather than regression because, for H2 in particular, there is no clear predictor or response variable, as H2 allows that some external factor may have driven simultaneous changes in genome size and habitat-type.
To gather and formulate the data for correlation analyses, we first estimated genome sizes at internal nodes of the genus-level tree containing 96 tips using FastAnc in phytools v. 0.7–70 under default settings. We used the average genome size (2C) of each genus, based on our own assessments and those reported in databases, for inference. Using averages reduces the effects of outliers, such as polyploids, that may be uncommon and evolutionarily derived in each genus. Nevertheless, we recognize that this is a crude approach that could be refined under a different experimental design. Results of FastAnc for ancestral genome sizes are shown in Supplemental File 3.
For all three hypotheses, we used custom scripts in Python and R (Supplemental Files 4 and 5) to obtain genome size and habitat-type data for each node. The genome size data was based on our measurements for terminal nodes or reconstructions using FastAnc for internal nodes. The data for the type of habitat comprised the information we obtained for extant species for terminal nodes and the most highly supported reconstructions using stochastic mapping for internal nodes. We also obtained lengths of the branches, in units of time, connecting each node within the phylogeny. For H1, we compared genome sizes one node forward (i.e., at each daughter node) with habitat types one node back. For H2, we compared genome sizes and habitat types at each node to those at one node back (i.e., at the ancestral node). For H3, we compared habitat types one node forward (i.e., at each daughter node) with genome sizes one node back. For H1 and H3, we treated each of the two daughter nodes independently. In the case of genome sizes, we subtracted the more ancient genome size (i.e., older node) from the more recent one. For habitat types, we sought cases where the more recent habitat type contained none of states of the older habitat type. Thus, initially, the data for habitat type were coded as binary with 1 indicating a shift in habitat type and 0 indicating no shift. After obtaining the differences in genome sizes and the shifts in habitat type, we divided each result by the branch length to account for time. While this represents a rough approach, the transformed genomic data may be generally considered to represent change in size per unit time, while the transformed shift results may represent probability of the shift occurring per unit time.
We removed instances from the pairings of habitat shifts and genomic change where there was no habitat type shift (i.e., where habitat shift = 0). We did this because our objectives were only concerned with cases where shifts occurred. For the remaining pairs, we performed correlation analyses and visualized results in R using the ggpubr library (Kassambara et al., 2020).
3. Results 3.1. Genome size profilesOur measurements of 2C values represent new reports for 228 species (242 samples, see Supplemental File 1 and Table 1). These new data increase the taxonomic coverage of ferns and lycophytes with known genome sizes by 2.0%. Among newly sampled species, 233 samples (more than 90%) belong to the fern order Polypodiales.
For the newly sampled species, we estimated mean, minimum, maximum, and x-fold of 2C values. The 2C values of species ranged from 1.45-fold in Cyatheales to 162.06-fold in Polypodiales. Overall, average 2C values were higher in Lycopodiales and Polypodiales than in other orders. The smallest genome size that we detected was in Microlepia hancei Prantl; however, with 2C = 0.64 pg, its genome is much larger than the smallest genome known among ferns and fern allies [i.e., the lycophyte, Selaginella apoda (L.) Spring, with 2C = 0.17 pg; Little et al., 2007]. Asplenium xinyiense Ching & S.H. Wu had the largest genome size among sampled species with 2C = 103.72 pg.
The mean 2C value (7.71 pg) of the 25 lycophyte species was smaller than that of the 330 fern species (17.87 pg). Species of Selaginellaceae had relatively small 2C values (0.17–0.48 pg) and chromosome numbers (c. 2n = 20), whereas genome size and chromosome numbers of the other lycophyte species were more similar to some fern species. Interestingly, species of Salviniaceae (ferns) also have small 2C values (0.95–1.53 pg).
3.2. Relationship between 2C value and phylogenetic signalWe detected significant phylogenetic signal for base chromosome numbers (K > 1, p < 0.01) and weak phylogenetic signal for 2C values (Fig. 1; Table 2). The Blomberg's K value for the base number of chromosomes was 1.21, indicating that the base number among related taxa is more similar than expected (Table 2). In contrast, the Blomberg's K for 2C was 0.81, indicating that there is less similarity among related taxa than is expected under a Brownian motion model of evolution. This finding may be the result of convergent evolution or stabilizing selection, which tend to return species to some optimal value of 2C over evolutionary time (David, 2009). Neither ploidy levels nor total number of chromosomes (2n) showed significant phylogenetic signal.
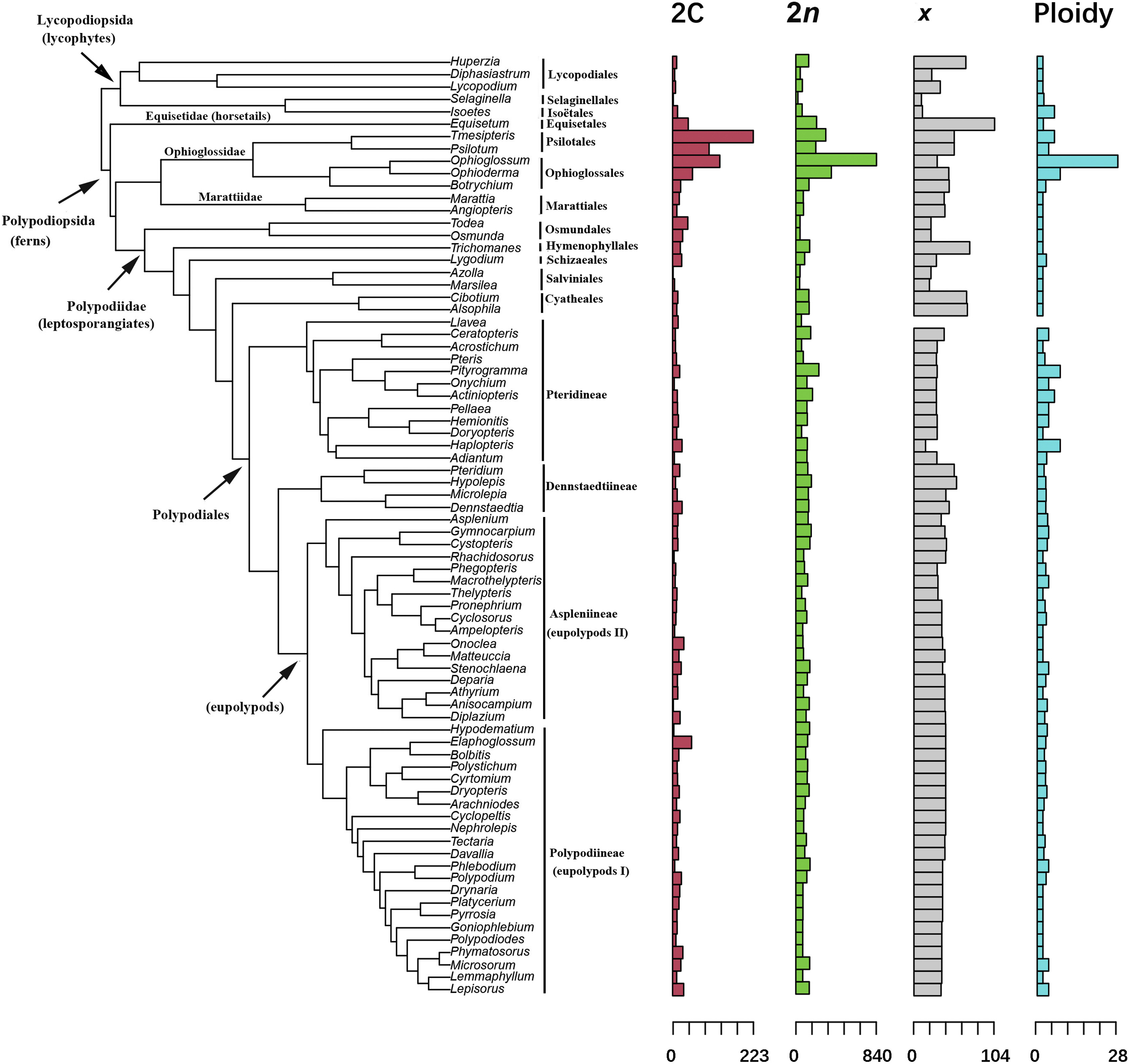
|
| Fig. 1 Phylogenetic distribution of 2C values, numbers of chromosomes (2n), base number of chromosomes (x), and ploidy levels of 96 genera of ferns and lycophytes. Data for each genus were averaged from among all sampled species and transformed into percentages by dividing the maximum value for the genus. Within Polypodiales, phylogenetic nomenclature (i.e., the eupolypods) follows Smith et al. (2006). |
| Traits | Blomberg's K | p |
| 2C | 0.81 | 0.012 |
| 2n | 0.53 | 0.091 |
| x | 1.1 | 0.001 |
| Ploidy | 0.41 | 0.241 |
In ferns and lycophytes, spore size and genome size are not related (see Fig. 2). Logarithmic regression analyses revealed that 2C values were not significantly correlated with polar axis length (P) (r = -0.084, p = 0.257), equatorial axis width (E) (r = 0.098, p = 0.186), the P/E ratio (r = -0.008, p = 0.911), or spore area (r = 0.063, p = 0.396). Our results differ from at least one previous study that showed spore length was significantly correlated with 2C values (all p < 0.001) for ferns of Ontario (Henry et al., 2014; though the exact meaning of “spore length” was not defined in that paper); however, our finding is consistent with Dyer et al. (2013), in which polar length was not correlated with genome size within the Asplenium monanthes L. complex after adjusting for intraspecific phylogenetic relatedness. The difference between our result and that found for ferns of Ontario may be due to taxonomic scale (i.e., 34 species in 12 families in Henry et al. versus 240 species in 27 families in this study), inclusion of lycophytes (i.e., not included in Henry et al. but included here), and/or geographic scale (i.e., Ontario in Henry et al. versus global in this study).

|
| Fig. 2 Logarithmic regression analysis comparing 2C values and spore sizes based on a, polar length (P); b, equatorial width (E); c, P/E ratio; and d, spore area. |
Logarithmic regression analyses showed that 2C values were significantly, positively correlated with ploidy levels (r = 0.259, p < 0.001), chromosome numbers (r = 0.483, p < 0.001), and base number of chromosomes (r = 0.490, p < 0.001) (Fig. 3). Chromosome number explained 40% of the variation in 2C values while the base number of chromosomes explained 41%. Ploidy levels explained only 6% of the variation (Fig. 4).
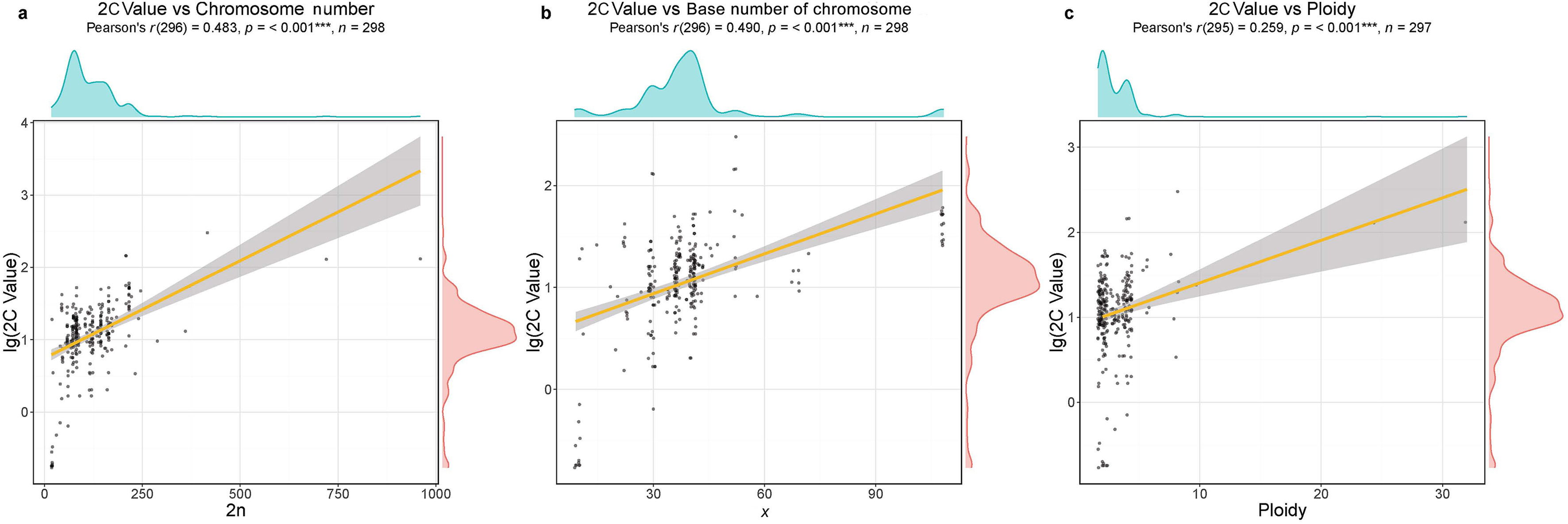
|
| Fig. 3 Correlation analysis between 2C values and chromosomal traits using logarithmic regression analysis. |
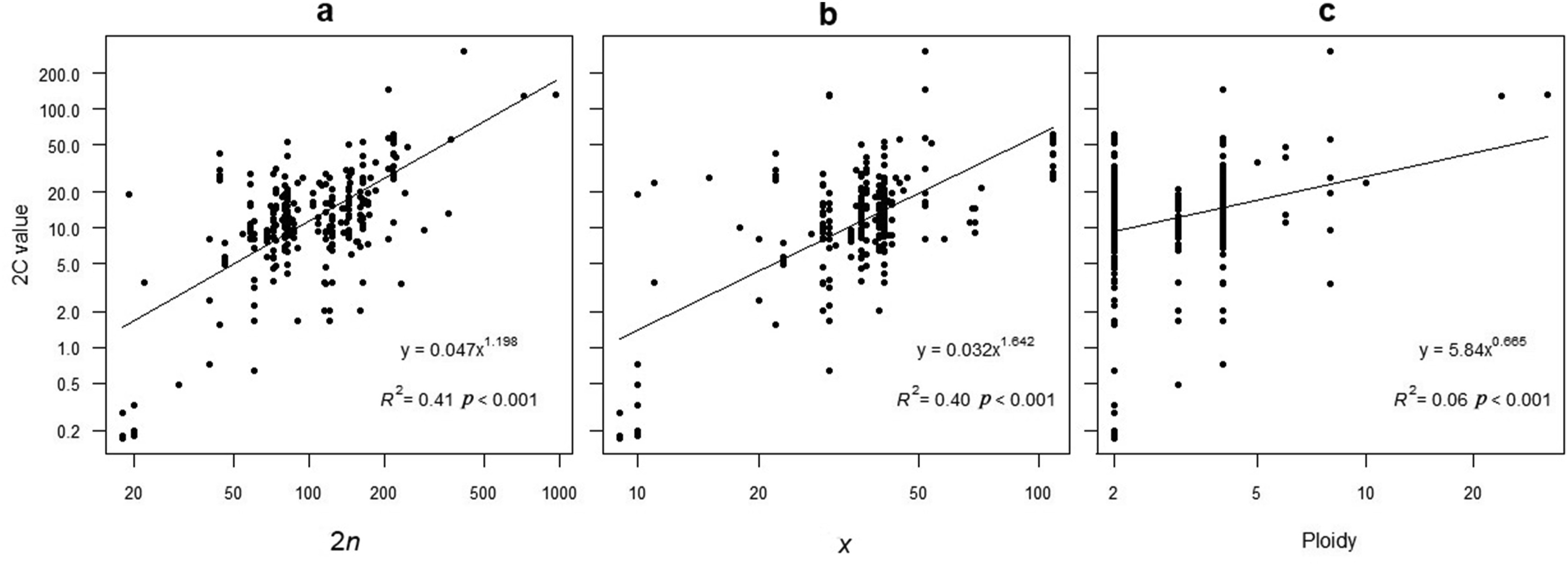
|
| Fig. 4 Relationships between genome size (2C) and a: chromosomenumber (2n); b: base number of chromosome (x); and c: ploidy level. |
ANOVA analyses showed that 2C values vary significantly among four different types of habitat (df = 4, F = 5.056, p = 0.000548, Fig. 5). Our use of multinomial logistic regression analysis with terrestrial habitat type as a benchmark indicated that ln P(epiphytic)/P(terrestrial) = 1.266 × lg 2C–2.413. From this, we calculated that if the 2C > 80.538 pg, the value of P(epiphytic)/P(terrestrial) will be > 1. Accordingly, when 2C value is in the interval [80.538, infinity], larger 2C values predict an epiphytic habitat, with a high degree of significance (p = 2.330 e−8). Similarly, for the following formulas, ln P(hygrophytic)/P(terrestrial) = 0.180 × lg 2C–2.855 and ln P(petrophytic)/P(terrestrial) = 1.839 × e−3 × lg 2C–1.284, we calculated that only when the 2C values are > 1015 and 1014 pg, respectively, P(hygrophytic)/P(terrestrial) and P(petrophytic)/P(terrestrial) will be > 1. Although 2C values this large have yet to be found, P(hygrophytic) and P(petrophytic) are always smaller than P(terrestrial); furthermore, they are positively correlated with 2C values (p = 7.496 e−6 and p = 6.065 e−5).

|
| Fig. 5 ANOVA and regression analysis comparing 2C values and habitat types.. |
Therefore, assuming that the last common ancestor of Lycophytes and Monilophytes was terrestrial (see Results below), increases in 2C values correspond to changes to epiphytic, hygrophytic, or petrophytic habitat types. Moreover, the probability that a species is epiphytic increases at higher 2C values.
3.5. Ancestral character reconstruction for the habitat typesAmong 96 genera examined, 64 (66.7%) grow in a single habitat type, with 46 genera being terrestrial (47.9%), 11 being epiphytic (11.5%), four being petrophytic (4.2%), and three being hygrophytic (aquatic, 3.1%). The other 32 genera grow in two or three different habitat types (33.3%). Of these genera, 12 are both terrestrial and petrophytic, ten are terrestrial, petrophytic, and epiphytic, and five are petrophytic and epiphytic. Thus, most genera that grown in more than one habitat type include the terrestrial type. Overall, terrestrial habitat appears to be the most common among fern and lycophyte genera, followed by epiphytic and petrophytic habitats. Our results indicate that the ancestral habitat type of lycophytes, all genera of true ferns (including tree ferns), and the leptosporangiate ferns of subclass Polypodiidae was terrestrial (Fig. 6). Among leptosporangiate ferns, the suborder Pteridineae exhibits several independent transitions to petrophyte, such as in the genera Pellaea Link and Doryopteris J. Sm., an independent transition to hygrophyte (aquatic) in Ceratopteris Brongn., and a transition to epiphyte in Haplopteris C. Presl. Primarily, transitions within Pteridineae occurred from the terrestrial habitat to petrophyte, whereas there were fewer transitions to hygrophyte and rarely to epiphyte. However, there were no transitions in the suborder Dennstaedtiineae of Polypodiales, in which all sampled species are terrestrial. Further, among leptosporangiate ferns of Aspleniineae (eupolypods II), few genera (three genera) have undergone a change in habitat type, whereas, in Polypodiineae (eupolypods I), there were frequent transitions from terrestrial to petrophyte or epiphyte. Our sampling contained only two genera of lycophytes and four genera of eusporangiate ferns; thus, analyses of trends at lower taxonomic ranks within these groups was not possible (Fig. 6).

|
| Fig. 6 Ancestral character reconstruction of different habitat types using a phylogeny representing 96 genera of lycophytes and ferns. Different colors represent different habitat types. The tree topology with node age constraints was generated in Phylocom v.4.2 based on Lehtonen et al. (2017). |
Correlation analysis designed to determine whether changes in genome size occur during, before, or after shifts in habitat type revealed no significant results and weak correlations (Fig. 7). This is unsurprising given the small sample sizes. The strongest correlation was observed for our hypothesis that shifts in habitat type occur before a change in genome size (R = -0.23, p = 0.66). However, the data for our hypotheses that shifts in habitat type predate changes in genome size (H1) and that shifts in habitat type accompany changes in genome size (H2) suggest a possible non-linear pattern, which cannot be properly assessed using correlation analysis.

|
| Fig. 7 Scatterplots showing the relationships between genome size (pg) per unit time (million years, Myr) and probability of habitat type change per Myr. Hypothesis one (H1), shifts in habitat type occur prior to a change in genome size; Hypothesis two (H2), genome size changes occur simultaneously with habitat type shifts; Hypothesis three (H3), habitat type shift occur after changes in genome size. All plots show Pearson's R, p, a trendline, and the 95% CI (gray) for the model. |
In eukaryotes, the evolution of genome size is largely attributed to genome expansion and gene losses or deletions (Grover and Wendel, 2010). The primary modes of genome expansion are polyploidization and the accumulation of transposable elements, while losses mainly arise from unequal homologous recombination and illegitimate recombination (Petrov, 2002; Wendel et al., 2002; Gregory, 2004; Hawkins et al., 2008). Losses and deletions constrain expansion of the genome. Genome sizes vary less in ferns and lycophytes than in angiosperms, but are generally much larger and possess higher average chromosome numbers (Clark et al., 2016). These findings are consistent with recent studies on ferns that have detected recurrent genome duplication events (19 WGD events) during fern evolution (Huang et al., 2020) and a higher proportion (31%) of polyploid species than in angiosperms (Wood et al., 2009). Taken together, these findings may be explained by Haufler's hypothesis that WGDs accompanied by subsequent diploidization involve gene silencing, but without apparent chromosome loss, so that high chromosome numbers are retained (Haufler 2002, 2014; Clark et al., 2016). Our results indicate that, in ferns and lycophytes, 2C values are significantly correlated with chromosome number, base number of chromosomes and ploidy levels. Such correlations are not found in any other plant lineages (Barker and Wolf, 2010; Szövényi et al., 2021). Reasons for this discrepancy may be related to two processes. First, ferns and lycophytes may differ from angiosperms in their post-polyploidization genomic processes (Szövényi et al., 2021). The predominantly small (yet historically polyploid) genomes of angiosperms imply that post-polyploidization genome fractionation and downsizing is effective and frequent (Szövényi et al., 2021); conversely, the diploidization process in ferns might be slower (Barker and Wolf, 2010). Second, it has been hypothesized that DNA content per chromosome is constrained in ferns and lycophytes, requiring more chromosomes to sustain a larger genome (Clark et al., 2016). Available data suggest that chromosomes of ferns and lycophytes are smaller and more uniform in size compared with those of angiosperms (Wagner and Wagner, 1979; Nakazato et al., 2008b; Liu et al., 2019).
Our results show great variation in the genome sizes of lycophytes and ferns based on 2C values, including, in some cases, two or three different 2C values within the same species (see Supplemental File 6). For example, our results identified two 2C values (7.26 pg and 16.36 pg) for Microlepia speluncae (L.) T. Moore, as well as a third (15.88 pg) according to publicly available data; in addition, the larger value is roughly two times that of the smaller one. This suggests that this species has undergone polyploidization or that it actually comprises two or more cryptic species. Similarly, we found three 2C values in Polypodium virginianum L. between our measurements and published data (10.54 pg, 20.66 pg, 30.97 pg). This suggests that there are diploid, tetraploid, and hexaploid variants in this species, which is known to possess such chromosomal variants, but which was previously reported as having only diploid, triploid, and tetraploid cytotypes (Kott and Britton, 1982).
We compared the 2C values of 12 species for which we obtained new 2C measurements from fresh material and values from databases. This comparison (Supplemental File 6) revealed that only three species had similar values between our study and the publicly available data: Microlepia speluncae (this study: 16.36 pg, database: 15.88 pg), Nephrolepis exaltata (L.) Schott (this study: 20.81 pg, database: 19.12 pg), and Pteridium aquilinum (L.) Kuhn (this study: 15.20 pg, database: 16.12 pg). Values for other species varied considerably between our measurements and those obtained from databases. These discrepancies may be related to a variety of factors, especially intraspecific chromosome diversity such as chromosome size diversity. This variation highlights the need for direct karyotype analysis in ferns and lycophytes. Observations in angiosperms suggest that above a certain chromosome arm/spindle length ratio, mitotic divisions fail (Schubert and Oud, 1997); however, the mechanisms that regulate chromosome size and chromosome structure in ferns and lycophytes remain unknown and need further study.
Not all ferns and lycophytes have large genomes. For example, Marattiales have small genomes, and their mean 2C value is 10.67 pg (Table 1), and species in this order have probably undergone substantial genome rearrangements (Clark et al., 2016). In fact, our data contained 161 diploids and137 polyploids (including 36 triploids, 87 tetraploids, one pentaploid, four hexaploids, six octaploids, one decaploid, one 24-ploid, and one 32-ploid) (Supplemental File 1). Extensive genome rearrangements associated with gene loss during the diploidization process of post-polyploidization might also have occurred for those diploids with small genomes, except for the Equisetaceae, which, although diploid, have larger genomes with a high base number of chromosomes (x) (Christenhusz et al., 2021). This may help explain why the base number of chromosomes has strong phylogenetic signal, whereas the phylogenetic signal of 2C values were weak in ferns and lycophytes. Another probable reason for weak phylogenetic signal of 2C values might be the diversity in chromosome size among different species in ferns.
Besides polyploidy, activity of transposable elements (TEs), especially long terminal repeat (LTR) transposons, are thought to contribute considerably to genome size variation in angiosperms and gymnosperms (Wendel et al., 2016). Proliferation of TEs has been thought to play less prominent roles in ferns and lycophytes. However, interestingly, Baniaga and Barker (2019) demonstrated that haploid nuclear genome size of ferns and lycophytes were correlated with the time of insertion of median long terminal repeat retrotransposons (LTR-RTs). They also found that LTR-RT insertions occurred more recently on average in species with small genomes (e.g., Salviniaceae), whereas they occurred much earlier in species with large genomes (e.g., homosporous ferns), suggesting that LTR-RT insertions may coincide with the process of polyploidization.
4.2. Genome size and habitat typesC values are well-known predictors of ecology and environment (Wakamiya et al., 1993). Previous studies found that genome size is associated with habitat type in green algae. Specifically, the sister algal species, Mesostigma viride Lauterborn (genome sizes: 329 Mb) and Chlorokybus atmophyticus Geitler (genome size: 85 Mb), which occur within the earliest-diverging clade of algae, dwell in benthic/freshwater and subaerial/terrestrial environments, respectively. M. viride is thought to have transitioned to a benthic, freshwater habitat, whereas the ancestral species occurred within a subaerial/terrestrial environment (Zhou et al., 2019). Thus, this transition to a new environment may be correlated with a large increase in genome size. Previous research on ferns found that several traits, including habitat type, are significantly associated with 2C values, and specifically, that wetland species tend to have larger genomes (Henry et al., 2014).
We found that the 2C values of lycophytes and ferns were significantly correlated with their habitat types. In particular, our results show that epiphytic species tend to have higher 2C values. For example, Asplenium xinyiense (2C = 103.72 pg) and A. scortechinii Bedd (2C = 69.46 pg), tend to be attached to the trunks of trees or wet rocks within forests (see Supplemental File 1 and Flora of China, 2013). In contrast, species with smaller 2C values tend to be aquatic, such as Ceratopteris thalictroides (L.) Brongniart and Azolla microphylla Kaulf. Additionally, the aquatic order Salviniales had the smallest mean 2C value among sampled orders (Table 1 and Fig. 7).
Our results indicate frequent transitions from the ancestral terrestrial habitat to other types of habitats. Of three hypotheses tested, correlational analysis provided the strongest support for our hypothesis that shifts in habitat type predate genome size changes (R = -0.23, p = 0.66), although data for this hypothesis and our hypothesis that shifts in habit type accompany changes in genome size suggest a possible non-linear pattern. Our findings indicate that these shifts in habitat type may be followed by a period of stability in genome size. Somewhat similarly, shifts in habitat may occur during periods of genomic stability. This is broadly consistent with a recent study in salamanders, which have some of the largest genomes among vertebrates. In salamanders, shifts in habitat types that occurred at different phases in its evolutionary history were associated with negligible changes in genome size (Bonett et al., 2020). However, genomes expanded in lineages that experienced long periods of conserved habitat preference during phases of evolutionary history (Bonett et al., 2020). Overall, our findings indicate that habitat shifts appear to be correlated with genome stability (i.e., cluster around 0; Fig. 7). When we tested whether shifts in habitat type occurred after changes in genome size, we failed to find a clear non-linear pattern, indicating that genome size changes are not a predictor for subsequent habitat shifts. Nevertheless, given the small sample size and lack of significant patterns in the data, denser taxonomic sampling of ferns and fern allies is needed to more rigorously investigate the temporal relationship between change in genome size and shifts in habitat type.
4.3. Correlation between habitat type and the phylogenyOur results show that epiphytic ferns are more likely to have larger genomes. Diversification in nearly all epiphytic fern clades appears restricted to the Cenozoic, and the preference for the epiphytic habitat arose independently many times within ferns, with several subsequent losses (Schuettpelz and Pryer, 2009). However, the origin of epiphytic ferns is still poorly understood, and may be traced to rupestral, climbing, or strictly terrestrial ancestors.
According to our reconstructions of habitat types for lycophytes and ferns across the phylogeny the terrestrial habitat is inferred to be ancestral and accounts for the largest proportion of sampled species (78.1%), followed by the epiphytic habitat (27.1%). Genera with an exclusively aquatic habitat are rare and their phylogenetic positions are primarily at the base of leptosporangiate ferns (Fig. 6). Among leptosporangiates, the evolutionary trajectory of habitat types within the order Polypodiales was well resolved: from terrestrial habitat to terrestrial/petrophyte, to petrophyte/epiphyte or petrophyte, to epiphyte (Fig. 6). Additionally, we observed that the numbers of habitat transitions away from terrestrial gradually increased from the early-diverging Dennstaedtiineae to the more derived eupolypods II and eupolypods I (Fig. 6).
4.4. Genome size and phylogenetic signalRecent studies of genome evolution in lycophytes and ferns have increasingly shed light on the complex evolutionary history of these lineages (Henry et al., 2014; Clark et al., 2016). However, evolutionary history can only be investigated within the context of a robust phylogeny.
We analyzed the phylogenetic signal of genome size (2C), chromosome number, base number of chromosomes (x), and ploidy levels (Fig. 1). Our results showed that both genome size (2C) and base number of chromosomes (x) had phylogenetic signal whereas chromosome number (2n) and ploidy level did not. Thus, our results differ from Clark et al. (2016), who found that there was a positive correlation between homoploid genome (1C) and chromosome number (2n) in ferns. According to the Blomberg's K values for both 2C (0.81) and x (1.21), we infer that genome size (2C) fluctuates within lineages, possibly due to differential gene loss events following a polyploidization event at the base of lineages, while this lability does not affect the base number of chromosomes (x), suggesting that the base number of chromosomes remains stable or is rarely lost in lineages despite gene loss or splitting into segments. Thus, the base number of chromosomes is likely under stabilizing selection, while ploidy varies according to polyploidization events, such as hybrid speciation, during evolutionary radiation.
Author contributionsFGW and AHW drafted the manuscript; DMJ, LYN, and AJ-H analyzed the data; LC, JJW performed the experiments; SYL, LX, HS, YFG, HS collected part materials and participated in data collection and analysis; LD and XCZ modified the manuscript and contributed to the supervision of the research; CKB, HFC, YHY designed the experiments, participated in interpretation for the data and results and discussions, contributed to the supervision of the research.
Declaration of competing interestThere are no conflicts of interest to declare.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.11.007.
AcknowledgementsWe thank Yu-Wen Cui for his assistance with data collection and analyses in R. This research was supported by the National Natural Science Foundation of China (grant number 31870188, 31800174, 31700172, 41571056) to Wang, Shen, Wang and Xing, Shanghai Landscaping and City Appearance Administrative Bureau of China, Scientific Research Grants (G182411) to Yan, the Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDA13020603, XDA13020500) to Chen and Jian, Guangdong Natural Science Foundation (grant number 2015A030308015) to Wang.
Bai, C.K., Alverson, W.S., Follansbee, A., et al., 2012. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann. Bot., 110: 1623-1629. DOI:10.1093/aob/mcs222 |
Baniaga, A.E., Barker, M.S., 2019. Nuclear genome size is positively correlated with median LTR-RT insertion time in fern and lycophyte genomes. Am. Fern J., 109: 248-266. DOI:10.1640/0002-8444-109.3.248 |
Baniaga, A.E., Arrigo, N., Barker, M.S., 2016. The small nuclear genomes of Selaginella are associated with a low rate of genome size evolution. Genome Biol. Evol., 8: 1516-1525. DOI:10.1093/gbe/evw091 |
Banks, J.A., Nishiyama, T., Hasebe, M., et al., 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science, 332: 960-963. DOI:10.1126/science.1203810 |
Barker, M.S., Wolf, P.G., 2010. Unfurling fern biology in the genomics age. Bioscience, 60: 177-185. DOI:10.1525/bio.2010.60.3.4 |
Barrington, D.S., Paris, C.A., Ranker, T.A., 1986. Systematic inferences from spore and stomate size in the ferns. Am. Fern J., 76: 149-159. DOI:10.2307/1547723 |
Beck, J.B., Windham, M.D., Yatskievych, G., et al., 2010. A diploids-first approach to species delimitation and interpreting polyploid evolution in the fern genus Astrolepis (Pteridaceae). Syst. Bot., 35: 223-234. DOI:10.1600/036364410791638388 |
Bennett, M.D., Leitch, I.J., 2001. Nuclear DNA amounts in pteridophytes. Ann. Bot., 87: 335-345. DOI:10.1006/anbo.2000.1339 |
Bennett, M.D., Leitch, I.J. 2012. Plant DNA C-values Database, Release 6.0. [Online] Royal Botanic Gardens, Kew. Available from http://www.kew.org/cvalues/
|
Biderre, C., Canning, E.U., Metenier, G., et al., 1999. Comparison of two isolates of Encephalitozoon hellem and E. intestinalis (Microspora) by pulsed field gel electrophoresis. Eur. J. Protistol., 35: 194-196. DOI:10.1016/S0932-4739(99)80037-6 |
Bonett, R.M., Hess, A.J., Ledbetter, N.M., 2020. Facultative transitions have trouble committing, but stable life cycles predict Salamander genome size evolution. Evol. Biol., 47: 111-122. DOI:10.1007/s11692-020-09497-8 |
Christenhusz, M.J.M., Chase, M.W., Fay, M.F., et al., 2021. Biogeography and genome size evolution of the oldest extant vascular plant genus, Equisetum (Equisetaceae). Ann. Bot., 127: 681-695. DOI:10.1093/aob/mcab005 |
Clark, J., Hidalgo, O., Pellicer, J., et al., 2016. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytol., 210: 1072-1082. DOI:10.1111/nph.13833 |
Dai, S.J., Wang, Q.X., Bao, W.M., 2002. Spore morphology of pteridophytes from China III. Thelypteridaceae 1. Cyclosorus Link. Acta Phytotaxon. Sin., 40: 334-344. |
Dai, S.J., Wang, Q.X., Bao, W.M., et al., 2005. Spore morphology of pteridophytes from China IV. Thelypteridaceae 2. Acta Phytotaxon. Sin., 43: 233-245. |
Dai, S.J., Wang, Q.X., Bao, W.M., 2005. Spore morphology of pteridophytes from China V. Aspleniaceae. Acta Phytotaxon. Sin., 43: 246-261. |
Dai, S.J., Wang, Q.X., Yu, J., et al., 2005. Spore morphology of pteridophytes from China VI. Pteridaceae. Acta Bot. Yunnan., 27: 489-500. |
David, A., 2009. Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proc. Natl. Acad. Sci. U.S.A., 106: 19699-19706. DOI:10.1073/pnas.0901635106 |
Dolezel, J., Bartos, J., 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot., 95: 99-110. DOI:10.1093/aob/mci005 |
Dolezel, J., Bartos, J., Voglmayr, H., et al., 2003. Nuclear DNA content and genome size of trout and human. Cytometry, 51: 127-128. |
Dyer, R.J., Pellicer, J., Savolainen, V., et al., 2013. Genome size expansion and the relationship between nuclear DNA content and spore size in the Asplenium monanthes fern complex (Aspleniaceae). BMC Plant Biol., 13: 219. DOI:10.1186/1471-2229-13-219 |
Fujiwara, T., Liu, H., Meza-Torres, E.I., et al., 2021. Evolution of genome space occupation in ferns: linking genome diversity and species richness. Ann. Bot., XX: 1-12. DOI:10.1093/aob/mcab094 |
Gregory, T.R., 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev., 76: 65-101. |
Gregory, T.R., 2004. Insertion-deletion biases and the evolution of genome size. Gene, 324: 15-34. DOI:10.1016/j.gene.2003.09.030 |
Gregory, T.R., 2005. Synergy between sequence and size in large-scale genomics. Nat. Rev. Genet., 6: 699-708. DOI:10.1038/nrg1674 |
Grover, C.E., Wendel, J.F., 2010. Recent insights into mechanisms of genome size change in plants. J. Bot., 2010: 382732. DOI:10.1155/2010/382732 |
Hanson, L., Leitch, I.J., 2002. DNA amounts for five pteridophyte species fill phylogenetic gaps in C-value data. Bot. J. Linn. Soc., 140: 169-173. DOI:10.1046/j.1095-8339.2002.00083.x |
Haufler, C.H., 2002. Homospory 2002: an odyssey of progress in pteridophyte genetics and evolutionary biology. Bioscience, 52: 1081-1093. DOI:10.1641/0006-3568(2002)052[1081:HAOOPI]2.0.CO;2 |
Haufler, C.H., 2014. Ever since Klekowski: testing a set of radical hypotheses revives the genetics of ferns and lycophytes. Am. J. Bot., 101: 2036-2042. DOI:10.3732/ajb.1400317 |
Hawkins, J.S., Grover, C.E., Wendel, J.F., 2008. Repeated big bangs and the expanding universe: directionality in plant genome size evolution. Plant Sci., 174: 557-562. DOI:10.1016/j.plantsci.2008.03.015 |
Henry, T.A., Bainard, J.D., Newmaster, S.G., 2014. Genome size evolution in Ontario ferns (Polypodiidae): evolutionary correlations with cell size, spore size, and habitat type and an absence of genome downsizing. Genome, 57: 555-566. DOI:10.1139/gen-2014-0090 |
Hidalgo, O., Pellicer, J., Christenhusz, M.J.M., et al., 2017. Is there an upper limit to genome size?. Trends Plant Sci., 22: 567-573. DOI:10.1016/j.tplants.2017.04.005 |
Hidalgo, O., Pellicer, J., Christenhusz, M.J.M., et al., 2017. Genomic gigantism in the whisk-fern family (Psilotaceae): Tmesipteris obliqua challenges record holder Paris japonica. Bot. J. Linn. Soc., 183: 509-514. DOI:10.1093/botlinnean/box003 |
Huang, C.H., Qi, X., Chen, D., et al., 2020. Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. J. Integr. Plant Biol., 62: 433-455. DOI:10.1111/jipb.12877 |
Ji, Y.Q., Qin, Z.Y., Wang, Q.X., et al., 2014. Development of gametophytes of Tectaria decurrens and Tectaria fauriei. Acta Bot. Bor.-Occid. Sin., 34: 689-694. |
Jiang, N., Dai, X.L., Cao, J.G., et al., 2010. Spore morphology of pteridophytes from China X. Polypodiaceae. Acta Bot. Bor.-Occid. Sin., 30: 2151-2163. |
Kang, M., Tao, J.J., Wang, J., et al., 2014. Adaptive and nonadaptive genome size evolution in Karst endemic flora of China. New Phytol., 202: 1371-1381. DOI:10.1111/nph.12726 |
Kassambara, A., Kassambara, M.A., 2020. Package ‘ggpubr’, pp. Klekowski, E.J., Baker, H.G., 1966. Evolutionary significance of polyploidy in pteridophyta. Science, 153: 305-307. |
Klekowski, E.J., Baker, H.G., 1966. Evolutionary significance of polyploidy in pteridophyta. Science, 153: 305-307. DOI:10.1126/science.153.3733.305 |
Kott, L.S., Britton, D.M., 1982. A comparative study of sporophyte morphology of three cytotypes of Polypodium virginianum in Ontario. Can. J. Bot., 60: 1360-1370. DOI:10.1139/b82-173 |
Lehtonen, S., Silvestro, D., Karger, D.N., et al., 2017. Environmentally driven extinction and opportunistic origination explain fern diversification patterns. Sci. Rep., 7: 4831. DOI:10.1038/s41598-017-05263-7 |
Leitch, A.R., Leitch, I.J., 2012. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol., 194: 629-646. DOI:10.1111/j.1469-8137.2012.04105.x |
10.1007/978-3-7091-1160-4_19 |
Li, Y.Q., Li, Y.J., Li, H., et al., 2010. A comparative study on the leaf blade structure and spore of seven Pyrrosia species in Guangxi. Guihaia, 30: 462-470. |
Little, D.P., Moran, R.C., Brenner, E.D., Stevenson, D.W., 2007. Nuclear genome size in Selaginella. Genome, 50: 351-356. DOI:10.1139/G06-138 |
Liu, J.X., 1997. Studies on the spore morphology of Equisetaceae from China. J. Guizhou Agric. Coll., 16: 31-33. |
Liu, H.M., Ekrt, L., Koutecky, P., et al., 2019. Polyploidy does not control all: lineage-specific average chromosome length constrains genome size evolution in ferns. J. Syst. Evol., 57: 418-430. DOI:10.1111/jse.12525 |
Loureiro, J., Rodriguez, E., Doležel, J., et al., 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann. Bot., 100: 875-888. DOI:10.1093/aob/mcm152 |
Lu, J.M., Li, D.Z., Wu, D., 2007. Spore morphology of the family Dryopteridaceae. Acta Bot. Yunnanica, 29: 397-408. |
Moran, R.C., 1982. The Asplenium trichomanes complex in the United States and adjacent Canada. Am. Fern J., 72: 5-11. DOI:10.2307/1547078 |
Nakazato, T., Bogonovich, M., Moyle, L.C., 2008. Environmental factors predict adaptive phenotypic differentiation within and between two wild Andean tomatoes. Evolution, 62: 774-792. DOI:10.1111/j.1558-5646.2008.00332.x |
Nakazato, T., Barker, M.S., Rieseberg, L.H., et al., 2008b. In: Ranker, T.A., Haufler, C.H., eds. Biology and Evolution of Ferns and Lycophytes. UK: Cambridge Univ. Press, 175-198
|
Obermayer, R., Leitch, I.J., Hanson, L., et al., 2002. Nuclear DNA C-values in 30 species double the familial representation in pteridophytes. Ann. Bot., 90: 209-217. DOI:10.1093/aob/mcf167 |
One Thousand Plant Transcriptomes Initiative, 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574: 679-685. DOI:10.1038/s41586-019-1693-2 |
Otto, F., 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z. eds. Methods in Cell Biology, Vol. vol. 33. New York: Academic Press, 105-110
|
Pellicer, J., Fay, M.F., Leitch, I.J., 2010. The largest eukaryotic genome of them all?. Bot. J. Linn. Soc., 164: 10-15. DOI:10.1111/j.1095-8339.2010.01072.x |
Petrov, D.A., 2002. Mutational equilibrium model of genome size evolution. Theor. Popul. Biol., 61: 531-544. DOI:10.1006/tpbi.2002.1605 |
R Core Team, 2018. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
|
R Core Team. 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org/
|
Rabinowicz, P.D., 2005. Differential methylation of genes and repeats in land plants. Genome Res., 15: 1431-1440. DOI:10.1101/gr.4100405 |
Revell, L.J., 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol., 3: 217-223. DOI:10.1111/j.2041-210X.2011.00169.x |
Schubert, I., Oud, J.L., 1997. There is an upper limit of chromosome size for normal development of an organism. Cell, 88: 515-520. DOI:10.1016/S0092-8674(00)81891-7 |
Schuettpelz, E., Pryer, K.M., 2009. Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc. Natl. Acad. Sci. U.S.A., 106: 11200-11205. DOI:10.1073/pnas.0811136106 |
Shi, X., Yang, L.H., Chen, Q., 2017. Spore morphology of genus Pyrrosia from China. Guihaia, 37: 1455-1462. |
Slijepcevic, P., 2018. Genome dynamics over evolutionary time: "C-value enigma" in light of chromosome structure. Mutat. Res-Gen. Tox. En., 836: 22-27. DOI:10.1016/j.mrgentox.2018.05.005 |
Smith, A.R.., Pryer, K.M., Schuettpelz, E., et al., 2006. A classification for extant ferns. Taxon, 55: 705-731. DOI:10.2307/25065646 |
Szövényi, P., Gunadi, A., Li, F.W., 2021. Charting the genomic landscape of seed-free plants. Nat. Plants, 7: 554-565. DOI:10.1038/s41477-021-00888-z |
The Pteridophyte Phylogeny Group, 2016. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol., 54: 563-603. DOI:10.1111/jse.12229 |
Thomas, C.A., 1971. The genetic organization of chromosomes. Annu. Rev. Genet., 5: 237-256. DOI:10.1146/annurev.ge.05.120171.001321 |
Tryon, A.F., Lugardon, B., 1991. Spores of the Pteridophyta. New York: Springer
|
Wagner, W.H.J., Wagner, F.S., 1979. Polyploidy in pteridophytes. Basic Life Sci., 13: 199-214. |
Wakamiya, I., Newton, R.J., Johnston, J.S., et al., 1993. Genome size and environmental-factors in the genus Pinus. Am. J. Bot., 80: 1235-1241. DOI:10.1002/j.1537-2197.1993.tb15360.x |
Wang, Q.X., Yu, J., Zhang, X.C., 2001. Spore morphology of pteridophytes from China I. Lygodiaceae. Acta Phytotaxon. Sin., 39: 38-44. |
Wang, R.X., Lu, S.G., Deng, X.C., et al., 2006. Spore morphology of three species of Adiantum L. from Guangxi, China. J. Guangxi Normal Univ. Nat. Sci. Ed.: J. Nat. Resour. Life Sci. Educ., 24: 79-81. |
Wang, R.X., Lu, S.G., Deng, X.C., et al., 2006. Spore morphology of pteridophytes from Guangxi I. Polypodiaceae. Guihaia, 26: 565-569. |
Wang, F.G., Liu, H.M., He, C.M., et al., 2015. Taxonomic and evolutionary implications of spore ornamentation in Davalliaceae. J. Syst. Evol., 53: 72-81. DOI:10.1111/jse.12115 |
Wang, J., Yu, J., Sun, P., et al., 2020. Paleo-polyploidization in lycophytes. Dev. Reprod. Biol., 18: 333-340. |
Webb, C.O., Ackerly, D.D., Kembel, S.W., 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics, 24: 2098-2100. DOI:10.1093/bioinformatics/btn358 |
Wendel, J.F., Cronn, R.C., Johnston, J.S., et al., 2002. Feast and famine in plant genomes. Genetica, 115: 37-47. DOI:10.1023/A:1016020030189 |
Wendel, J.F., Jackson, S.A., Meyers, B.C., et al., 2016. Evolution of plant genome architecture. Genome Biol., 17: 37. DOI:10.1186/s13059-016-0908-1 |
Wood, T.E., Takebayashi, N., Barker, M.S., et al., 2009. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. U.S.A., 106: 13875-13879. DOI:10.1073/pnas.0811575106 |
Xu, Y., Dai, X.L., Cao, J.G., 2012. Spore morphology of pteridophytes from China XI. Huperziaceae. Acta Bot. Bor.-Occid. Sin., 32: 1140-1147. |
Yang, D.M., He, R.R., Xing, F.W., et al., 2013. Study on spore and leaf epidermis morphology of Pteris (Pteridaceae) from China. Guihaia, 33: 1-19. |
Zanne, A.E., Tank, D.C., Cornwell, W.K., et al., 2013. Three keys to the radiation of angiosperms into freezing environments. Nature, 506: 89-92. |
Zhang, Y.L., Xi, Y.Z., Zhang, C.T., et al., 1976. Sporae Pteridophytorum Sinicorum. Beijing: Science Press
|
Zhang, J.M., Liu, Y.F., Dong, K., et al., 2002. Studies on the spores morphology of Psilotaceae. J. Cap. Normal Univ., 23: 49-51. |
Zhang, Y.J., Liu, Y.J., Zhou, X.M., et al., 2012. Spore morphology of Arachniodes (Dryopteridaceae) from Yunnan. Acta Bot. Bor.-Occid. Sin., 32: 2215-2223. |
Zhou, K., Liu, B.B., Wang, Y.L., et al., 2019. Evolutionary mechanism of genome duplication enhancing natural autotetraploid sea barley adaptability to drought stress. Environ. Exp. Bot., 159: 44-54. DOI:10.1016/j.envexpbot.2018.12.005 |



