b. State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, 430062, China;
c. Haiyan Engineering & Technology Center, Kunming Institute of Botany, Chinese Academy of Science, Kunming, Yunnan, 650201, China;
d. University of Chinese Academy of Sciences, Beijing, 100049, China
Drought is an extreme abiotic stress condition for plants that results in a staggering 40.8% loss in crop production, based on an analysis of the last 40 years of agronomic data in the USA (Boyer, 1982). Therefore, enhancing drought tolerance in crops is a very desirable target for agriculture (Bailey-Serres et al., 2019) to help meet the demands of the worldwide human population (FAO et al., 2020).
Plants may exhibit a degree of drought memory in response to their exposure to recurring or chronic drought stress, which may increase their drought tolerance and survival under water scarcity conditions (Avramova, 2015; Li et al., 2019). Several studies have investigated drought stress memory by transcriptome deep sequencing (RNA-seq) in Arabidopsis (Arabidopsis thaliana) (Ding et al., 2012, 2013; Liu et al., 2016), maize (Zea mays) (Ding et al., 2014; Virlouvet et al., 2018), and rice (Oryza sativa) (Li et al., 2019). However, the molecular mechanisms underlying drought memory and how it is regulated remain poorly understood.
It is, however, clear that drought stress is associated with alternative splicing (AS) in plants, such as in maize (Thatcher et al., 2016), rice (Wei et al., 2017; Zhang and Xiao, 2018), and wheat (Triticum aestivum) (Liu et al., 2018). Alternative splicing, first discovered in 1977, is the process by which one pre-mRNA generates two or more distinct transcripts, and constitutes one of the most significant post-transcriptional mechanisms controlling gene expression (Lee and Rio, 2015; Zhang et al., 2015). Alternative splicing can be classified into five basic/simple types: intron retention (IR), alternative donor (A5SS), alternative acceptor (A3SS), exon skipping (ES), and mutual exon skipping (MXE) (Shen et al., 2014; Shi et al., 2019). Most previous reports have indicated that IR is the most frequent AS type in plants, based on genome-wide analyses (Filichkin et al., 2010, 2015; Reddy et al., 2013; Thatcher et al., 2016; Wei et al., 2017; Wang et al., 2019). By contrast, in animals the most frequent AS event is ES (Reddy et al., 2013; Zhang et al., 2017b; Wang et al., 2019; Jia et al., 2020). A handful of reports in plants have identified A3SS (Tang et al., 2016; Shi et al., 2019) or ES (Xu et al., 2019; Xia et al., 2020) as the most frequent AS event, when considering large datasets comprising over 1000 AS events. Among many software tools exist for AS detection, AStalavista and rMATS are widely accepted in plants (Song et al., 2019).
The canonical GT-AG dinucleotide sequence is highly over-represented at splicing sites, and non-canonical dinucleotides are rare (Sharp and Burge, 1997). While the U2-type spliceosome commonly uses GT-AG intron sites, AT-AC can be used by the U12-type spliceosome (Tarn and Steitz, 1996; Sharp and Burge, 1997). U12 and U2 spliceosomes are conserved between animals and plants (Lorkovic et al., 2005; Reddy et al., 2013), as are the U12- and U2-type intron positions (Basu et al., 2008). In Arabidopsis, U12-type introns are enriched in genes related to DNA/RNA metabolism (Marquez et al., 2012). The conserved motif at the 5′ splice site is AAG|GTATGTT, AAG|GTAAGTT, and CAG|GTAAGTA in budding yeast (Saccharomyces cerevisiae), fission yeast (Schizosaccharomyces pombe), human (Homo sapiens), and mouse (Mus musculus), respectively (Ast, 2004). Moso bamboo (Phyllostachys edulis) shows the conserved 5’ motif (A)nG|GTAAG(T)n (Wang et al., 2017).
Here, we reveal the connection between alternative splicing events and the molecular and functional characterization of drought memory genes in rice. We discovered that ES events may be induced by drought memory. Our results may provide new targets for breeding crops with enhanced drought tolerance in the future.
2. Material and methods 2.1. Plant materials and samplingSeeds of the rice cultivar “Zhonghua 11” (Oryza sativa L. ssp. japonica) were surface sterilized with 2.5% NaClO for 15 min, rinsed with sterilized water, and then allowed to soak in sterilized water at 37 °C in an incubator. After germination, the seedlings were transferred to culture vessels containing one-quarter-strength modified Hoagland solution (Jones, 1982). Seedlings were grown in this solution for four weeks under a 12 h light: 12 h dark photoperiod at 28 °C. The Hoagland solution was changed every 2 days. We then selected four-week-old plants for the well-watered control samples (R0). We followed previously described procedures for drought memory experiments (Ding et al., 2012; Li et al., 2019) by exposing plants to air drying for 80 min at 28 °C; these samples were labeled as first drought stress treatment (S1). Plants were then re-watered after drought treatment for 22 h 40 min at 28 °C, constituting samples for first recovery treatment (R1). We repeated these stress/recovery cycles three times to enhance drought memory, annotating each sample stage as S2, R2, S3, R3, and S4 for the 2nd drought stress, 2nd recovery, 3rd drought stress, 3rd recovery, and 4th drought stress, respectively (Ding et al., 2012, 2013; Li et al., 2019).
2.2. Library preparations for RNA-seqTotal RNA from the leaves of R0, S1, R3, and S4 stage with three biological replicates was isolated with TRIzol reagent, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, United States), and then treated with RNase-free DNase I to remove trace DNA contamination. Total RNA was quantified on an Agilent Bioanalyzer 2100 instrument, and the libraries were constructed and sequenced on an Illumina HiSeq 2500 sequencing platform by Genedenovo Biotechnology, Co., Ltd. (Guangzhou, China) to generate 150-bp paired-end reads.
2.3. Data filtering, mapping of reads and differentially expressed genes (DEGs) analysisThe raw reads were filtered using Trimmomatic v.0.36 (Bolger et al., 2014) to remove adaptor sequences and low-quality reads. The filtered reads were then mapped with Hisat2 (Kim et al., 2015) to the rice japonica reference genome (ftp://ftp.ensemblgenomes.org/pub/plants/release-43/gtf/oryza_sativa/Oryza_sativa.IRGSP-1.0.43.gtf.gz, ftp://ftp.ensemblgenomes.org/pub/plants/release-43/fasta/oryza_sativa/dna/Oryza_sativa.IRGSP-1.0.dna.toplevel.fa.gz). The resulting output files were in SAM (Sequence Alignment/Map) format. We used SAMtools v.1.3.1 (Li et al., 2009) to convert SAM files into BAM (binary format of SAM file) format, followed by StringTie v.1.3.6 (Pertea et al., 2015) software to generate assembled GTF format files. We used DESeq2 v.1.32.0 (Love et al., 2014) to identify differentially expressed genes (DEGs) with log2 (|fold change|)>1 and adjusted P value < 0.05.
2.4. Analysis of AS eventsWe used AStalavista v.4.0 software (Foissac and Sammeth, 2007) with the parameters (-t asta -i) to quantify the type of AS events from the merged GTF file for all assembled transcripts. We investigated the codes for AS events according to the notes from the software author. For simple AS events, AStalavista software defined AS code “0, 1–2ˆ” for exon skipping (ES), “1ˆ, 2ˆ” for alternative donor (A5SS), “1-, 2-” for alternative acceptor (A3SS), “'0, 1ˆ2-” for intron retention (IR), and “'1–2ˆ, 3–4ˆ” for mutual exon skipping (MXE). For transcripts with more than one AS event, AStalavista uses more complex codes (Sammeth et al., 2008; Foissac and Sammeth, 2015). The AStalavista webserver (v.2.2) uses “Others” for such complex events (http://astalavista.sammeth.net/). Thus, we extracted five basic AS events types (IR, A5SS, A3SS, MXE, and ES) and the remained complex AS events were grouped as “Others” type, as described previously (Foissac and Sammeth, 2007; Min et al., 2015; Li et al., 2017).
We also used rMATS v.4.0.2 software (Shen et al., 2014) to detect differential AS events with parameters --nthread 8 --cstat 0.0001 --tstat 6 --readLength 150. Splicing events were represented with rmats2sashimiplot (https://github.com/Xinglab/rmats2sashimiplot). Dinucleotides at splicing sites were counted by comparing our GTF files with the genome fasta file using in-house scripts. Flanking sequences of splicing sites were extracted with in-house scripts, then uploaded to WebLogo v.3 (http://weblogo.threeplusone.com/) to obtain conserved sequences.
We identified transcription factors (TFs) from genes showing differential alternative splicing (DAS) by querying the PlantTFDB 4.0 database (Jin et al., 2017).
2.5. Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analysisThe agriGO (Du et al., 2010) software was used to identify enriched Gene Ontology (GO) terms. ClusterProfiler (Yu et al., 2012), and in-house scripts were employed to perform KEGG analysis. Protein–protein interaction (PPI) analysis was performed with the STRING database (Szklarczyk et al., 2019). Cytoscape 3.6.1 (Shannon et al., 2003) and TBtools v.0.66813 (Chen et al., 2018) software were used to generate figures.
2.6. RT-qPCR analysisWe initiated first-strand cDNA synthesis from 1 μg total RNA with oligo (dT) primer using the PrimeScript™ RT Reagent Kit (Takara, Japan). The relative expression level of each gene was measured with gene-specific primers (Supplementary Table S1). Real-time quantitative PCR (RT-qPCR) was performed with SYBR premix Ex Taq II kit (TaKaRa, Japan) in a total reaction volume of 20 μl on a CFX96™ system (Bio-Rad, CA, USA). The internal control was the rice ELONGATION FACTOR-1 (EF-1a, LOC_Os03g08020) gene (Li et al., 2019).
3. Results 3.1. Quality of the RNA-seq dataTo identify alternative splicing (AS) events during drought stress, we collected rice samples from well-watered control plants (R0), plants exposed to drought stress once (S1) or four times (S4), and plants re-watered after the third drought stress cycle (R3). In total, we obtained 509 million raw reads corresponding to about 76.4 Gb data across all 12 samples. After quality-trimming, we retained 486 million reads (~65.3 Gb), with an average of 40 million reads per sample. We then aligned these clean reads to the rice reference genome (IRGSP-1.0) downloaded from the Ensembl website. On average, 96% of reads mapped to the rice genome, confirming the relative high quality of the RNA-seq data (Supplementary Table S2). Principal component analysis (PCA) profiling transcript patterns in rice leaf samples between well-watered and drought stress conditions revealed large contributions of repeated stress (50% of standing variance) and drought (12% of variance) to the total observed variance (Fig. 1A).
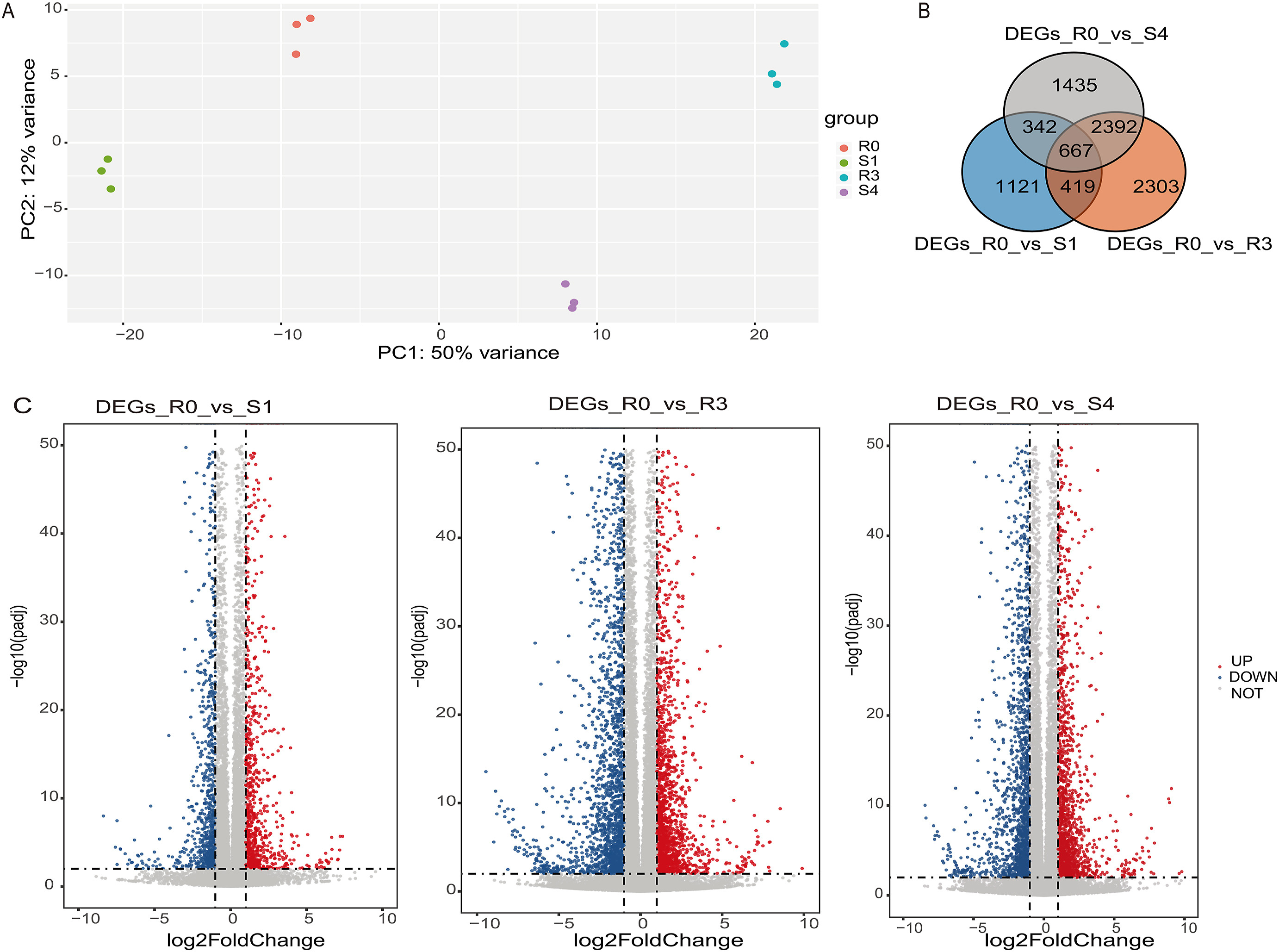
|
| Fig. 1 An overview of reference-based transcriptome assembly. A. PCA of relative isoform percentages for all samples, drought-stressed samples (S1, S4) were clearly separable from water condition samples (R0, R3). B. The results of DEGs was presented by Venn diagram, in the analysis R0 was treated as control condition. C. Volcano plot for DEGs of R0_vs_S1, R0_vs_R3 and R0_vs_S4. |
We identified 8679 DEGs when using well-watered samples (R0) as control (Fig. 1B and C). The number of DEGs almost doubled between the first and fourth drought-stress treatments (S1: 2549 DEGs; R3: 5781 DEGs; and S4: 4836 DEGs) (Supplementary Tables S3, S4 and S5). This large difference indicates that repeated cycles of drought and recovery influence gene expression in a more pronounced manner, presumably through drought memory.
To further investigate the functions associated with the three groups of DEGs mentioned above, we performed GO and KEGG enrichment analysis. We identified the GO terms GO:0019748 (“secondary metabolic process”), GO:0009875 (“pollen-pistil interaction”), GO:0007049 (“cell cycle”), GO:0009856 (“pollination”), GO:0051704 (“multi-organism process”), GO:0006259 (“DNA metabolic process”), GO:0006950 (“response to stress”), and GO:0006629 (“lipid metabolic process”) as being enriched in different drought conditions (Fig. 2A). After the 1st drought stress (R0_vs_S1), the enrichment levels of GO terms such as “secondary metabolic process” and “lipid metabolic process” significantly changed, suggesting that these two processes may be important for drought memory (Fig. 2A). KEGG enrichment analysis revealed a significant enrichment for “phenylpropanoid biosynthesis” (dosa00940), “plant hormone signal transduction” (dosa04075), “diterpenoid biosynthesis” (dosa00904), “MAPK signaling pathway – plant” (dosa04016), “DNA replication” (dosa03030), “zeatin biosynthesis” (dosa00908), “linoleic acid metabolism” (dosa00591), “starch and sucrose metabolism” (dosa00500), “galactose metabolism” (dosa00052), “brassinosteroid biosynthesis” (dosa00905), and “carotenoid biosynthesis” (dosa00906) (Fig. 2B, Supplementary Table S6). Of these KEGG terms, “phenylpropanoid biosynthesis” and “plant hormone signal transduction” showed the highest enrichment, suggesting that the associated pathways are highly affected by drought stress.
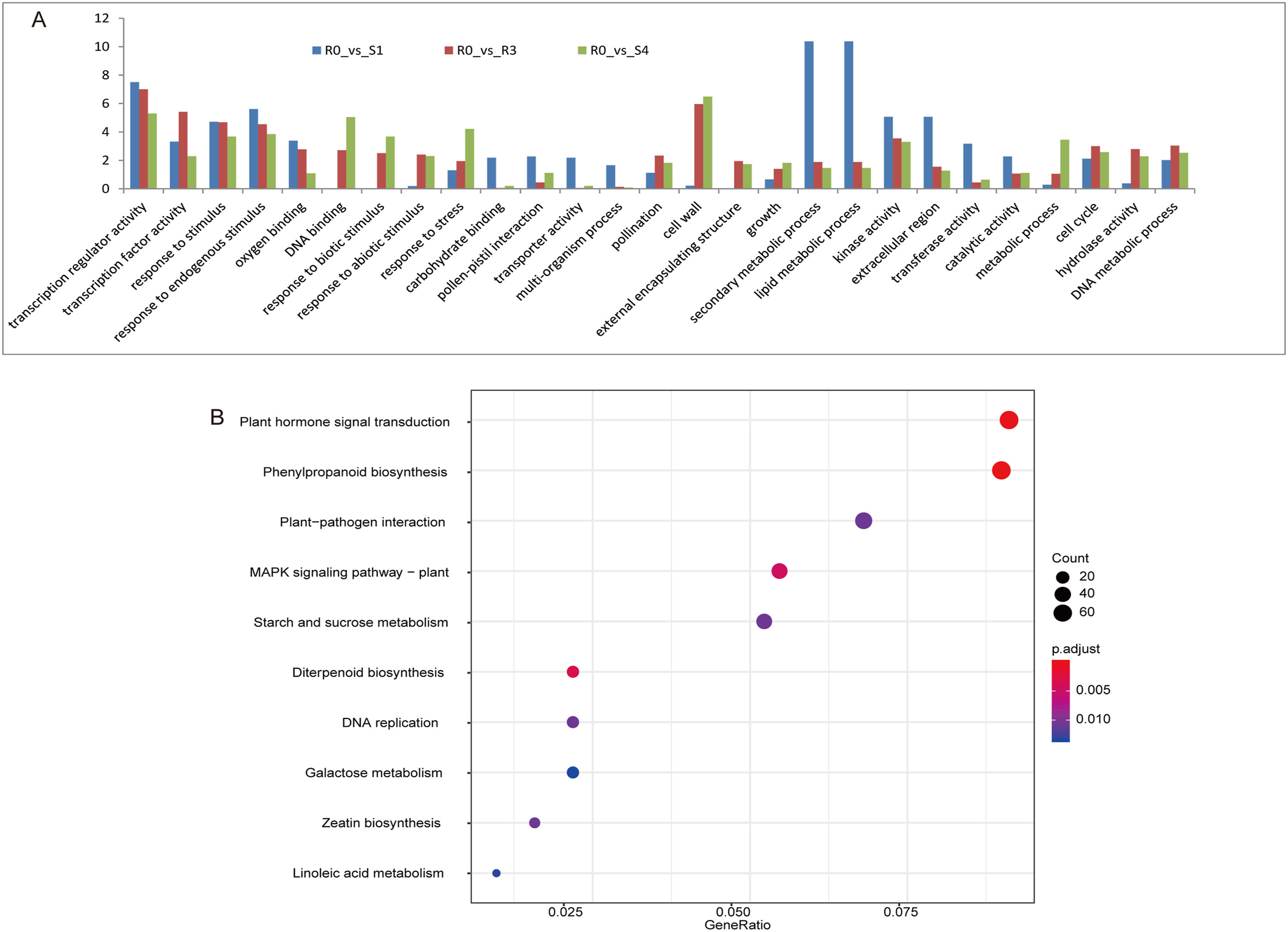
|
| Fig. 2 GO and KEGG of DEGs. A. Significant GO terms of DEGs in early stress (S1), recovery (R3) and re-stress (S4) condition when compare to the control (R0), respectively. Y-axis is -log10 (p_value). B. Top 10 KEGG classification of DEGs. |
We detected various types of AS events among our assembled transcripts, as determined by the AStalavista software. In this study, we extracted 24,558 AS events (Supplementary Table S7), which fell into six types: IR, A5SS, ES, A3SS, MXE, and Others. The numbers for each AS event type are listed in Table 1.
| AS type | Events number | Freq. (%) |
| IR | 8138 | 33.14% |
| A5SS | 3045 | 12.40% |
| ES | 2592 | 10.55% |
| A3SS | 5566 | 22.66% |
| MXE | 54 | 0.22% |
| Others | 5163 | 21.02% |
To investigate splicing events from another angle, we used the software rMATS. While AStalavista calculates the splicing events based on the GTF file of assembled transcripts at the whole-genome level, rMATS calculates differential AS events between two samples based on aligned BAM files. We thus subjected our samples to rMATS analysis to extract splicing events. Notably, the ES type was the predominant form of AS identified by rMATS. In addition, drought stress conditions (R0_vs_S1 and R0_vs_S4) resulted in more AS events (8456 and 9,586, respectively) than the stress recovery condition (R0_vs_R3, with 7265 AS events) (Fig. 3A, Table 2). This result indicates that drought stress induces differential alternative splicing.
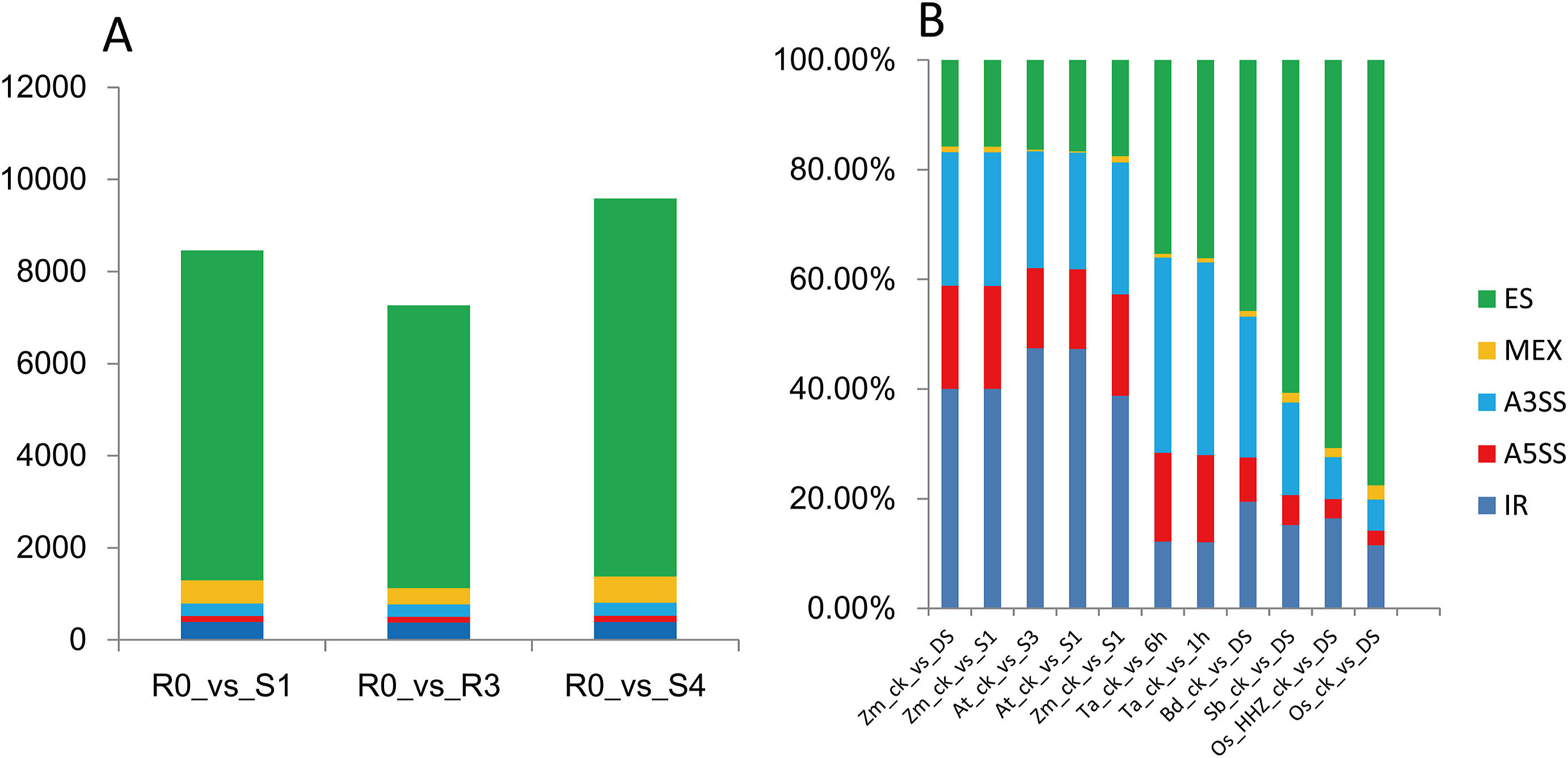
|
| Fig. 3 Differential AS events in every two-condition calculated by rMATS. A. DAS events in our 12 rice RNA-seq datasets. B. Using rMATS with raw data in different species from public database. Zm, At, Hv, Ta, Bd, Sb and Os means Zea mays, Arabidopsis thaliana, Hordeum vulgare, Brachypodium distachyon, Sorghum bicolor, Triticum aestivum and Oryza sativa, respectively. All the water condition of each sample was described in Supplementary Table S8. |
| IR | A5SS | A3SS | MXE | ES | Total | ||||||||||||||||||||||||
| R0_vs_S1 | 388 | 128 | 271 | 504 | 7165 | 8456 | |||||||||||||||||||||||
| R0_vs_R3 | 379 | 124 | 269 | 349 | 6144 | 7265 | |||||||||||||||||||||||
| R0_vs_S4 | 388 | 134 | 284 | 568 | 8212 | 9586 | |||||||||||||||||||||||
| Events_merged | 414 | 146 | 298 | 1419 | 10003 | 12280∗ | |||||||||||||||||||||||
| Gene_merged | 359 | 262 | 135 | 655 | 5835 | 6034∗ | |||||||||||||||||||||||
| Note: ∗ means 6034 was merged total genes without duplication. AS events with same gene id and same AS site position was treated as duplicated event. | |||||||||||||||||||||||||||||
To validate these results, we expanded our analysis of AS events to additional plant species exposed to drought stress with rMATS. We downloaded RNA-seq datasets from seven species from the Sequence Read Archive (SRA) database at the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/sra) (Supplementary Table S8), and processed the reads with the same pipeline used for our rice data to detect differential AS events. We discovered that ES was the largest AS type not only in rice (>70.73%), but also in sorghum (Sorghum bicolor, with 60.70%) and purple false brome (Brachypodium distachyon, with 45.76%). By contrast, IR was the largest class of AS events in Arabidopsis (47.31%) and maize (40.05%) (Fig. 3B). In wheat, ES (35.33%) and A3SS (35.61%) were the dominant AS types, while IR accounted for less than 13% of all AS events. We randomly selected one gene for each AS type to illustrate the expression and splicing events from AS genes (Fig. 4, Supplementary Fig. S1).
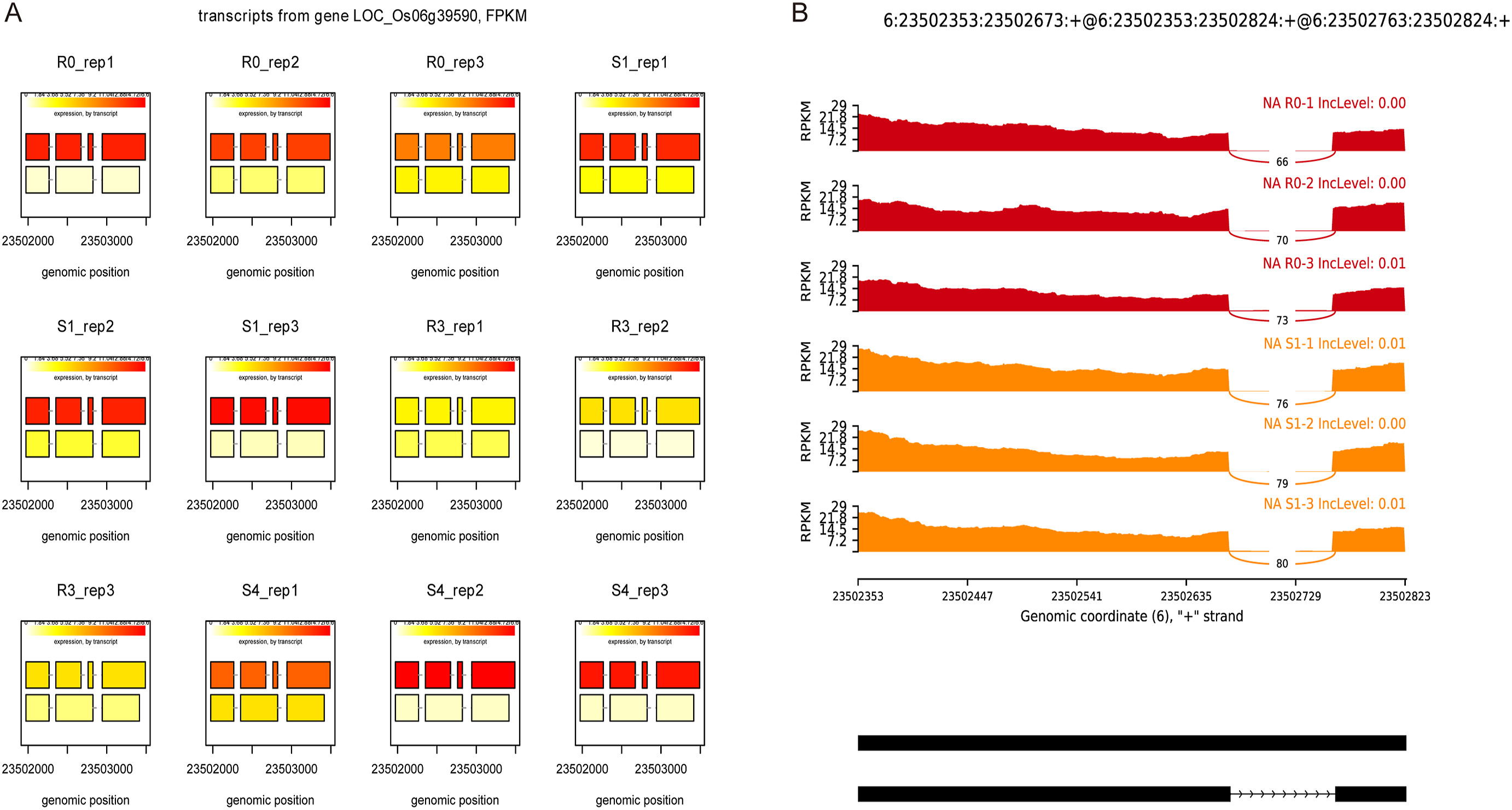
|
| Fig. 4 AS gene expression and reads on splice site. A. Expression of DAS gene LOC_Os06g39590. B. Reads distribution IR type DAS gene LOC_Os06g39590 derived from RNA-Seq data of R0 and S1. The horizontal and vertical axis represents per-base expression and genomic coordinates, respectively. The mRNA isoforms quantified has been given at the bottom; black boxes and lines with arrow heads represent exons and introns, respectively. |
We investigated the dinucleotides at splicing sites: we observed that the canonical GT-AG dinucleotide had a frequency of 92% in our RNA-seq data, which was slightly lower than the genome-wide frequency of 95% in rice (Fig. 5B). The non-canonical dinucleotide sequences GC-AG and AT-AC both increased (Fig. 5A). AT-AC is recognized by U12-type splicing, which has been reported to participate in stress transduction (Bai et al., 2019). This observation indicates that drought stress may employ more non-canonical dinucleotide splicing sites. We further extracted flanking sequence (30 bp) around splicing sites and determined that the consensus for the 5′ splicing donor site was G|GTAAGT, while the 3’ acceptor site was (T)nGCAG|G (Fig. 5C). Similar results were obtained in bamboo (Wang et al., 2017), suggesting a slight enrichment in conserved sequences at splicing sites for plants.

|
| Fig. 5 Dinucleotides at the intron border. A. Statistical analysis of noncanonical dinucleotides of splicing site between the reference genome (ref) and our assembled transcriptome (our); B, for the canonical dinucleotide. C, Conservation analysis using flanking sequences of 5′ splicing site and 3′ splicing site. |
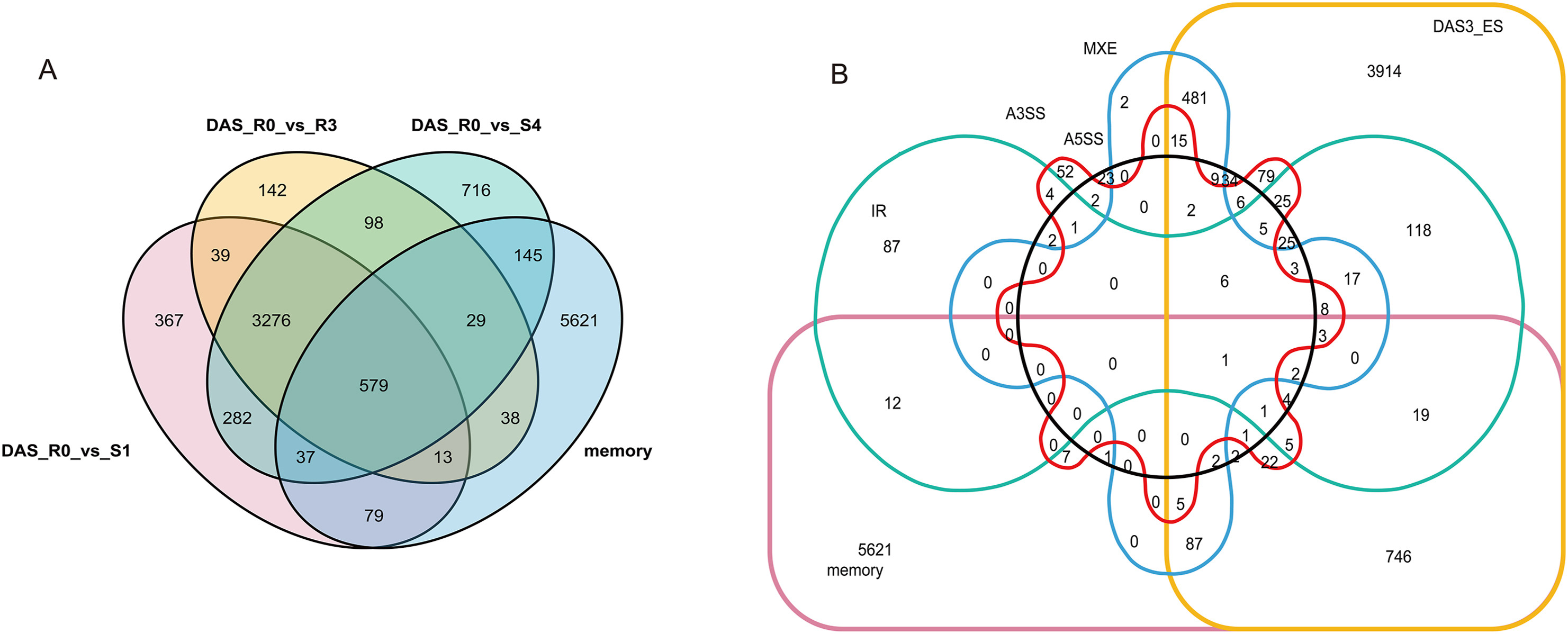
We previously identified 6885 drought memory genes (Li et al., 2019). In this study, 920 drought memory genes exhibited interesting patterns of differential alternative splicing events (DM-DAS) based on rMATS analysis (Fig. 6). GO and KEGG enrichment analysis showed different results compared to the initial set of 8679 DEGs mentioned above (Fig. 7).
|
| Fig. 6 Venn diagram for differential AS genes in comparison with drought memory genes. Venn diagram for differential AS genes in comparison with drought memory genes. Show 920 AS genes belong to drought memory genes. A. Compared by drought groups. B. Compared by alternative splicing types of DAS. |

|
| Fig. 7 Top 10 KEGG classifications of DAS genes in different compare groups. A. R0_S1; B. R0_R3; C. R0_S4; D. merged DAS genes from 3 above groups. |
Using these 920 DM-DAS genes as seeds, we performed a search in the STRING database, resulting in the identification of three well-known genes as hubs involved in drought memory: OS03T0738400-01, encoding serinehydroxymethyltransferase 1 (OsSHMT1, LOC_Os03g52840), OsJ_35887, encoding chloroplast stem-loop-binding protein of 41 kDa b (CSP41b, also called Light-Green Leaf 1 [LGL1], LOC_Os12g23180), and Phosphoinositide-dependent protein kinase 1 (PPDK1, LOC_Os05g33570) (Supplementary Fig. S2).
In order to better understand the transcriptional mechanisms underlying drought memory, we interrogated our DM-DAS genes for transcription factors (TFs), leading to the identification of 20 TFs. Two TFs belonged to the auxin response factor (ARF) family: LOC_Os05g48870 (OsETTIN1/OsARF15) and LOC_Os06g46410 (OsARF17). Another three genes encoded members of the plant-specific B3 superfamily (B3) of TFs: LOC_Os01g13300, LOC_Os07g37610 (GERMINATION DEFECTIVE 1, [GD1]), and LOC_Os10g17630. Four genes coded for members of the basic/helix-loop-helix (bHLH) superfamily: LOC_Os01g72370 (IRON-RELATED TRANSCRIPTION FACTOR 2, OsIRO2/OsbHLH056), LOC_Os03g43810 (PHYTOCHROME INTERACTING FACTOR 12, OsPIL12/OsbHLH103), LOC_Os07g43530 (OsbHLH1), and LOC_Os11g38870 (OsbHLH061). Three genes encoded bZIP TFs: LOC_Os01g64730 (ABSCISIC ACID [ABA] RESPONSIVE ELEMENT-BINDING FACTOR 1, ABF1), LOC_Os02g52780 (OsbZIP23), and LOC_Os11g05640 (OsbZIP80/OsZIP-2a). Three genes encoded members of the MYB TF family: LOC_Os12g13570, LOC_Os08g06110 (LATE ELONGATED HYPOCOTYL, OsLHY), and LOC_Os12g41920 (TELOMERE REPEAT-BINDING FACTOR 2, OsTRBF2). Finally, we identified one member each from the TF families G2-like, GRAS, NFYA, and YABBY, respectively: LOC_Os06g24070 (GOLDEN-LIKE 1, OsGLK1), LOC_Os03g48450 (OsGRAS17), LOC_Os02g53620 (NUCLEAR FACTOR-YA11, OsNF-YA11), and LOC_Os02g42950 (OsYABBY4) (Table 3).
| geneID | TF family | Symbol | Description | Reference |
| LOC_Os05g48870 | ARF | OsETTIN1 , OsARF15 | auxin response, carpel differentiation | Khanday etal. (2013) |
| LOC_Os06g46410 | ARF | OsARF17 | auxin response, disease resistance | Zhang etal. (2020) |
| LOC_Os01g13300 | B3 | |||
| LOC_Os07g37610 | B3 | GD1 | regulating GA and carbohydrate homeostasis | Guo etal. (2013) |
| LOC_Os10g17630 | B3 | |||
| LOC_Os01g72370 | bHLH | OsIRO2,OsbHLH056 | Fe relationed | Ogo etal. (2007) |
| LOC_Os03g43810 | bHLH | OsPIL12,OsBP-5,OsbHLH103 | regulate the rice Wx gene | Zhu etal. (2003) |
| LOC_Os07g43530 | bHLH | OsbHLH1 | enhancing cold tolerance | Wang etal. (2003) |
| LOC_Os11g38870 | bHLH | OsbHLH061 | drought tolerance | Li etal. (2006) |
| LOC_Os01g64730 | bZIP | OsABF1 | drought tolerance | Zhang etal. (2017a) |
| LOC_Os02g52780 | bZIP | OsbZIP23 | drought tolerance | Xiang etal. (2008) |
| LOC_Os11g05640 | bZIP | OsbZIP80,OsZIP-2a | inactivate some members of bZIP family | Nijhawan etal. (2008) |
| LOC_Os06g24070 | G2-like | OsGLK1 | activator of nuclear photosynthetic genes | Nakamura etal. (2009) |
| LOC_Os03g48450 | GRAS | OsGRAS17, OsSCL14 | regulating GA signaling pathway | Zhou etal. (2015) |
| LOC_Os04g51190 | GRF | OsGRF3 | regulating gibberellin-induced stem elongation | Kuijt etal. (2014) |
| LOC_Os12g13570 | MYB | heat stress | Chen etal. (2016) | |
| LOC_Os08g06110 | MYB_related | OsLHY | circadian clock CCA1 ortholog; ABA biosynthsis | Adams etal. (2018) |
| LOC_Os12g41920 | MYB_related | OsTRBF2 | relatived with telomeres | Byun etal. (2008) |
| LOC_Os02g53620 | NF-YA | OsNF-YA11 | down-regulated under drought stress | Yang etal. (2017) |
| LOC_Os02g42950 | YABBY | OsYABBY4 | associated with the GA signaling pathway | Liu etal. (2007) |
We selected six genes for RT-qPCR analysis of their transcript levels. Our results suggested a good correlation between RNA-seq data and RT-qPCR data (Supplementary Fig. S3, Supplementary Table S1).
4. Discussion 4.1. DM-DAS genes might have very important functionDespite the identification of drought memory genes in multiple plant species such as rice (Li et al., 2019), Arabidopsis (Ding et al., 2012), and maize (Virlouvet et al., 2018), the molecular landscape of plant drought memory has remained unclear. Previous studies have suggested that drought memory may rely on alternative splicing of specific genes, for example CBL-INTERACTING PROTEIN KINASE (CIPK) (Wan et al., 2019), DEHYDRATION RESPONSE ELEMENT BINDING 2B (OsDREB2B) (Matsukura et al., 2010), and DROUGHT RESPONSIVE ANKYRIN 1 (DRA1) (Guerra et al., 2015). Here, we reveal a much larger complement of 920 DM-DAS genes in rice, providing a large candidate gene list for future analysis of drought resistance mechanisms (Fig. 6).
We also detected several hub DM-DAS genes which could be widely connected to other genes: for example, the protein encoded by LOC_Os03g52840 (OsSHMT1, OS03T0738400-01) was shown to enhance chilling tolerance and interacted with the heat shock protein Hsp70 (Fang et al., 2020). Transgenic rice plants overexpressing OsSHMT1 exhibited increased photosynthetic efficiency and improved plant productivity (Wu et al., 2015). The gene ZmPPDK1 encodes an essential photosynthetic enzyme in maize, a C4 plant. By contrast, the rice ortholog OsPPDK1 (LOC_Os05g33570, Os05g0405000) has no role in photosynthesis but rather plays an important role in starch biosynthesis and during seed development (Kang et al., 2005; Zhang et al., 2018; Wang et al., 2020).
It has been shown that TF genes also undergo AS events during stress response (Mastrangelo et al., 2012). In this study, we identified 20 TFs out of the set of 920 DM-DAS, some of which have previously been linked to drought stress response (Table 3). For example, OsbZIP23 is a key gene conferring ABA-dependent drought tolerance (Xiang et al., 2008). OsbZIP23 is also associated with histone H3K4me3 modification at the chromatin around dehydrin genes to regulate their expression (Zong et al., 2020). Likewise, OsABF1 regulates the expression of PROTEIN PHOSPHATASE 2C (PP2C) and bZIP23, which then feed back onto the drought/ABA signaling pathway (Zhang et al., 2017a). We therefore hypothesize that DM-DAS genes have great potential as drought-related genes, and provide a list of high-confidence candidates for future analysis.
4.2. Why is ES the most frequent AS events type under drought stress conditions in rice?It is thought that intron retention (IR) constitutes the main type of AS event in plants, while exon skipping (ES) is the most prominent AS type in animals (Reddy et al., 2013; Zhang et al., 2015). However, our study suggests that ES is the dominant AS type among DAS events in rice plants under drought stress (Fig. 3A).
One possible reason to explain discrepancies between earlier analysis and our own was software bias, since rMATS and AStalavista were both designed to analyze animal data where ES is predominant. To eliminate this possibility, we investigated other plant datasets using rMATS. Notably, the reanalysis of these publicly available datasets showed the same high ES frequency in sorghum (60.70%), whereas ES frequency in maize, Arabidopsis and barley (Hordeum vulgare) were much lower (9.94%–16.67%), and the frequency in wheat and purple false brome was intermediate (35.33% and 45.76%, respectively) (Fig. 3B). These variable frequencies did not support a systematic bias caused by the software platform used.
A second potential reason for the differences in outcome called upon missing codes for complex AS events in AStalavista in a subset of earlier studies, as not all reports included complex/Others AS types when using AStalavista on plant samples. For example, when using only the simplest AStalavista code “0, 1–2ˆ” to extract ES events (Foissac and Sammeth, 2015), many ES events including under the “Others” category would be missed. A previous study in Oryza sativa ssp. japonica, Oryza sativa ssp. indica, sorghum and maize using AStalavista stated that IR constituted the “major common AS type” in these four species/subspecies, while the frequency of complex/Others types of AS events were 37.8%, 16.9%, 61.7%, and 20.4%, respectively (Min et al., 2015). In Arabidopsis, simple IR code: “0, 1ˆ2-” from AStalavista extracted just a little over half of all IR events, yielding a frequency of 24.21% when the total IR frequency is in fact ~40% (Marquez et al., 2012). In cabbage (Brassica oleracea), A3SS (10.31%) was the most frequent simple AS event, while complex AS (69.86%) was predominant (Xu et al., 2019). Thus, ES frequencies in plants may have been under-estimated. Since the rMATS software was released in 2014, few have paid attention to the relative frequencies of AS types, and our understandings of this area are under the ongoing development. When we applied AStalavista to all the data previously analyzed by rMATS, IR was the most prevalent type, while the frequency of complex/Others type varieties from 15.07% to 48.77% (Supplementary Table S9). These results therefore indicate that many ES events that belong to the complex/Others type may be under-represented in several earlier studies.
Last, another possible reason for the relative frequencies of AS events observed here was annotation bias in each genome. However, the majority of AS events corresponded to IR (83%) in 141 rice samples under different nutrient conditions that were analyzed by rMATS with two different annotations (MSU and RAPDB) of the rice genome (Dong et al., 2018). This study saw no annotation bias when using rMATS.
We thus hypothesize that 1) the high frequency of ES events during drought memory conditions in rice may be a direct consequence of drought stress on AS, and 2) the relative frequency of ES events is species specific. We propose the term “drought-stress related ES” for this hypothesis.
5. ConclusionThis study may be the first report to show that ES is the predominant form of alternative splicing in rice plants subjected to drought stress. This result differs greatly from those of most previous studies in plants, and may challenge currently accepted knowledge in the field. Based on our analysis and reasoning, we propose the hypothesis — “drought-stress related ES” — to explain why ES is the main AS event during drought stress in rice. This hypothesis should be further examined experimentally and computationally.
Declaration of competing interestThe author declares no conflict of interest.
AcknowledgementsThis study was funded by National Natural Science Foundation of China (31571311 to CZ and 31971410 to LL) and the CAS Pioneer Hundred Talents Program (CZ and LL).
Appendix A Supplementary dataThe following are the Supplementary data to this article: https://doi.org/10.1016/j.pld.2020.11.004.
Author contributionsCJ, LL, GJ and HY designed the experiments and analysis of data, drafting or revising the article. PL, DG and HY plant culture and performed the experiments. All authors reviewed and approved the manuscript.
Adams, S., Grundy, J., Veflingstad, S.R., et al., 2018. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol., 220: 893-907. DOI:10.1111/nph.15415 |
Ast, G., 2004. How did alternative splicing evolve?. Nat. Rev. Genet., 5: 773-782. |
Avramova, Z., 2015. Transcriptional 'memory' of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J., 83: 149-159. DOI:10.1111/tpj.12832 |
Bai, F., Corll, J., Shodja, D.N., et al., 2019. RNA binding motif protein 48 Is required for U12 splicing and maize endosperm differentiation. Plant Cell, 31: 715-733. DOI:10.1105/tpc.18.00754 |
Bailey-Serres, J., Parker, J.E., Ainsworth, E.A., et al., 2019. Genetic strategies for improving crop yields. Nature, 575: 109-118. DOI:10.1038/s41586-019-1679-0 |
Basu, M.K., Makalowski, W., Rogozin, I.B., et al., 2008. U12 intron positions are more strongly conserved between animals and plants than U2 intron positions. Biol. Direct, 3: 19. DOI:10.1186/1745-6150-3-19 |
Bolger, A.M., Lohse, M., Usadel, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30: 2114-2120. DOI:10.1093/bioinformatics/btu170 |
Boyer, J.S., 1982. Plant productivity and environment. Science, 218: 443-448. DOI:10.1126/science.218.4571.443 |
Byun, M.Y., Hong, J.P., Kim, W.T., 2008. Identification and characterization of three telomere repeat-binding factors in rice. Biochem. Biophys. Res. Commun., 372: 85-90. DOI:10.1016/j.bbrc.2008.04.181 |
Chen, C., Begcy, K., Liu, K., et al., 2016. Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant. Physiol., 171: 606-622. DOI:10.1104/pp.15.01992 |
Chen, C., Xia, R., Chen, H., et al., 2018. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv: 10.1101/289660. DOI:10.1101/289660 |
Ding, Y., Fromm, M., Avramova, Z., 2012. Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nat. Commun., 3: 740. DOI:10.1038/ncomms1732 |
Ding, Y., Liu, N., Virlouvet, L., et al., 2013. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol., 13: 229. DOI:10.1186/1471-2229-13-229 |
Ding, Y., Virlouvet, L., Liu, N., et al., 2014. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol., 14: 141. DOI:10.1007/s00228-013-1604-7 |
Dong, C., He, F., Berkowitz, O., et al., 2018. Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell, 30: 2267-2285. DOI:10.1105/tpc.18.00051 |
Du, Z., Zhou, X., Ling, Y., et al., 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res., 38: W64-W70. DOI:10.1093/nar/gkq310 |
Fang, C., Zhang, P., Li, L., et al., 2020. Serine hydroxymethyltransferase localised in the endoplasmic reticulum plays a role in scavenging H2O2 to enhance rice chilling tolerance. BMC Plant Biol., 20: 236. DOI:10.1186/s12870-020-02446-9 |
FAO, IFAD, UNICEF, et al. 2020. In Brief to The State of Food Security and Nutrition in the World 2020. Rome: FAO
|
Filichkin, S.A., Priest, H.D., Givan, S.A., et al., 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res., 20: 45-58. DOI:10.1101/gr.093302.109 |
Filichkin, S., Priest, H.D., Megraw, M., et al., 2015. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol., 24: 125-135. DOI:10.1016/j.pbi.2015.02.008 |
Foissac, S., Sammeth, M., 2007. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res., 35: W297-W299. DOI:10.1093/nar/gkm311 |
Foissac, S., Sammeth, M., 2015. Analysis of alternative splicing events in custom gene datasets by AStalavista. Methods Mol. Biol., 1269: 379-392. DOI:10.1007/978-1-4939-2291-8_24 |
Guerra, D., Crosatti, C., Khoshro, H.H., et al., 2015. Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider's web of mechanisms. Front. Plant Sci., 6: 57. |
Guo, X., Hou, X., Fang, J., et al., 2013. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J., 75: 403-416. DOI:10.1111/tpj.12209 |
Jia, J., Long, Y., Zhang, H., et al., 2020. Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat Plants, 6: 780-788. DOI:10.1038/s41477-020-0688-1 |
Jin, J., Tian, F., Yang, D.C., et al., 2017. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res., 45: D1040-D1045. DOI:10.1093/nar/gkw982 |
1982. Hydroponics: its history and use in plant nutrition studies. J. Plant Nutr., 5: 1003-1030. DOI:10.1080/01904168209363035 |
Kang, H.G., Park, S., Matsuoka, M., et al., 2005. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J., 42: 901-911. DOI:10.1111/j.1365-313X.2005.02423.x |
Khanday, I., Yadav, S.R., Vijayraghavan, U., 2013. Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant. Physiol., 161: 1970-1983. DOI:10.1104/pp.112.212423 |
Kim, D., Langmead, B., Salzberg, S.L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12: 357-360. DOI:10.1038/nmeth.3317 |
Kuijt, S.J., Greco, R., Agalou, A., et al., 2014. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant. Physiol., 164: 1952-1966. DOI:10.1104/pp.113.222836 |
Lee, Y., Rio, D.C., 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem., 84: 291-323. DOI:10.1146/annurev-biochem-060614-034316 |
Li, X., Duan, X., Jiang, H., et al., 2006. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant. Physiol., 141: 1167-1184. DOI:10.1104/pp.106.080580 |
Li, H., Handsaker, B., Wysoker, A., et al., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25: 2078-2079. DOI:10.1093/bioinformatics/btp352 |
Li, Y., Dai, C., Hu, C., et al., 2017. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J., 90: 164-176. DOI:10.1111/tpj.13462 |
Li, P., Yang, H., Wang, L., et al., 2019. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet., 10: 55. DOI:10.3390/nano10010055 |
Liu, H.L., Xu, Y.Y., Xu, Z.H., et al., 2007. A rice YABBY gene, OsYABBY4, preferentially expresses in developing vascular tissue. Dev. Gene. Evol., 217: 629-637. DOI:10.1007/s00427-007-0173-0 |
Liu, N., Staswick, P.E., Avramova, Z., 2016. Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ., 39: 2515-2529. DOI:10.1111/pce.12806 |
Liu, Z., Qin, J., Tian, X., et al., 2018. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J, 16: 714-726. DOI:10.1111/pbi.12822 |
Lorkovic, Z.J., Lehner, R., Forstner, C., et al., 2005. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA, 11: 1095-1107. DOI:10.1261/rna.2440305 |
Love, M.I., Huber, W., Anders, S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15: 550. DOI:10.1186/s13059-014-0550-8 |
Marquez, Y., Brown, J.W., Simpson, C., et al., 2012. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res., 22: 1184-1195. DOI:10.1101/gr.134106.111 |
Mastrangelo, A.M., Marone, D., Laido, G., et al., 2012. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci., 185–186: 40-49. |
Matsukura, S., Mizoi, J., Yoshida, T., et al., 2010. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom., 283: 185-196. DOI:10.1007/s00438-009-0506-y |
Min, X.J., Powell, B., Braessler, J., et al., 2015. Genome-wide cataloging and analysis of alternatively spliced genes in cereal crops. BMC Genom., 16: 721. DOI:10.1186/s12864-015-1914-5 |
Nakamura, H., Muramatsu, M., Hakata, M., et al., 2009. Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol., 50: 1933-1949. DOI:10.1093/pcp/pcp138 |
Nijhawan, A., Jain, M., Tyagi, A.K., et al., 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant. Physiol., 146: 333-350. |
Ogo, Y., Itai, R.N., Nakanishi, H., et al., 2007. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J., 51: 366-377. DOI:10.1111/j.1365-313X.2007.03149.x |
Pertea, M., Pertea, G.M., Antonescu, C.M., et al., 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol., 33: 290-295. DOI:10.1038/nbt.3122 |
Reddy, A.S., Marquez, Y., Kalyna, M., et al., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell, 25: 3657-3683. DOI:10.1105/tpc.113.117523 |
Sammeth, M., Foissac, S., Guigo, R., 2008. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol., 4: e1000147. DOI:10.1371/journal.pcbi.1000147 |
Shannon, P., Markiel, A., Ozier, O., et al., 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13: 2498-2504. DOI:10.1101/gr.1239303 |
Sharp, P.A., Burge, C.B., 1997. Classification of introns: U2-type or U12-type. Cell, 91: 875-879. DOI:10.1016/S0092-8674(00)80479-1 |
Shen, S., Park, J.W., Lu, Z.X., et al., 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A., 111: E5593-E5601. |
Shi, Y., Su, Z., Yang, H., et al., 2019. Alternative splicing coupled to nonsense-mediated mRNA decay contributes to the high-altitude adaptation of maca (Lepidium meyenii). Gene, 694: 7-18. DOI:10.1016/j.gene.2018.12.082 |
Song, Q.A., Catlin, N.S., Brad Barbazuk, W., et al., 2019. Computational analysis of alternative splicing in plant genomes. Gene, 685: 186-195. DOI:10.1016/j.gene.2018.10.026 |
Szklarczyk, D., Gable, A.L., Lyon, D., et al., 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res., 47: D607-D613. DOI:10.1093/nar/gky1131 |
Tang, W., Zheng, Y., Dong, J., et al., 2016. Comprehensive transcriptome profiling reveals long noncoding RNA expression and alternative splicing regulation during fruit development and ripening in kiwifruit (Actinidia chinensis). Front. Plant Sci., 7: 335. |
Tarn, W.Y., Steitz, J.A., 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell, 84: 801-811. DOI:10.1016/S0092-8674(00)81057-0 |
Thatcher, S.R., Danilevskaya, O.N., Meng, X., et al., 2016. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant. Physiol., 170: 586-599. DOI:10.1104/pp.15.01267 |
Virlouvet, L., Avenson, T.J., Du, Q., et al., 2018. Dehydration stress memory: gene networks linked to physiological responses during repeated stresses of Zea mays. Front. Plant Sci., 9: 1058. DOI:10.3389/fpls.2018.01058 |
Wan, X., Zou, L.H., Zheng, B.Q., et al., Circadian regulation of alternative splicing of drought-associated CIPK genes in Dendrobium catenatum (Orchidaceae). Int. J. Mol. Sci.. |
Wang, Y.J., Zhang, Z.G., He, X.J., et al., 2003. A rice transcription factor OsbHLH1 is involved in cold stress response. Theor. Appl. Genet., 107: 1402-1409. DOI:10.1007/s00122-003-1378-x |
Wang, T., Wang, H., Cai, D., et al., 2017. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J., 91: 684-699. DOI:10.1111/tpj.13597 |
Wang, X., Yang, M., Ren, D., et al., 2019. Cis-regulated alternative splicing divergence and its potential contribution to environmental responses in Arabidopsis. Plant J., 97: 555-570. |
Wang, H., Ham, T.H., Im, D.E., et al., A new SNP in rice gene encoding pyruvate phosphate dikinase (PPDK) associated with floury endosperm. Genes. |
Wei, H., Lou, Q., Xu, K., et al., 2017. Alternative splicing complexity contributes to genetic improvement of drought resistance in the rice maintainer HuHan2B. Sci. Rep., 7: 11686. DOI:10.1038/s41598-017-12020-3 |
Wu, J., Zhang, Z., Zhang, Q., et al., 2015. The molecular cloning and clarification of a photorespiratory mutant, oscdm1, using enhancer trapping. Front. Genet., 6: 226. |
Xia, W., Liu, R., Zhang, J., et al., 2020. Alternative splicing of flowering time gene FT is associated with halving of time to flowering in coconut. Sci. Rep., 10: 11640. DOI:10.1038/s41598-020-68431-2 |
Xiang, Y., Tang, N., Du, H., et al., 2008. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant. Physiol., 148: 1938-1952. DOI:10.1104/pp.108.128199 |
Xu, Y., Zeng, A., Song, L., et al., 2019. Comparative transcriptomics analysis uncovers alternative splicing events and molecular markers in cabbage (Brassica oleracea L.). Planta, 249: 1599-1615. DOI:10.1007/s00425-019-03108-3 |
Yang, W., Lu, Z., Xiong, Y., et al., 2017. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crops J, 5: 21-31. DOI:10.1016/j.cj.2016.06.014 |
Yu, G., Wang, L.G., Han, Y., et al., 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16: 284-287. DOI:10.1089/omi.2011.0118 |
Zhang, Z., Xiao, B., 2018. Comparative alternative splicing analysis of two contrasting rice cultivars under drought stress and association of differential splicing genes with drought response QTLs. Euphytica, 214: 73. DOI:10.1145/3297730.3297732 |
Zhang, C., Yang, H., Yang, H., 2015. Evolutionary character of alternative splicing in plants. Bioinf. Biol. Insights, 9: 47-52. |
Zhang, C., Li, C., Liu, J., et al., 2017. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot., 68: 4695-4707. DOI:10.1093/jxb/erx260 |
Zhang, R., Calixto, C.P.G., Marquez, Y., et al., 2017. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res., 45: 5061-5073. DOI:10.1093/nar/gkx267 |
Zhang, L., Zhao, L., Lin, L., et al., A novel mutation of OsPPDKB, encoding pyruvate orthophosphate dikinase, affects metabolism and structure of starch in the rice endosperm. Int. J. Mol. Sci.. |
Zhang, H., Li, L., He, Y., et al., 2020. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. U. S. A., 117: 9112-9121. DOI:10.1073/pnas.1918254117 |
Zhou, W., Malabanan, P.B., Abrigo, E., 2015. OsHox4 regulates GA signaling by interacting with DELLA-like genes and GA oxidase genes in rice. Euphytica, 201: 97-107. DOI:10.1007/s10681-014-1191-4 |
Zhu, Y., Cai, X.L., Wang, Z.Y., et al., 2003. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J. Biol. Chem., 278: 47803-47811. DOI:10.1074/jbc.M302806200 |
Zong, W., Yang, J., Fu, J., et al., 2020. Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice. J. Integr. Plant Biol., 62: 723-729. DOI:10.1111/jipb.12850 |



