b. Key Laboratory of Tobacco Biotechnological Breeding, Kunming, Yunnan, China;
c. National Tobacco Genetic Engineering Research Center, Kunming, Yunnan, China;
d. National Tobacco Gene Research Centre, Zhengzhou Tobacco Research Institute, Zhengzhou, Henan, China;
e. Institute ofBioengineering, Guangdong Academy of Sciences, Guangzhou, Guangdong, 510316, China
As one of three largest gene families, the basic helixeloopehelix (bHLH) proteins are widely distributed in eukaryotes (Jones 2004; Pires and Dolan 2010; Moore et al., 2000; Riechmann et al., 2000). Numerous studies revealed that the bHLH genes were participated in plant growth and development, and secondary metabolism as well as the responses to various stresses (Oh et al., 2004; Groszmann et al., 2010; Farquharson 2016; Nakata et al., 2013; Shoji and Hashimoto 2011; Zhang et al., 2012). The bHLH gene, MyoD, was first identified in the bacterium (Murre et al., 1989). The bHLH gene family members contain a bHLH domain, which is consisted of 50—60 amino acids (AA) including a basic region and an HLH region with diverse functions (Murre et al., 1989; Toledo-Ortiz et al., 2003). The basic region was used to bind to the DNA which contains 17 hydrophilic and basic amino acids at the N terminus of bHLH domain. Actually, the bHLH proteins were usually bind to the E-box (CANNTG) (Atchley et al., 1999; Massari and Murre 2000; Ferre-D'Amare et al., 1994; Ledent and Vervoort 2001). While the HLH region comprises two amphipathic helices and a loop between two helices at the C terminus of bHLH domain, which might result in the bHLH protein homodimerize with itself or heterodimerize with other bHLH proteins. Therefore, the bHLH proteins play diverse functions in eukaryotes which might due to their formation of many dimers (Murre et al., 1989; Atchley et al., 1999; Massari and Murre 2000).
The bHLH gene family was divided into six groups (from A to F) according to their binding motifs in the bHLH proteins in eukaryotes (Ledent and Vervoort 2001). The bHLH proteins from group A mainly bound to the E-box variant CAGCTG or CACCTG, such as MyoD, Twist and Net genes. The bHLH proteins from group B could bind to the CACGTG or CATGTTG motif, and the CACGTG motif which is commonly known as G-box, including the Mad, Max and Myc genes. The bHLH proteins from group C had a PAS domain which could bind to the ACGTG or GCGTG motif. The bHLH proteins from group D did not have a basic region which resulted in their failing to bind the DNA. However, the bHLH proteins from group D could antagonize with group A proteins by forming the dimers. The bHLH proteins from group E were usually bound to the CACGCG or CACGAG (N-box) motif. The bHLH proteins from group F which contained more than one COE domain are involved in the COE (Col/Olf-1/EBF) dimerization and DNA binding. However, the bHLH proteins from group F were not identified in plants, and the proteins from group B were appraised as the main bHLH proteins in plants.
The bHLH gene family has been widely studied in plants including Arabidopsis thaliala (Toledo-Ortiz et al., 2003; Bailey et al., 2003), Solanum lycopersicum (Sun et al., 2015), Chinese jujube (Li et al. 2019, Brachypodium distachyon (Niu et al. 2017, poplar (Zhao et al., 2018a), otton (Lu et al., 2018), Dendrobium officinale (Wang and Liu 2020), and Moso bamboo (Cheng et al., 2018). Numerous studies revealed that the bHLH genes showed multiple roles in the process of plant life, and identification of candidate members is the first step to investigate their function. Tobacco (Nicotiana tabacum L.) is an allotetraploid plant, and has been served as a typical model plant for analyzing the gene function for plant growth, development, and response to environmental stress. The NtMYC2 gene is a classical bHLH transcription factor, and the NtMYC2 gene is involved in the jasmonic acid (JA) signaling pathway which might regulate the content of nicotine, one of typical alkaloid in tobacco. NtMYC2 could bind to the G-box motif in the promoters of NtPMT and NtQPT gene for activating their expression, respectively (Shoji and Hashimoto 2011; Zhang et al., 2012). Previous study revealed that there were 190 NtbHLH genes in tobacco (Rushton et al., 2008), however, there might be some missing NtbHLH genes due to the low quality of tobacco genome, and a more comprehensive investigation is required with newly obtained high quality tobacco genome. In the current study, we identified 309 NtbHLH genes whose amount is far more than previous study. Moreover, the NtbHLH proteins were conserved during evolution through analyzing the amino acids, gene structures and motifs. Transcriptome profiles of 309 NtbHLH genes were then explored in 23 tobacco tissues from gene-chip data, and expression patterns of 20 randomly selected NtbHLH genes were further confirmed by quantitative real-time PCR, revealing that the NtbHLH genes might be participated in the regulation of plant growth and development. Therefore, our study would provide valuable information for further investigating their function of NtbHLH genes in tobacco.
2. Materials and methods 2.1. Plant materials and growth conditionsSeeds of tobacco cv. Yunyan87 were obtained from the Yunnan Academy of Tobacco Agricultural Sciences (Yunnan, China). The plant material was identified by Dr. Yongping Li, a researcher of Tobacco Breeding and Biotechnology Research Center, Yunnan Academy of Tobacco Agricultural Sciences. The voucher specimens were deposited at Tobacco Breeding and Biotechnology Research Center, Yunnan Academy of Tobacco Agricultural Sciences. Surfacesterilized seeds were directly sowed into the soil in pots. The Nicotiana young seedlings were grown in the plant growth chamber with a 16-h-light/8-h-dark photoperiod under continuous white light (~75 mol m—2 s—1) at 28℃-day/23℃-night. All plants were kept well-watered after sowing. Tobacco samples were collected from plants in the field and flash-frozen in liquid nitrogen. Field management was performed according to regular agricultural practices. The collected samples included 23 different tissues were harvested as described before (Bai et al., 2019).
2.2. Phylogenetic and gene structure analysesThe protein sequences were obtained from China tobacco genome database V2.0 and NtbHLH proteins were predicted by HMMER (Finn et al., 2015). The sequences of AtbHLH proteins were obtained from the Arabidopsis TAIR. The NtbHLH proteins were identified by HMMER software in tobacco with the newest Hidden Markov Model of the bHLH domain (PF00010) that was obtained from the Pfam database (http://pfam.xfam.org) (El-Gebali et al., 2019).The redundant protein sequences were refined by ElimeDupes (https://hcv.lanl.gov/content/sequence/ELIMDUPES/elimdupes.html) and the SMART database (http://smart.embl-heidelberg.de) (Letunic and Bork 2018).
The NtbHLH, AtbHLH and SIbHLH proteins were aligned with ClustalW software (McWilliam et al., 2013), and an unrooted phylogenetic tree was constructed using MEGA 7.0 (https://www.megasoftware.net/) with the neighbor-joining method and 1000 replicates of bootstrap (Kumar et al., 2016).
The gene structure of each NtbHLH was analyzed by comparing the cDNA and genomic DNA sequences (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). The conserved motifs of NtbHLH proteins were predicted with MEME (http://meme.nbcr.net/meme3/mme.html) (Bailey et al., 2009). The conserved motifs were further identified in the InterPro database (http://www.ebi.ac.uk/interpro). The protein isoelectric point and molecular weight of NtbHLH proteins were analyzed by ProtParam tool (http://web.expasy.org/protparam/).
2.3. RNA extraction, cDNA preparation and gene chipTotal RNA was extracted with the SuperPure Plantpoly RNA Kit (GeneAnswer, BeiJing, China). All RNA samples were treated with RNase-free DNase I (GeneAnswer, BeiJing, China) and analyzed for integrity on a Bioanalyzer 2100 (Agilent technologies, USA). About 33.3 ng total RNA was used for amplification with the Amplification Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). A total of 5.5 μg of the amplified product was fragmented by uracilDNA glycosylase and apurinic/apyrimidinic endonuclease 1 (Thermo Fisher Scientific, USA). The expression of NtbHLH gene was detected by microarray (Bai et al., 2019).
2.4. Chromosomal location and gene duplicationBased on the physical annotation files downloaded from the China tobacco genome database V2.0, the chromosomal locations of NtbHLH genes were analyzed. The interaction network was carried out with Circos software (Krzywinski et al., 2009).
2.5. qRT-PCR analysisRandomly selected 20 NtbHLH genes were examined via qPCR and the method of qPCR were previously described (Bai et al., 2019). Briefly, A total of 2 μg of total RNA in a 20 μL reaction was converted to cDNA with a SuperScript III Reverse Transcriptase (Invitrogen, Waltham, Massachusetts, USA) by the manufacturer's instructions on an Eppendorf Mastercycler thermocycler (Eppendorf AG, Germany). qPCR reactions were made with a SuperReal PreMix Plus SYBR Green Kit (TIANGEN Biotech, BeiJing, China) following manufacturer's instructions in a 20 μL volume. qPCR was done on an Applied Biosystems™ QuantStudio™ 6 Flex Real-Time PCR System (ThemoFisher Scientific, Waltham, Massachusetts, USA). The log2fold change was calculated by the 2—ΔΔCT method using 26S as a reference gene. The CT values represent the average of three technical replicates. The sequences of primers used for qRT-PCR are listed in Table S6.
2.6. Plasmid construction and tobacco transgenic plantTotal RNA was purified from the tobacco leaf and cDNA was obtained with the First Strand cDNA Synthesis kit (Qiagen, Hilden, Germany). The full-length sequences of NtbHLH86 CDS were amplified with two primers. The CDS sequences were cloned into pDONR-zeo vector by BP reaction (Invitrogen, USA) and then cloned into pB2GW7 by LR reaction (Invitrogen, USA). The pB2GW7 contained NtbHLH86 gene was transformed into the tobacco leaves via Agrobacteria.
2.7. Drought treatmentTo detect the expression of NtbHLH86 gene in response to drought stress, the plants were grown for 7—8 weeks with 6—7 leaves. The plants were moved out from the pots carefully without disturbing the root, and the surface soil was washed out gently. Then the plants were put on the bench for air drying which termed as drought stress treatment. The whole seedlings were collected at the indicated time after treatment, and were immediately frozen in liquid nitrogen for RNA extraction for qRT-PCR. Five biological replicates were used for sample harvesting at each indicated time of the treatment. The transgenic plants were grown in small flower pots, and plants with 6—7 leaves were selected for drought treatment, the wild-type tobacco plants as controls. The phenotype was observed after 14 days, and the survival rate was calculated after 20 days of water loss.
3. Results 3.1. Identification and classification of tobacco NtbHLH genesThe protein sequences of Arabidopsis bHLH family proteins were retrieved from the Arabidopsis TAIR. A total of 309 NtbHLH genes were identified by HMMER software in tobacco with the newest Hidden Markov Model of the bHLH domain (PF00010) that was obtained from the Pfam database (http://pfam.xfam.org) (El-Gebali et al., 2019). The 309 NtbHLH genes were renamed from NtbHLH1 to NtbHLH309 according their phylogenetic tree with Arabidopsis bHLH genes (Table S1). The coding sequence (CDS) lengths of NtbHLH genes range from 264 bp to 2637 bp, and the genome lengths of NtbHLH genes range from 343 bp to 32759 bp (Table S2), with protein molecular weights from 9.8 kDa to 98.9 kDa and isoelectric point from 4.3 to 11.08 (Table S3). The number of NtbHLH genes in tobacco is more than that in Arabidopsis, tomato, poplar, wheat, maize, Chinese Jujube and B. distachyon (Toledo-Ortiz et al., 2003; Bailey et al., 2003; Sun et al., 2015; Zhao et al., 2018a; Guo and Wang 2017; Zhang et al., 2018; Li et al., 2019; Niu et al., 2017), but less than that in cotton, Moso bamboo and Brassica napus (Lu et al., 2018; Cheng et al., 2018; Shen et al., 2019), indicating that divergence of NtbHLH genes might lead to the differentiated function in tobacco. To investigate the phylogenetic relationship of bHLH proteins, an unrooted phylogenetic tree was constructed by MEGA 7 with 309 tobacco NtbHLH proteins, 159 tomato SlbHLH proteins (Sun et al., 2015) and 162 Arabidopsis AtbHLH proteins (Toledo-Ortiz et al., 2003; Bailey et al., 2003). It was found that there are 23 clades in the tree, and each clade contains 2 to 40 genes in tobacco (Fig. 1 and Fig. S1). Among them, the clade 17 harbored as many as 40 genes, which accounts for 13% of total tobacco NtbHLH genes, while the clade 13 only harbors two genes (Fig. S1). Although the bHLH genes were well-distributed among three species, the NtbHLH proteins were highly homologous to the tobacco proteins, and then to the proteins from tomato and Arabidopsis.
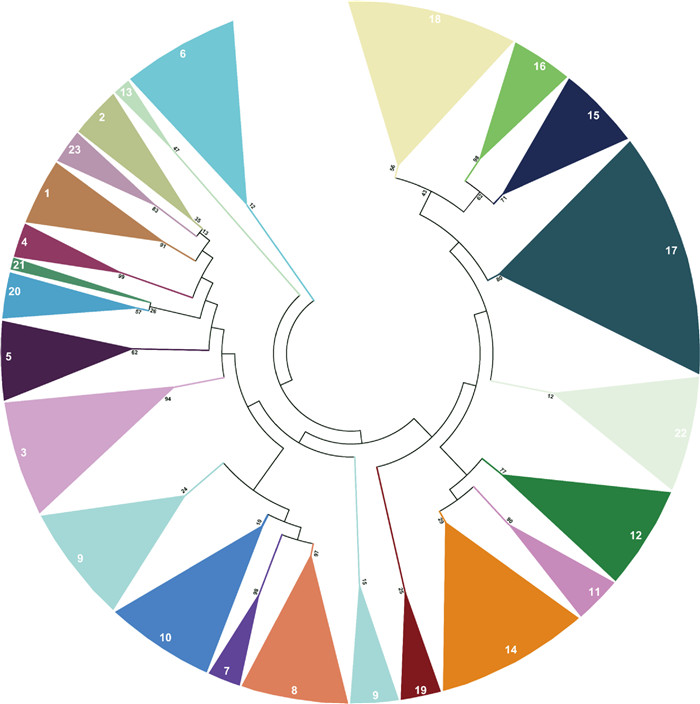
|
| Fig. 1 Phylogenetic analysis of NtbHLH proteins from cultivated tobacco. A total of 309 NtbHLH proteins from cultivated tobacco (Nicotiana tabacum) were used to generate the unrooted neighbor-joining (NJ) tree with 1000 bootstrap replicates. The bHLH proteins were classified into 23 subfamilies and distinguished by different colors. |
The Gene Structure Display Server 2.0 was used to predict the gene structures of tobacco NtbHLH genes. It was found that the number of introns genes was varied among the NtbHLH genes (Fig. 2 and Fig. S2). In the clade 8, most members did not contain intron except of NtbLHL82, NtbHLH83 and NtbHLH84 genes which contained one or two introns. Moreover, none of intron was existed in the clade 12, and the members from clade 10 and clade 12 had only one intron. Members from clade 1, clade 2, clade 3, clade 4, clade 5, clade 9, clade 19, clade 20 and clade 23 contained two or three introns except that the NtbHLH275 and NtbHLH276 genes did not have intron. Furthermore, most members from clade 6 contained four introns, while members of clade 7, clade 11, clade 12, clade 13, clade 14, clade 15, clade 16 and clade 17 contained five introns. Interestingly, the number of introns in the clade 18 showed distinct patterns in which the NtbHLH248 to NtbHLH261 and NtbHLH264 genes contain one intron or none of intron, while the NtbHLH262, NtbHLH263 and NtbHLH266 to NtbHLH274 genes contained four introns. Besides, the intron lengths of NtbHLH genes were largely varied from 0.08 Kb to 9.21 Kb, and the NtbHLH63 gene had the longest intron (Fig. 2 and Fig. S2).
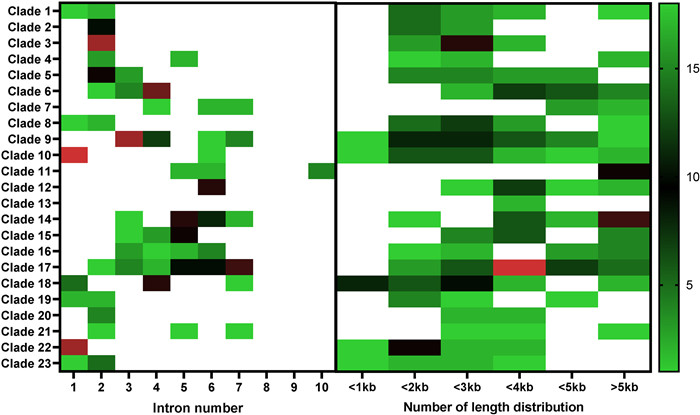
|
| Fig. 2 Intron number and Number of length distribution of NtbHLH gene family in tobacco. Exon-intron analyses of identical tobacco NtbHLH genes were performed with GSDS 2.0. Introns number and number of length distribution were calculated for 23 clades in tobacco, respectively. |
The motifs in the NtbHLH proteins were analyzed by the online software Multiple EM for Motif Elicitation (MEME, http://memesuite.org/index.html). There were ten motifs among the NtbHLH proteins (Fig. 3 and Fig. S3). Consistent with previous studies, the motif 1 and motif 2 were the most conserved motif which could form the bHLH domain in the bHLH proteins (Toledo-Ortiz et al., 2003; Li et al., 2006). The motif 1 and motif 2 were widely distributed in the members of clade 1, clade 3, clade 5, clade 7 and clade 10 to clade 21. It was showed that most members contained the motif 2 in the clade 2, clade 4, clade 8, clade 9, clade 22 and clade 23, however, the motif 1 was only present in some NtbHLH members, such as NtbHLH13, NtbHLH14, and NtbHLH20 from the clade 2, NtbHLH38 from the clade 4, NtbHLH84 and NtbHLH95 from the clade 8, NtbHLH107 from the clade 9, NtbHLH290 and NtbHLH292 to NtbHLH295 from the clade 22 and NtbHLH309 from the clade 23, respectively (Fig. S3).
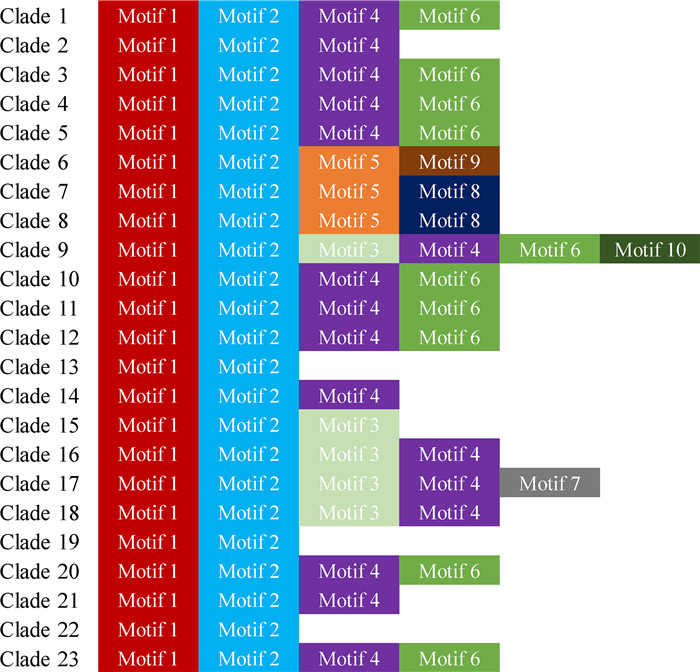
|
| Fig. 3 Conserved motifs of tobacco NtbHLH proteins were predicted by MEME. Colored boxes indicate different motifs. |
The motif 3 was imbedded in the NtbHLH111 to NtbHLH114 proteins which belong to the clade 9, and NtbHLH184 to NtbHLH197 proteins from the clade 15, NtbHLH265 to NtbHLH274 proteins from the clade 18, and all members from the clade 16 and clade 17. The motif 4 and motif 6 were widely presented in multiple clades and were closely linked together in many proteins including members from the clade 3 and clade 4, NtbHLH42 to NtbHLH45 and NtbHLH47 to NtbHLH54 proteins from the clade 5, NtbHLH78 to NtbHLH79 proteins from the clade 7, NtbHLH80 to NtbHLH83, NtbHLH85 to NtbHLH88, NtbHLH91, NtbHLH93 and NtbHLH94 proteins from the clade 8, NtbHLH101 to NtbHLH106, NtbHLH108 to NtbHLH110 and NtbHLH115 to NtbHLH122 proteins from the clade 9. The motif 5 was confined to the members of clade 6 except of NtbHLH68 protein. The motif 7 was existed in the NtbHLH208 to NtbHLH220, NtbHLH222 to NtbHLH236 and NtbHLH244 to NtbHLH247 proteins from the clade 17. The motif 7 was closely to motif 1 and motif 2. The motif 8 was presented in the members from the clade 7, NtbHLH80 to NtbHLH87 and NtbHLH89 to NtbHLH94 proteins from the clade 8, and NtbHLH103 to NtbHLH106 proteins from the clade 9. The motif 9 was restricted in the NtbHLH56 to NtbHLH63 proteins from the clade 6, while the motif 10 was confined in the NtbHLH96 to NtbHLH100 proteins from the clade 9 (Fig. S3).
3.4. Conserved amino acid residues in the bHLH domains and DNAbinding ability of bHLH domainIt is well known that the bHLH domain is an essential domain in the bHLHs protein family. To get the detail information on the bHLH domain of NtbHLH proteins, the amino acids of bHLH domain from 309 NtbHLH proteins were aligned by MEGA 7.0 (Fig. S4). It was showed that the bHLH domain was consisted of 54 AAs in tobacco which was similar to that in Arabidopsis but was lower than that in tomato and animals (Table 1). Moreover, the conservation rates of 22 AAs were more than 50% (Fig. 4), and seven AAs were more conserved in plants, such as Ile-20, Leu-24, Gln-28, Lys-36, Met-43, Ile-48, Val-51 and Leu-54, indicating their essential role in plants. More importantly, the site of Lys-36 was more conserved in tobacco than corresponding residue in Arabidopsis and tomato (Table 1). Furthermore, three residues were extremely conserved in both plants and animals including Arg-16, Leu-27 and Leu-61, implying that these residues might be the core sites in the bHLH domain (Table 1).
| Atchley et al. | Toledo-Ortiz et al. | Hua Sun et al. | this study | ||||||||
| Position in the alignment | Consensus motif amino acid frequency within the bHLH domain | Position in the alignment | Amino acid frequency within theArabidopsis bHLH domains | Position in the alignment | Amino acid frequency within the tomato bHLH domains | Position in the alignment | Amino acid frequency within the tobacco bHLH domains | ||||
| Basic | 1 | R (61%), K (27%) | 1 | R (24%), K (22%) | 1 | K (28%), R (25%), N (11%) | 1 | K (27%), R (30%) | |||
| 2 | R (77%), K (16%) | 2 | R (35%) | 2 | R (32%), K (11%) | 2 | R (37%) | ||||
| 9 | E (93%) | 13 | E (76%), A (10%) | 13 | E (75%), A (11%) | 13 | E (77%), A (10%) | ||||
| 10 | R (81%), K (14%) | 14 | R (74%), K (14%) | 14 | R (76%), K (18%) | 14 | R (80%), K (15%) | ||||
| 12 | R (91%) | 16 | R (91%) | 16 | R (94%) | 16 | R (92%) | ||||
| Helix | 16 | I (35%), L (33%), V (23%) | 20 | I (52%), L (27%), M (12%) | 20 | I (53%), L (28%), M(17%) | 20 | I (53%), L (26%), M(17%) | |||
| 17 | N (74%) | 21 | N (51%), S (19%) | 21 | N (45%), S (26%) | 21 | N (51%), S (25%) | ||||
| 20 | F (72%), L (14%), I (9%) | 24 | F (26%), L (26%), M (20%), I (14%) | 24 | L (28%), F (26%), M(19%), I (16%) | 24 | L (27%), F (22%), M(21%), I (11%) | ||||
| 23 | L (98%) | 27 | L (100%) | 27 | L (99%) | 27 | L (97%) | ||||
| 24 | R (44%), K (35%) | 28 | Q (42%), R (35%) | 28 | Q (41%), R (37%) | 28 | Q (44%), R (36%) | ||||
| Loop | 47 | K (58%), R (24%) | 39 | K (66%) | 36 | K (68%) | 36 | K (85%) | |||
| 50 | K (93%) | 42 | K (45%), T (13%) | 47 | K (45%), T (21%) | 40 | K (47%), T (18%), R (10%) | ||||
| 53 | I (74%), T (15%), V (7%) | 45 | M (33%), I (27%), V (16%), L (14%) | 50 | M(33%), I (28%), V (15%), L (14%) | 43 | M(35%), I (27%), V (15%), L (11%) | ||||
| 54 | L (98%) | 46 | L (76%), V (14%) | 51 | L (78%), I (11%) | 44 | L (80%), I (10%), V (7%) | ||||
| 57 | A (76%) | 49 | A (60%), I (16%), V (12%) | 54 | A (60%), I (18%), V (11%), T (10%) | 47 | A (55%), I (17%), V (13%), T (14%) | ||||
| 58 | I (31%), V (27%), T (23%) | 50 | I (63%), V (22%) | 55 | I (60%), V (25%) | 48 | I (60%), V (22%) | ||||
| 60 | Y (77%) | 52 | Y (78%) | 57 | Y (74%), H (13%) | 50 | Y (78%), H (8%) | ||||
| 61 | I (69%), L (16%), V 8%) | 53 | I (40%), V (33%), L (13%) | 58 | I (43%), V (38%), L (13%) | 51 | I (38%), V (41%), L (15%) | ||||
| 64 | L (80%), M (7%) | 56 | L (93%) | 61 | L (97%) | 54 | L (95%) | ||||
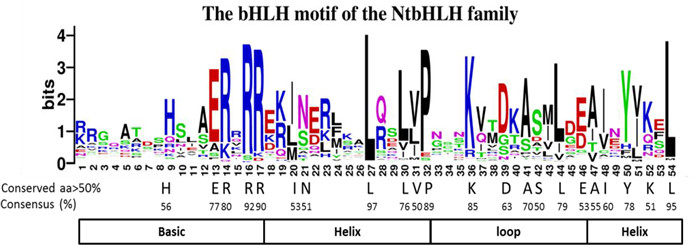
|
| Fig. 4 The bHLH domain is highly conserved across all NtbHLH proteins. The overall height of each stack represents the conservation of sequence at that position. Capital letters indicate over 50% conservation of amino acids among 309 NtbHLH domains. |
Obviously, the most important function of bHLH protein family was attributed to their binding ability to the promoter regions of downstream genes. Consisted with previous studies, the bHLH proteins could be divided into two categories according to their DNA binding, which are DNA binding bHLHs and non-DNA binding bHLHs (Toledo-Ortiz et al., 2003). Actually, the binding ability was largely depended on the number of basic residues in the 1 to 17 residues of bHLH domain. For example, the bHLH protein could bind to the DNA motif if the number of basic residues is more than six. However, the bHLH protein could not bind to the DNA motif if the number of basic residues is less than six. Therefore, there were 286 DNA binding proteins and 23 non-DNA binding proteins among 309 NtbHLH proteins (Table 2). The DNA binding bHLH proteins could be divided into E-box binders and non-E-box binders according to the binding motifs. Moreover, the types of binding style were largely depended on two basic regions. The bHLH proteins could bind to the E-box when two sites are Glu-13 and Arg-16.241 NtbHLH proteins contained the Glu-13 and Arg-16 residues which accounted for 78% of total NtbHLH proteins. Furthermore, the E-box binders can be classified into G-box binders and non-G-box binders according to the residues at 13, 16 and 17 sites. The bHLH proteins are required to recognize the classic G-box (CACGTG) motif when three residues are His/Lys, Glu and Arg. Subsequently, there were 191 G-box binders and 50 non-G-box binders in tobacco NtbHLH proteins, which accounted for 61.8% and 16.1% of total NtbHLH proteins, respectively (Table 2). In contrast, there were 45 NtbHLH proteins which can bind to the DNA due to the presence of 5—8 basic residues in the HLH domain, which were accounted for 14.6% of total NtbHLH proteins (Table 2). It was showed that the G-box binders were mainly distributed in the clade 1 to clade 8, clade 10 and clade 11, clade 14, clade 16 and clade 17, clade 20 and clade 21, while non-G-box binders were mainly distributed in the clade 9, clade 15 and clade 23, and NtbHLH11, NtbHLH12, NtbHLH78 and NtbHLH79 proteins. Moreover, 23 NtbHLH proteins were belonged to the none DNA binding proteins due to lack of enough basic residues, including NtbHLH4, NtbHLH64, NtbHLH161, NtbHLH256 to NtbHLH264, NtbHLH277, NtbHLH278, NtbHLH289 to NtbHLH293 and NtbHLH303 (Table S4).
| Predicted activity | Predicted motif | Number of AtbHLHs (Toledo-Ortiz) | Number of SlbHLHs | This study | |||
| E-box | bHLH | 237 | 74.15% | 98 | 61.63% | 241 | 78.00% |
| G-box | bHLH | 187 | 60.54% | 72 | 45.28% | 191 | 61.81% |
| Non-G-box | bHLH | 50 | 13.61% | 26 | 16.35% | 50 | 16.18% |
| Non-E-box | bHLH | 11 | 7.48% | 12 | 7.55% | 45 | 14.56% |
| Total | 120 | 81.63% | 110 | 69.18% | 286 | 92.55% | |
| Non-DNA binding | HLH | 27 | 18.37% | 49 | 30.82% | 23 | 7.40% |
Expression patterns of NtbHLH genes were analyzed with genechip data from 23 tobacco tissues (Bai et al., 2019), and the followed number was showed as the FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) value (Table S5). It was showed that the highest expression level of NtbHLHs genes in all the tissues was 11.54 while the lowest level was 0.98, respectively (Fig. 5). Twenty genes were highly expressed in all tested tissues, including NtbHLH216, NtbHLH 217, NtbHLH188, NtbHLH189, NtbHLH199, NtbHLH56, NtbHLH57, NtbHLH86, NtbHLH88, NtbHLH146, NtbHLH97, NtbHLH198, NtbHLH203, NtbHLH204, NtbHLH206, NtbHLH207, NtbHLH153, NtbHLH154, NtbHLH60 and NtbHLH61, while 78 genes showed the lower expression levels (Fig. 5). Moreover, expression levels of 99 genes were less than 4.0 in most tested tissues, while some genes showed more than 6.0 in certain tissues (Fig. 5). Besides, there were 106 NtbHLH genes whose expression levels were ranged between 4.0 and 7.0 (Fig. 5). Unexpectedly, six genes cannot be detected by the gene-chip from 23 tobacco tissues. There were five pairs of genes that showed the same expression levels, including NtbHLH188 and NtbHLH189, NtbHLH56 and NtbHLH57, NtbHLH97 and NtbHLH198, NtbHLH203 and NtbHLH204, NtbHLH153 and NtbHLH154, respectively (Fig. 5). The possible explanation is that these pair genes had the high similarity in sequences which was hard to discriminate them from each other (Fig. 5). Notably, NtbHLH216 and NtbHLH217 had extremely higher expression levels in flowers including corolla, filament, ovary, anther, calyx and style, and had the highest levels in corolla of 11.54 and 11.48, respectively (Fig. 5). Furthermore, the NtbHLH216 and NtbHLH217 genes had the higher expression in ten true leaf and root which was up to 9.41 and 9.37, respectively. The NtbHLH180, NtbHLH181, NtbHLH171 and NtbHLH239 genes showed specific expression patterns which were mainly expressed in dry seeds and germination seeds, and the expression levels of these genes were much higher in dry seeds than that in germination seeds. In addition, the NtbHLH237 and NtbHLH238 genes were specifically expressed in calyx and style whose expression levels were up to 9.68 and 8.63, respectively. The expression levels of NtbHLH233, NtbHLH234, NtbHLH218 and NtbHLH219 were higher in corolla, filament, ovary, anther, calyx and style (Fig. 5).
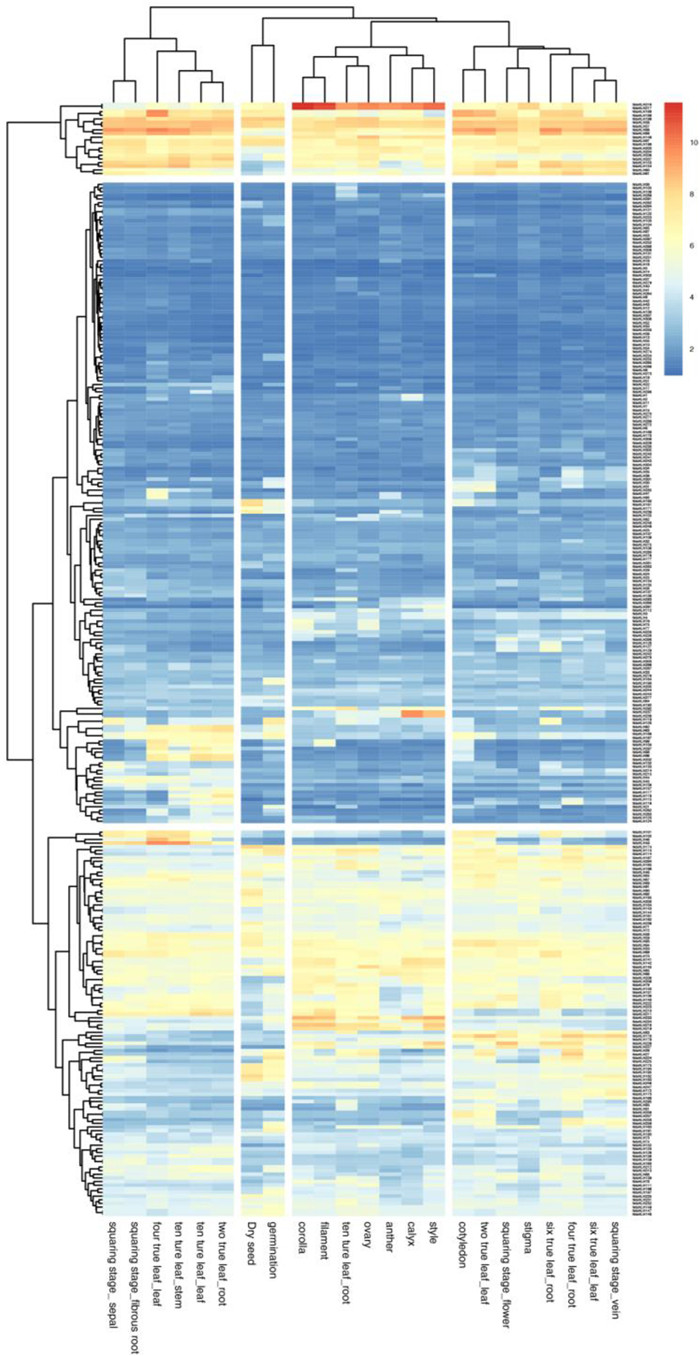
|
| Fig. 5 Expression profile of 303 NtbHLH genes in tissues at different developmental stages. The relative transcript abundances of 303 NtbHLH genes were examined via microarray and visualized as a heatmap. The expression profiles of NtbHLH genes in 23 different samples, including dry seeds, germination seeds, cotyledons, leaves from two-true leaf stage (labeled as two true leaf_leaf), roots from two-true leaf stage (two true leaf_root), leaves from four-true leaf stage (four true leaf_leaf), roots from fourtrue leaf stage (four true leaf_root), leaves from six-true leaf stage (six true leaf_- leaf), roots from six-true leaf stage (six true leaf_root), leaves from ten-true leaf stage (ten ture leaf_leaf), roots from ten-true leaf stage (ten ture leaf_root), and flowers at squaring stage (squaring stage_flower). The X axis is the samples in tissues at different developmental stages. The color scale represents Log2 expression values. |
To further confirm the expression patterns of NtbHLHs genes, 20 randomly selected NtbHLH genes were chosen to detect their expression levels by quantitative real-time PCR (qPCR). It was showed that 13 genes were highly expressed, six genes showed tissue-specific expression and one was lowly expressed (Fig. 6), which was consistent with microarray data. The NtbHLH146, NtbHLH216 and NtbHLH217 genes that had the higher expression levels showed the flowerspecific expression patterns, and NtbHLH216 and NtbHLH217 genes showed the highest expression levels among all the detected genes. Moreover, the NtbHLH188 and NtbHLH199 genes were highly expressed in the root, stem and leaves but were lowly expressed in flower (Fig. 6). In addition, the NtbHLH206, NtbHLH207 and NtbHLH154 genes showed the root-specific expression patterns, and the NtbHLH237, NtbHLH238, NtbHLH233, NtbHLH219 and NtbHLH212 genes were specifically expressed in flowers (Fig. 6). Besides, expression levels of NtbHLH237 and NtbHLH 238 genes were lower while the NtbHLH233 and NtbHLH219 genes showed the higher expression levels in the root, stem and leaf tissues (Fig. 6). The NtbHLH212 genewas highly expressed in the root and leaves but lower in the stem and flower. Furthermore, the NtbHLH87 had the lowest expression levels among all tissues (Figs. 5 and 6).
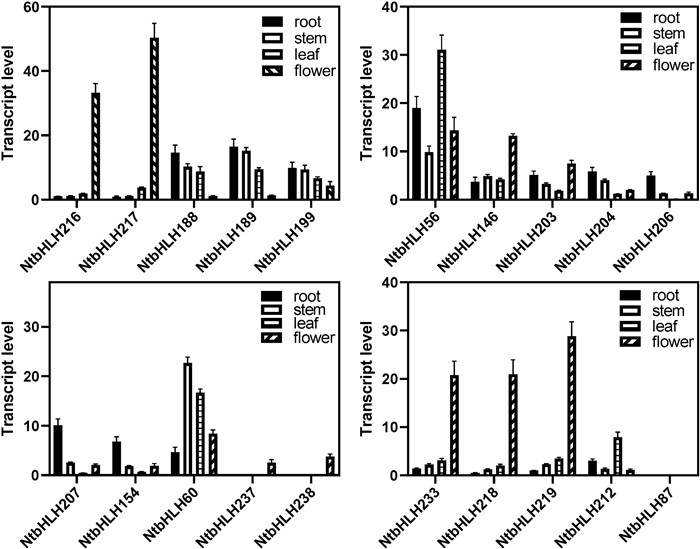
|
| Fig. 6 Expression patterns of 20 randomly selected NtbHLH genes in tabcco. The relative transcript abundances of 20 randomly selected NtbHLH genes were examined via qPCR and visualized as a histogram. Tobacco flower and 6e7 weeks old seedings grown in the soil were collected for RNA extraction and qPCR analysis. 26S was used as an internal control. Error bars represent SD (n = 3). |
There were 265 NtbHLH genes that were widely distributed among 24 chromosomes, and 44 NtbHLH genes could not be located in the chromosome due to the unanchored scaffolds (Fig. 7). Furthermore, the number of NtbHLH genes were varied greatly among 24 chromosomes, from 3 to 22. It was showed that the chromosome 21 had the least genes NtbHLHs while the chromosome 23 contained the most. Moreover, most chromosomes had more than eight NtbHLHs genes, however, the number of genes were less than eight in the chromosome 3, chromosome11, chromosome16 and chromosome24 (Fig. 7). Actually, there were 238 gene pairs which had the higher homologous, however, none of gene cluster were identified for the NtbHLH genes. Previous studies revealed that gene duplications had been involved in the course of evolution by positive selection (Kondrashov et al., 2002; Flagel and Wendel 2009). There were 20 segment duplications of NtbHLH genes, for example NtbHLH33 to NtbHLH36; NtbHLH56 to NtbHLH59; NtbHLH67 to NtbHLH70; NtbHLH71 to NtbHLH74; NtbHLH84, NtbHLH92 to NtbHLH95; NtbHLH86, NtbHLH88, NtbHLH89, NtbHLH91; NtbHLH96 to NtbHLH100; NtbHLH108 to NtbHLH110; NtbHLH111 to NtbHLH113; NtbHLH119 to NtbHLH121; NtbHLH128 to NtbHLH130; NtbHLH137 to NtbHLH141; NtbHLH163 to NtbHLH166; NtbHLH174 to NtbHLH177; NtbHLH184, NtbHLH185, NtbHLH196, NtbHLH197; NtbHLH198, NtbHLH200 to NtbHLH202; NtbHLH228 to NtbHLH230; NtbHLH237 to NtbHLH239, NtbHLH242; NtbHLH244 to NtbHLH247; NtbHLH289 to NtbHLH303 (Fig. 7), indicating that these duplication might be participated in the selection during the evolution.
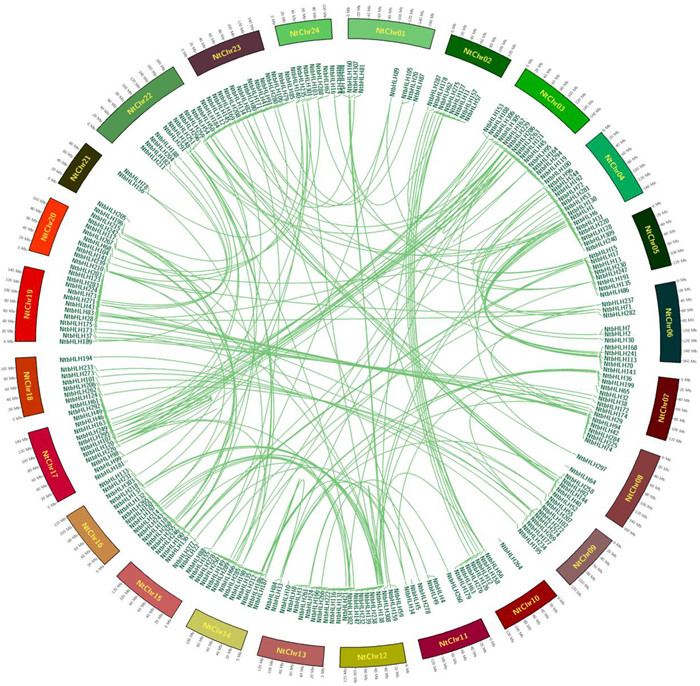
|
| Fig. 7 Collinear analysis for the NtbHLH gene family in tobacco. The annulus represents chromosomes of tobacco (Nicotiana tabacum), and scale on the annulus is labeled in megabases (Mb). Homoeologous genes are linked by lines. The figure was generated and modified using the Circus program. |
Previous studies revealed that the NtMYC2b was mainly focused in the regulation of nicotine in tobacco (Shoji and Hashimoto 2011; Zhang et al., 2012). Moreover, AtMYC2 was involved in the ABA signaling pathway and its overexpression increased the sensitivity to abscisic acid (ABA) (Abe et al., 2003) (Abe et al., 2003). We therefore identified a NtbHLH86 gene which is homologue to NtMYC2 in tobacco. To explore the function of NtbHLH86 gene, gene expression of NtbHLH86 was investigated by qPCR under drought stress condition. It was found that expression of NtbHLH86 was induced by drought stress (Fig. 8A), indicating that it might be involved in the regulation of drought stress. Then five independent NtbHLH86 overexpressors were obtained and confirmed by qPCR (Fig. 8B). Under normal growth conditions, there was no significant difference in the growth between the wild-type plants and NtbHLH86 overexpressors. However, the wild-type plants showed wilted phenotype under drought stress for 14 days, but with lesser in the NtbHLH86 overexpressors (Fig. 8C). Meantime, the survival ratio of NtbHLH86 overexpressors was higher than that in the wild type plants under drought stress for 20 days (Fig. 8D), demonstrated that the NtbHLH86 overexpressors were more resistant to drought than that in the wild-type plants in response to drought stress.
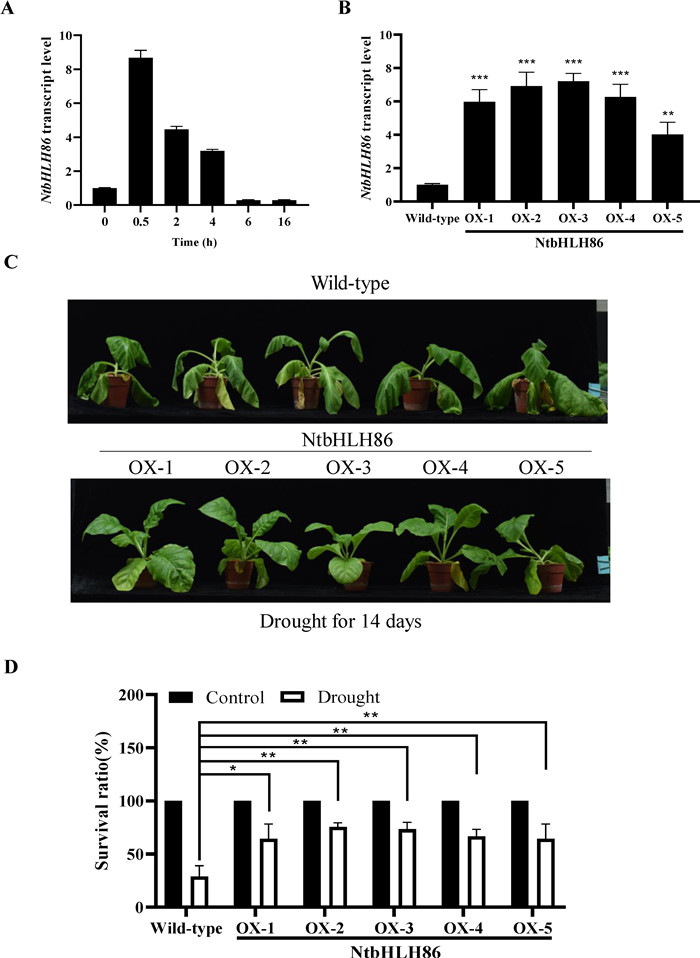
|
| Fig. 8 Phenotypes of NtbHLH86 overexpression transgenic lines in tobacco. (A) Expression of NtbHLH86 was induced by drought treatment. (B) Five independent NtbHLH86 overexpression plants were obtained and analyzed by qPCR. (C) Represented picture of wild-type and five independent NtbHLH86 overexpressors under drought treatment for 14 days. (D) Survival ratio of wild-type and five independent NtbHLH86 overexpressors seedlings under drought treatment for 20 days. For (A) and (B), 26S was used as an internal control. For (A), (B) and (D), error bars represent SD (n = 3). Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) as determined by a two-tailed paired Student's t-test. |
Transcription factors play an important role in plant various processes, including plant growth and development, improvement in stress tolerance, regulation of secondary metabolites (Shoji and Hashimoto 2011; Zhang et al., 2012; Zhao et al. 2018a, 2018b). The bHLH genes are the second largest type of plant transcription factors, which had been widely studied in many plant species (Jones 2004; Pires and Dolan 2010; Moore et al., 2000; Riechmann et al., 2000). Previous studies showed that AtMYC2 (AtbHLH006), AtMYC3 (AtbHLH005) and AtMYC4 (AtbHLH004) were participated in the development of roots, production of secondary metabolites and resistance to insects in Arabidopsis (Dombrecht et al., 2007; Schweizer et al., 2013). AtTT8 (AtbHLH42) and AtGL3 (AtbHLH001) genes were involved in the biosynthesis of anthocyanin and development of trichomes in Arabidopsis (Gonzalez et al., 2008). AtICE1 (AtbHLH116) and AtICE2 (AtbHLH33) were the mainly regulator in response to cold stress (Chinnusamy et al., 2003; Fursova et al., 2009). Therefore, identification of the bHLH gene family would provide more comprehensive information on the function of specific bHLH genes in diverse plant species. In tobacco, two bHLH genes, NtMYC2a and NtMYC2b, were involved in the wounding, topping and biting, and their expression levels were up-regulated after wounding, topping and biting (Li et al., 2016). NtMYC2 active the expression of NtMPO and NtPMT genes through binding to their G-box sequences in the promoters regions, and then regulate the biosynthesis of nicotine (Zhang et al., 2012; Shoji and Hashimoto 2011). The bHLH gene family has been well identified in multiple plants (Li et al., 2006; Toledo-Ortiz et al., 2003; Sun et al., 2015; Guo and Wang 2017; Zhang et al., 2018), and initially identification of bHLH genes has been carried out by Timko group (Rushton et al., 2008). However, there might be some missing bHLH genes due to the incompletely tobacco genome, and it would be necessary to perform a more comprehensive identification of bHLH genes in tobacco with our unpublished high-quality genome.
A total of 309 NtbHLH genes were identified in tobacco, and their phylogenetic relationship, gene structures, conserved amino acids, protein motifs and expression patterns were further analyzed. The number of NtbHLH proteins was more than that in other plants, such as Arabidopsis, rice and tomato (Toledo-Ortiz et al., 2003; Li et al., 2006; Bailey et al., 2003). A possible explanation is that N. tabacum L. is allotetraploid plant, and the NtbHLH genes might have multiple functions than expected. Phylogenetic analysis of bHLH proteins showed that the bHLH proteins can be divided into 23 subfamilies in tobacco (Fig. 1 and Fig. S1). However, the bHLH proteins from Arabidopsis and tomato were formed into 21 subfamilies while bHLH proteins from rice are aligned into 22 subfamilies (Li et al., 2006). Therefore, the number of subfamilies in tobacco bHLH gene families was more than that in Arabidopsis and tomato, which might be due to contain more genes in tobacco. The bHLH genes had a bHLH domain which contains four regions including a basic region, two helices and a loop connecting the helices (Toledo-Ortiz et al., 2003; Li et al., 2006; Sun et al., 2015; Bailey et al., 2003). The most conserved amino acids are the Leu-27 and Leu-54, and these two residues were in the two helices regions which are important for the dimerization of bHLH proteins (Fig. 4). The bHLH genes can bind to the promoter regions in order to regulate the expression of target gene. It was showed that the ratio of NtbHLH genes that can bind to the DNA was up to 92.6% in tobacco, which was higher than that in Arabidopsis, tomato and rice (Table 2). In addition, the ratio of NtbHLH genes that can bind to the G-box DNA in tobacco was similar to that in Arabidopsis but was higher than that in tomato, and the ratio of non-E-box binders in tobacco was much higher than that in Arabidopsis and tomato (Table 2). These results revealed that NtbHLH genes might have more regulatory functions for binding the DNA motif in tobacco. The ratio of most conserved amino acids in tobacco was similar to that in tomato and Arabidopsis (Toledo-Ortiz et al., 2003; Sun et al., 2015), except that the ratio of site Lys-36 in the loop region of bHLH domain in tobacco was much higher than that in Arabidopsis and tomato (Toledo-Ortiz et al., 2003; Sun et al., 2015). These results implied that the Lys-36 would have essential role in the function of NtbHLH proteins that requires further investigation. Moreover, most members in the same subfamily shared the same type of the DNA binding and non-DNA binding which were already observed in Arabidopsis and tomato (Toledo-Ortiz et al., 2003; Sun et al., 2015). Notably, the bHLHs protein from tobacco, Arabidopsis and tomato that were clustered in the same subfamily showed the similar patterns on DNA binding (Toledo-Ortiz et al., 2003), suggesting that bHLHs protein might have conservative functions in plants (Sun et al., 2015; Toledo-Ortiz et al., 2003).
The gene structures of NtbHLH gene family were highly conserved in plants. It was showed that the NtbHLH genes which contained the same number of introns were clustered together (Figs. 2 and S2), that can also be found in other species, such as Nelumbo nucifera, apple, cotton and rice (Lu et al., 2018; Yang et al., 2017; Mao et al., 2019). These results indicated that the bHLH gene were conserved during evolution in the plant kingdom (Fedorov et al., 2002; Rogozin et al., 2003). Moreover, the most conserved motifs of NtbHLH proteins were the motif 1 and motif 2 which are consisted of bHLH domain (Fig. 3), and the bHLH domain was highly conserved in species (Toledo-Ortiz et al., 2003; Li et al., 2006; Sun et al., 2015). Furthermore, large number of NtbHLH genes were widely located in 24 chromosomes although 44 NtbHLH genes could not be located in the chromosome due to the unanchored scaffolds (Fig. 7). The possible reason was mainly due to the incompletely assembly by the complexity of tobacco genome. Surprisingly, there were 308 gene pairs that had high homologous with more than 70%, however, none of gene tandem duplication can form the gene cluster in tobacco, which was different from in Arabidopsis and tomato (Sun et al., 2015; Toledo-Ortiz et al., 2003). Besides, most NtbHLH genes were segment duplicated in tobacco, indicating that gene duplication of NtbHLH genes might be one of mechanism in genomic adaptation to the changing environment (Kondrashov et al., 2002).
It is well known that gene function could be predicted based on their expression (Smaczniak et al., 2012). A comprehensive analysis of gene expression in the NtbHLH genes were performed which would provide the foundation for their function investigation (Figs. 5 and 6). Among 309 NtbHLH genes, the NtbHLH216 and NtbHLH217 genes showed the flower-specific expression patterns with higher expression levels (Fig. 6), respectively. Notably, these two genes were highly homologous with the AtbHLH31 and AtbHLH79 genes in the phylogenetic tree that were abundantly expressed in flowers (Szecsi et al., 2006; Brioudes et al., 2009; Mandaokar et al., 2003). These results suggested that the NtbHLH216 and NtbHLH217 genes would be involved in the regulation of flower development in tobacco. Moreover, the NtbHLH180, NtbHLH181 and NtbHLH171 genes were specifically expressed in the seeds, and NtbHLH182 and NtbhLH183 genes had the higher expression levels in the seeds and germinated seeds, respectively (Fig. 5). The NtbHLH180, NtbHLH181, NtbHLH182 and NtbHLH183 genes were highly homologues to the AtbHLH15 (PIL5) gene which identified as a negative regulator of phytochrome-mediated seed germination (Oh et al., 2004, 2006, 2007). Besides, the NtbHLH171 gene was highly homologues to the AtbHLH16 (PIF8) gene which could inhibit the phyA-induced seed germination in Arabidopsis (Tepperman et al., 2004). Importantly, the NtMYC2 homologue gene, NtbHLH86, was induced by drought stress (Fig. 8A), and the NtbHLH86 overexpressors were more resistant to drought than that in the wild-type plants in response to drought stress (Fig. 8B—D), demonstrated that NtbHLH86 gene might be involved in the regulation of drought stress. Similarly, overexpressed AtMYC2 in Arabidopsis confer hypersensitive to ABA, and AtMYC2 was involved in the ABA signaling pathway (Abe et al., 2003). Therefore, these results suggesting that gene function might be predicted based on their expression, and comprehensively transcriptome analysis of the NtbHLH genes would provide the insights information on elucidating the function of NtbHLH genes in the tobacco development.
In the present study, a comprehensive identification of the NtbHLH gene family members was performed, and their phylogenetic relationship and forms of DNA binding were then analyzed. Totals of 309 NtbHLH proteins were identified and can be divided into 23 subfamilies. Meantime, the conserved amino acids in the bHLH domain and DNA binding for these NtbHLH proteins were predicted which are essential for their specific function. Moreover, 265 NtbHLH genes were mapped to 24 chromosomes and 44 NtbHLH genes were aligned to the scaffolds due to the complexity of tobacco genome. Importantly, the NtbHLH86 gene was involved in the regulation of drought stress, and transcriptome profiles of NtbHLH genes revealed their tissue-specific expression that might be contributed to their potential function in tobacco. Therefore, our study provides an insight for further investigation of gene function in tobacco NtbHLH genes.
Author contributionsConceptualization, Jun Yang; Data curation, Ge Bai; Formal analysis, Dahai Yang and Feng Li; Funding acquisition, Dahai Yang and He Xie; Investigation, Ge Bai, MingLiang Fei and Bingguang Xiao; Methodology, Peijian Cao, Heng Yao and Feng Li; Project administration, He Xie; Resources, Peijian Cao, MingLiang Fei, Yihan Zhang, Xuejun Chen, Bingguang Xiao and Feng Li; Software, Heng Yao, Yihan Zhang and Xuejun Chen; Supervision, Jun Yang; Writing e original draft, Ge Bai and He Xie; Writing e review & editing, Dahai Yang and Zhenyu Wang.
Availability of data and materialsThe original data that support the findings of this study are available from National Tobacco Gene Research Centre at Zhengzhou Tobacco Research Institute, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are available from the authors upon reasonable request and with permission of National Tobacco Gene Research Centre at Zhengzhou Tobacco Research Institute.
Declaration of competing interestThe authors declare no conflict of interest.
AcknowledgementsThis work was funded by the National Natural Science Foundation of China (grant number 31760072 to G. Bai, and grant number 31860413 to H. Xie) and Yunnan Applied Basic Research Project (grant number 202001AT070010 to G. Bai) and the Yunnan Academy of Tobacco Agricultural Sciences (grant numbers YNTC-2016YN22 and CNTC-110202001025(JY08) to H. Xie, YNTC-2016YN24, YNTC-2015YN02, YNTC-2018530000241002, and YNTC-2019530000241003 to D.-H. Yang).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.10.004.
Abe H., Urao T., Ito T., et al, 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 15: 68-78. |
Atchley W.R., Terhalle W., Dress A., 1999. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol, 48: 501-516. DOI:10.1007/PL00006494 |
Bai G., Yang D.H., Cao P., et al, 2019. Genome-Wide identification, gene structure and expression analysis of the MADS-box gene family indicate their function in the development of tobacco (Nicotiana tabacum L. ). Int. J. Mol. Sci, 20. |
Bailey T.L., Boden M., Buske F.A., et al, 2009. Meme suite: tools for motif discovery and searching. Nucleic Acids Res, 37: W202-W208. DOI:10.1093/nar/gkp335 |
Bailey P.C., Martin C., Toledo-Ortiz G., et al, 2003. Update on the basic helix-loophelix transcription factor gene family in Arabidopsis thaliana. Plant Cell, 15: 2497-2502. DOI:10.1105/tpc.151140 |
Brioudes F., Joly C., Szecsi J., et al, 2009. Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J. : Cell Mole Bio, 60: 1070-1080. DOI:10.1111/j.1365-313X.2009.04023.x |
Cheng X., Xiong R., Liu H., et al, 2018. Basic helix-loop-helix gene family: genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem, 132: 104-119. DOI:10.1016/j.plaphy.2018.08.036 |
Chinnusamy V., Ohta M., Kanrar S., et al, 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev, 17: 1043-1054. DOI:10.1101/gad.1077503 |
Dombrecht B., Xue G.P., Sprague S.J., et al, 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell, 19: 2225-2245. DOI:10.1105/tpc.106.048017 |
El-Gebali S., Mistry J., Bateman A., et al, 2019. The Pfam protein families database in 2019. Nucleic Acids Res, 47: D427-D432. DOI:10.1093/nar/gky995 |
Farquharson K.L., 2016. A domain in the bHLH transcription factor DYT1 is critical for anther development. Plant Cell, 28: 997-998. DOI:10.1105/tpc.16.00331 |
Fedorov A., Merican A.F., Gilbert W., 2002. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl. Acad. Sci. U.S. A, 99: 16128-16133. DOI:10.1073/pnas.242624899 |
Ferre-D'Amare A.R., Pognonec P., Roeder R.G., et al, 1994. Structure and function of the b/HLH/Z domain of USF. EMBO J, 13: 180-189. DOI:10.1002/j.1460-2075.1994.tb06247.x |
Finn R.D., Clements J., Arndt W., et al, 2015. HMMER web server: 2015 update. Nucleic Acids Res, 43: W30-W38. DOI:10.1093/nar/gkv397 |
Flagel L.E., Wendel J.F., 2009. Gene duplication and evolutionary novelty in plants. New Phytol, 183: 557-564. DOI:10.1111/j.1469-8137.2009.02923.x |
Fursova O.V., Pogorelko G.V., Tarasov V.A., 2009. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene, 429: 98-103. DOI:10.1016/j.gene.2008.10.016 |
Gonzalez A., Zhao M., Leavitt J.M., et al, 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. : Cell Mole Bio, 53: 814-827. DOI:10.1111/j.1365-313X.2007.03373.x |
Groszmann M., Bylstra Y., Lampugnani E.R., et al, 2010. Regulation of tissuespecific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot, 61: 1495-1508. DOI:10.1093/jxb/erq015 |
Guo X.J., Wang J.R., 2017. Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biol, 17: 70. DOI:10.1186/s12870-017-1021-7 |
Hu B., Jin J., Guo A.Y., et al, 2015. Gsds 2. 0: an upgraded gene feature visualization server. Bioinformatics, 31: 1296-1297. DOI:10.1093/bioinformatics/btu817 |
Jones S., 2004. An overview of the basic helix-loop-helix proteins. Genome Biol, 5: 226. DOI:10.1186/gb-2004-5-6-226 |
Kondrashov F.A., Rogozin I.B., Wolf Y.I., et al, 2002. Selection in the evolution of gene duplications. Genome Biol, 3. |
Krzywinski M., Schein J., Birol I., et al, 2009. Circos: an information aesthetic for comparative genomics. Genome Res, 19: 1639-1645. DOI:10.1101/gr.092759.109 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol. Biol. Evol, 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Ledent V., Vervoort M., 2001. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res, 11: 754-770. DOI:10.1101/gr.177001 |
Letunic I., Bork P., 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res, 46: D493-D496. DOI:10.1093/nar/gkx922 |
Li H., Gao W., Xue C., Zhang Y., Liu Z., Zhang Y., Meng X., Liu M., Zhao J., 2019. Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba Mill.) and wild jujube. BMC Genom, 20: 568. DOI:10.1186/s12864-019-5936-2 |
Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J., Ma H., Wang J., Zhang D., 2006. Genome-wide analysis of basic/helix-loophelix transcription factor family in rice and Arabidopsis. Plant Physiol, 141: 1167-1184. DOI:10.1104/pp.106.080580 |
Li F., Zhang H., Wang S., et al, 2016. Identification of topping responsive proteins in tobacco roots. Front. Plant Sci, 7: 582. |
Lu R., Zhang J., Liu D., et al, 2018. Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton(Gossypium hirsutum). BMC Plant Biol, 18: 304. DOI:10.1186/s12870-018-1523-y |
Mandaokar A., Kumar V.D., Amway M., et al, 2003. Microarray and differential display identify genes involved in jasmonate-dependent anther development. Plant Mol. Biol, 52: 775-786. DOI:10.1023/A:1025045217859 |
Mao T.Y., Liu Y.Y., Zhu H.H., et al, 2019. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ, 7: e7153. DOI:10.7717/peerj.7153 |
Massari M.E., Murre C., 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell Biol, 20: 429-440. DOI:10.1128/MCB.20.2.429-440.2000 |
McWilliam H., Li W., Uludag M., et al, 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res, 41: W597-W600. DOI:10.1093/nar/gkt376 |
Moore A.W., Barbel S., Jan L.Y., et al, 2000. A genomewide survey of basic helixloop-helix factors in Drosophila. Proc. Natl. Acad. Sci. U.S.A, 97: 10436-10441. DOI:10.1073/pnas.170301897 |
Murre C., McCaw P.S., Baltimore D., 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell, 56: 777-783. DOI:10.1016/0092-8674(89)90682-X |
Nakata M., Mitsuda N., Herde M., et al, 2013. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell, 25: 1641-1656. DOI:10.1105/tpc.113.111112 |
Niu X., Guan Y., Chen S., Li H., 2017. Genome-wide analysis of basic helix-loophelix (bHLH) transcription factors in Brachypodium distachyon. BMC Genom, 18: 619. DOI:10.1186/s12864-017-4044-4 |
Oh E., Kim J., Park E., et al, 2004. PIL5, a phytochrome-interacting basic helix-loophelix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell, 16: 3045-3058. DOI:10.1105/tpc.104.025163 |
Oh E., Yamaguchi S., Hu J., et al, 2007. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell, 19: 1192-1208. DOI:10.1105/tpc.107.050153 |
Oh E., Yamaguchi S., Kamiya Y., et al, 2006. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J, 47: 124-139. DOI:10.1111/j.1365-313X.2006.02773.x |
Pires N., Dolan L., 2010. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol, 27: 862-874. DOI:10.1093/molbev/msp288 |
Riechmann J.L., Heard J., Martin G., et al, 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science, 290: 2105-2110. DOI:10.1126/science.290.5499.2105 |
Rogozin I.B., Wolf Y.I., Sorokin A.V., et al, 2003. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol, 13: 1512-1517. DOI:10.1016/S0960-9822(03)00558-X |
Rushton P.J., Bokowiec M.T., Han S., et al, 2008. Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol, 147: 280-295. DOI:10.1104/pp.107.114041 |
Schweizer F., Fernandez-Calvo P., Zander M., et al, 2013. Arabidopsis basic helixloop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell, 25: 3117-3132. DOI:10.1105/tpc.113.115139 |
Shen W., Cui X., Li H., et al, 2019. Genome-wide identification and analyses of bHLH family genes in Brassica napus. Can. J. Plant Sci, 99: 589-598. DOI:10.1139/cjps-2018-0230 |
Shoji T., Hashimoto T., 2011. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol, 52: 1117-1130. DOI:10.1093/pcp/pcr063 |
Smaczniak C., Immink R.G., Angenent G.C., et al, 2012. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development, 139: 3081-3098. DOI:10.1242/dev.074674 |
Sun H., Fan H.J., Ling H.Q., 2015. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom, 16: 9. DOI:10.1186/s12864-014-1209-2 |
Szecsi J., Joly C., Bordji K., et al, 2006. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J, 25(16): 3912-3920. DOI:10.1038/sj.emboj.7601270 |
Tepperman J.M., Hudson M.E., Khanna R., et al, 2004. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J, 38: 725-739. DOI:10.1111/j.1365-313X.2004.02084.x |
Toledo-Ortiz G., Huq E., Quail P.H., 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell, 15: 1749-1770. DOI:10.1105/tpc.013839 |
Wang Y., Liu A., 2020. Genomic characterization and expression analysis of basic helix-loop-helix (bHLH) family genes in traditional Chinese herb Dendrobium officinale. Plants, 9. |
Yang J., Gao M., Huang L., et al, 2017. Identification and expression analysis of the apple (Malus x domestica) basic helix-loop-helix transcription factor family. Sci. Rep, 7: 28. DOI:10.1038/s41598-017-00040-y |
Zhang H.B., Bokowiec M.T., Rushton P.J., et al, 2012. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant, 5: 73-84. DOI:10.1093/mp/ssr056 |
Zhang T., Lv W., Zhang H., et al, 2018. Genome-wide analysis of the basic HelixLoop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol, 18: 235. DOI:10.1186/s12870-018-1441-z |
Zhao K., Li S., Yao W., et al, 2018a. Characterization of the basic helix-loop-helix gene family and its tissue-differential expression in response to salt stress in poplar. PeerJ, 6: e4502. DOI:10.7717/peerj.4502 |
Zhao Q., Xiang X., Liu D., et al, 2018b. Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front. Plant Sci, 9: 381. DOI:10.3389/fpls.2018.00381 |



