b. College of Forestry, Nanjing Forestry University, Nanjing, 210037, Jiangsu, China;
c. Yunnan Key Laboratory for Integrative Conservation of Plant Species with Extremely Small Populations, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, Yunnan, China;
d. Yunnan Key Laboratory of Plant Reproductive Adaption and Evolutionary Ecology, School of Ecology and Environmental Science, Yunnan University, Kunming 650504, Yunnan, China
The genetic diversity of a species, which is the product of long-term evolution, is the basis of species survival, adaptation, and development (Meffe and Carroll, 1997). Accordingly, protecting the genetic diversity of a species ensures that species maintain the ability to adapt to change (Huang et al., 2016). Thus, it is critical for researchers to understand the genetic structure and evolutionary history of endangered species in order to provide guidance for ex situ conservation, introduction, and cultivation (Groom et al., 2006).
Rhododendron L. (Ericaceae), which consists of over 1000 species around the world, is the largest genus of angiosperms in China (Goetsch et al., 2005). Rhododendron meddianum Forrest is a perennial evergreen plant of Rhododendron, subg. Hymenanthes, sect. Ponticum, subsect. Thomsonia, distributed in southwestern China and northeastern Myanmar (Fig. 1; Fang et al., 2005). R. meddianum has high ornamental value, but is listed as critically endangered (Qin et al., 2017). Recently, our field surveys discovered three wild populations of R. meddianum with a narrow area of distribution in western Yunnan, China (Fig. 1a). In this narrow region, R. meddianum habitats may be severely disturbed by anthropogenic activities (e.g., wind power stations and roads) (Fig. 1b). This is consistent with studies that indicate human-induced environmental change and habitat fragmentation threaten about 60% of wild Rhododendrons (Gibbs et al., 2011). Thus, it is urgent to carry out studies on the conservation biology of R. meddianum. Importantly, understanding the population genetics (e.g., genetic diversity, population structure) of these populations will help researchers set appropriate conservation strategies.

|
| Fig. 1 Geographic locations for three populations of Rhododendron meddianum analyzed in the present study (a, see Table S1 for population codes), habitat of R. meddianum in Caojian Town (b), flowers (c). |

|
| Fig. 2 Distribution of mean LnP (K) (a) and Delta K (b). Model-based population assignment by STRUCTURE analysis for K = 2–4 (c). Principal components analysis (PCA) of all 45 Rhododendron meddianum samples with the proportion of the variance explained being 3.12% for PC1 and 2.73% for PC2 (d). Neighbor-joining (NJ) phylogenetic tree of R. meddianum samples (e). |
|
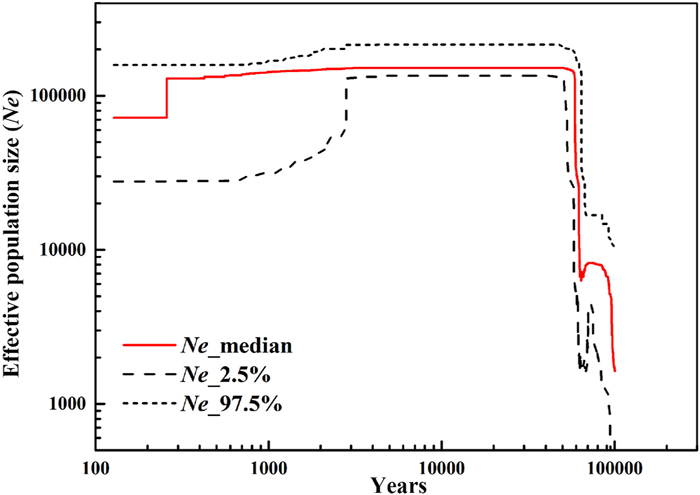
| Fig. 3 Estimates of the effective population size (Ne) of Rhododendron meddianum through time based on nuclear SNPs using stairway plot. Take the median of these estimates as the final result (red line), as well as the upper (97.5%) and lower (2.5%) bounds as the pseudo-confidence interval. |
Population genetics of plants have long been assessed using molecular markers, including simple sequence repeat (SSR), amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), and expressed sequence tag SSR (Yilmaz, 2016; Stone et al., 2019; Everaert et al., 2020; Wagutu et al., 2020). Recently, single nucleotide polymorphism (SNPs) based on next-generation sequencing (NGS) have become the markers of choice for determining population structure because they are abundant, stable in the genome, and can be accurately scored (Deschamps et al., 2012). One sample, low cost, high throughput approach to discovering hundreds to thousands of SNPs in non-model species is restriction-site associated DNA sequencing (RAD-seq). RAD-seq methods have been used on plant communities to reveal their evolutionary history and local adaptability (Jiang et al., 2019), identify the genetic structure of populations (Yamashita et al., 2019), and determine how environmental and topographical elements affect genetic diversity and differentiation (Xiao et al., 2020). RAD-seq methods have also been used to identify genetic lineages of plants, which has rendered references for the conservation of rare species (Amor et al., 2020).
In this study, neutral loci generated by double digest restriction-site-associated DNA sequencing (ddRAD) were used to determine the genetic diversity, population structure and demographic history of Rhododendron meddianum in southwestern China. In addition, we elucidate the evolutionary history between R. meddianum and its close relative Rhododendron cyanocarpum (Franch.) W.W. Smith (a threatened species endemic to Dali Cangshan Mountain, Yunnan province). The findings of this study will contribute to a better understanding of the evolutionary history of R. meddianum and provide a scientific basis for its conservation and management.
2. Materials and methods 2.1. Experimental materialExperimental material was taken from 45 samples of three wild Rhododendron meddianum populations from Yunnan province, southwestern China (Table S1, Fig. 1). The distance between sampling plants within each population was greater than 50 m. After collection, the young, tender leaves were quickly dried and preserved with silica gel.
2.2. DNA extraction and ddRAD library constructionGenomic DNA was extracted for each sample of Rhododendron meddianum by the CTAB method (Doyle and Doyle, 1987). DNA quality was assessed visually on 1.2% agarose gels and subsequently quantified using a Qubit 3.0 Fluorometer. Library construction was constructed following a previously reported ddRAD protocol (Peterson et al., 2012). Briefly, 100 ng of genomic DNA of each sample was digested with two restriction enzymes, EcoRI and MseI (New England Biolabs, Beverly, USA) at 37 ℃ for 5 h. Restriction enzymes were then inactivated by heating at 65 ℃ for 20 min. After ligation with individually barcoded EcoRI and MseI adapters with T4 DNA ligase (New England Biolabs, Beverly, USA) for each sample at 16 ℃ for 4 h, the reaction was stopped by heating at 65 ℃ for 20 min, and finally the product was maintained at 12 ℃. The fragments (300–500 bp) were recovered using a gel extraction kit (Omega Inc., USA). The purified product was amplified by PCR to the expected concentration, and then 150 bp paired-end sequencing was performed on the Illumina HiSeq X-ten platform. Sequencing was completed by Guangzhou Jierui Biotechnology Co., Ltd. Each sample generated about 0.5 Gb of data. The sequencing reads of R. meddianum in this study have been deposited in the NCBI sequence read archive (SRA) under bioproject accession PRJNA706564.
2.3. SNP miningRaw sequence data was analyzed using Stacks v.2.0 (Catchen et al., 2011, 2013). First, the module process_radtags was used to demultiplex and sort the raw data based on the barcodes used in each sample; low-quality reads with missing restriction sites or containing ambiguous barcodes were eliminated. Sequence quality was verified by Fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The ustacks module was used to cluster single sample sequences to form loci, with parameters were set as follows: minimum depth of coverage required to create a stack (-m), 2; maximum distance (in nucleotides) allowed between stacks (-M), 2; maximum distance allowed to align secondary reads to primary stacks, 4; max number of stacks allowed per de novo locus, 3; minimum alignment length, 0.8; alpha significance level for model, 0.01. Loci data of all of samples were merged into a catalog using cstacks module with default parameters. The sstacks module was run to compare the single sample loci to the catalog so as to obtain the SNPs, as well as allele and tag information of the sample. Finally, thepopulations module was used to screen SNPs, with parameters set as follows: minimum number of populations a locus must be present in to process a locus (-r), 6; minimum percentage of individuals in a population required to process a locus for that population (-p), 0.75; minimum minor allele frequency required to process a nucleotide site at a locus, 0.01; maximum observed heterozygosity required to process a nucleotide site at a locus, 0.6. One random SNP per locus was kept (-write_random_snp) to reduce linkage disequilibrium (Falush et al., 2003; Kaeuffer et al., 2007). The output result was exported in VCF format using the populations module.
2.4. Data analysis 2.4.1. Analysis of genetic diversity and structureTajima'D values were estimated to detect the selection effect (Tajima, 1989). We calculate Tajima'D using VCFtools v.0.1.15 (Danecek et al., 2011); the sliding window size of the operation was 3000 bp. The populations module (Catchen et al., 2013) of Stacks software was utilized to calculate genetic diversity parameters, including nucleotide diversity (π), the number of private alleles (Private), expected heterozygosity (HE), observed heterozygosity (HO) and inbreeding coefficient (FIS). Outlier loci (False Discovery Rate (q) > 0.05) was detected by BayeScan v.2.1 (Foll and Gaggiotti, 2008) with default parameters to guarantee that neutral SNPs were generated for subsequent analysis. The format conversion was implemented using PGDSpider v.2.1.1.5 (Lischer and Excoffier, 2012).
Arlequin v.3.5.2 was used to estimate pairwise differentiation coefficient (FST) between populations (1000 permutations) (Excoffier et al., 2007) and to calculate the genetic variation between and within populations with analysis of molecular variance (AMOVA). STRUCTURE v.2.3.5 (Pritchard et al., 2000) was used to analyze the genetic relationship between samples via speculating the proportion of a certain hypothetical ancestor of the test samples, with the number of genetic clusters (K) set to 1–8. Each K was run with 10 independent iterations, each with a burn-in period of 20, 000 iterations and 100, 000 Markov chain Monte Carlo (MCMC) iterations by assuming the admixture model. The output result of LnP (D) under different K values was subsequently visualized by Structure Harvester v.0.6.94 (http://taylor0.biology.ucla.edu/struct_harvest/). Optimal K values were determined by maximum delta K and the mean LnP (K) value (Evanno et al., 2005). Principal components analysis (PCA) was performed via Plink v.1.90 (Purcell et al., 2007). The top 10 principal components (PCs) were extracted with 'pca' parameters. The top two PCs (PC1, PC2) were visualized in R v.3.3.1 (R Core Team, 2016) to confirm the subsection structure of all 45 samples. MEGA X (Kumar et al., 2018) software was used to construct Neighbor Joining (NJ) trees, and finally, EvolView online tool (http://www.evolgenius.info/evolview) was edited for visualization.
2.4.2. Historical demographic inference of Rhododendron meddianumStairway Plot v.2.1 was used to infer recent historical dynamics of effective population size of Rhododendron meddianum based on nuclear SNP data (Liu and Fu, 2015). One-dimensional unfolded SFS (1D-SFS) was generated from posterior probabilities of sample allele frequencies using the python script easySFS (https://github.com/isaacovercast/easySFS). To estimate the trend of effective population size (Ne) over time, 67% of all sites was randomly selected for training. The number of random breakpoints (nrand) in each test was set at 29–90. These break points may be thought of as points at which Ne changes in time. Using the median value of these estimates as final values, Morimoto et al. (2003) observed that Rhododendron reticulatum D. Don ex G. Don and Rhododendron macrosepalum Maxim. began to flower at about 10 years old. The Rhododendron ponticum L. began to flower at about 10 years (Cross, 1975). Yoichi et al. (2016) gave the mutation rate for Rhododendron weyrichii Maxim. as 1.581e-9. We therefore set the generation time and mutation rate to 10 years and 1.581e-9 for R. meddianum.
2.4.3. Evolutionary history of Rhododendron meddianum and R. cyanocarpumBecause we used ddRAD sequencing in both species (Liu et al., 2020a), we assumed SNPs were consistent with the 2.3 section analysis and estimated population structure the same as 2.4.1 analysis (BayeScan, STRUCTURE, PCA and NJ trees analysis). Raw sequence data of R. cyanocarpum was obtained from SRA in the NCBI (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA640883).
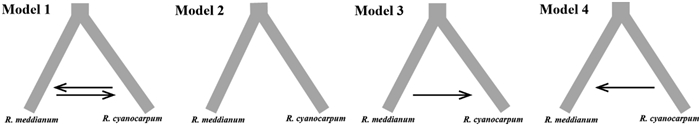
The results of our analysis of the population genetic structure (see the results section) of both Rhododendron meddianum and R. cyanocarpum indicated that these species are clearly divided into two genetic groups (Caojian (CJ), Pianma (PM) and Houqiao (HQ) for R. meddianum group; Huadianba (HDB), Xiaohuadianba (XHD), Dianshita (DST) and Dahuayuan (DHY) for R. cyanocarpum group, Fig. S1). To infer the divergence time and migration events of these two groups, we used Fastsimcoal2 v.2.6 (Excoffier et al., 2013) to test four demography models according to 1D-SFS (Model 1–4, Fig. 4). The mutation rate and generation time were set as 1.581e-9 and 10 years. Each model was run 50 times with 100, 000 coalescent simulations and 40 optimization cycles. Model comparison was based on the maximum likelihood value (Max log (Est_hood), Table 4) using the Akaike Information Criteria (AIC) and AIC weight of evidence. The maximum AIC weight value was selected for the optimal model (Excoffier et al., 2013). Finally, Parameter-confidence intervals (CIs) of the optimal model were calculated from 100 para-metric bootstrap replicates by simulating SFS from the maximum composite likelihood estimates (_maxL.par file), with 50 independent runs in each bootstrap.
|
| Fig. 4 Four basic demographic models analyzed with Fastsimcoal2. Between-species gene flow were indicated by the arrows. |
| Population | Private | P | HO | HE | π | FIS | D |
| CJ | 946 | 0.9491±0.0017 | 0.0440±0.0015 | 0.0781±0.0022 | 0.0813±0.0023 | 0.1428±0.0130 | −0.4151 |
| PM | 954 | 0.9533±0.0016 | 0.0578±0.0020 | 0.0711±0.0021 | 0.0738±0.0022 | 0.0654±0.0173 | −0.5230 |
| HQ | 902 | 0.9561±0.0017 | 0.0496±0.0018 | 0.0646±0.0021 | 0.0671±0.0022 | 0.0620±0.0172 | −0.5057 |
| Mean | 934 | 0.9528±0.0017 | 0.0505±0.0018 | 0.0713±0.0021 | 0.0741±0.0022 | 0.0901±0.0158 | −0.6740 |
| Private, number of private alleles; P, the average frequency of the major allele; HO, observed heterozygosity; HE, expected heterozygosity; π, Nucleotide diversity; FIS, inbreeding coefficient; D, Tajima'D test statistic. | |||||||
| CJ | PM | HQ | |
| CJ | – | 0.1582** | 0.2189** |
| PM | 51.88 | – | 0.2388** |
| HQ | 88.27 | 62.89 | – |
| Significance: **, P < 0.01. | |||
| Source of variation | d.f. | Sum of squares | Variance components | Percentage variation | Fixation index (FST) |
| Among populations | 2 | 1562.600 | 22.267 | 19.88 | 0.1988** |
| Within populations | 87 | 7737.267 | 89.746 | 80.12 | |
| Total | 89 | 9299.867 | 112.013 | ||
| Significance: **, P < 0.01; d.f., degree of freedom. | |||||
| Model | Max log (Est_hood) | No. estimated parameters | AIC | ΔAIC | AIC weight |
| 1 | −1047 | 5 | 2104 | 22 | 1.6701e-5 |
| 2 | −1049 | 5 | 2108 | 26 | 2.2602e-6 |
| 3 | −1036 | 5 | 2082 | 0 | 0.9999 |
| 4 | −1048 | 5 | 2106 | 24 | 6.1440e-6 |
ddRAD sequencing was performed on 45 individuals of Rhododendron meddianum, a total of 268, 549, 498 raw reads were obtained (sample reads range: 1, 221, 942–1, 484, 088), and 265, 045, 933 reads were retained (per sample, 1, 210, 660–14, 707, 008) after quality control. A total of 3, 503, 562 reads were eliminated due to low quality and no rad tag. The average GC content per sample was 41.56% (40%–47%). Using ustacks module, 7, 252, 426 loci were retained. The mean coverage depth of locus of sample were different, which might be due to the degradation of sample DNA (Table S2). A total of 4, 204, 450 catalogs were obtained after merging sample loci. After mapping each sample loci to the generated catalogs, 3377 loci with 482, 194 variant loci were gained. After retaining one SNP per locus, 3377 SNPs were finally obtained for further analysis.
3.2. Genetic diversity and structure of Rhododendron meddianumTajima'D statistics were significantly negative for the three populations of Rhododendron meddianum (CJ, PM and HQ), with values ranging from −0.4151 to −0.5057 (Table 1), supporting the hypothesis that R. meddianum populations underwent a population expansion. Outlier loci with a significant elevated ofFST values (q < 0.05) were not found via BayeScan analysis, indicating that ddRAD-seq loci were not under strong selection (Fig. S2a).
The genetic diversity of 3377 neutral loci is shown in Table 1. The CJ population showed the highest genetic diversity (HE = 0.0781 ± 0.0022, π = 0.0813 ± 0.0023). In contrast, the HQ population showed the lowest genetic diversity (HE = 0.0646 ± 0.0021, π = 0.0671 ± 0.0022). The PM population had the largest number of private alleles (952), followed by the CJ population (946), and the HQ population had the fewest private alleles (902). The inbreeding coefficient (FIS) of each population were positive (0.0620 ± 0.0172–0.1428 ± 0.0130), indicating a deficit of heterozygotes (Wright, 1922).
All the pairwise FST values between the populations were statistically significant (P < 0.01, Table 2). Moderate degrees of differentiation were found between the three populations (0.1582–0.2388). AMOVA indicated that 19.88% (P < 0.01) of the genetic variation occurred among populations, and 80.12% (P < 0.01) of the variation occurred within populations (Table 3). STRUCTURE analysis supported the best genetic cluster (K = 3) based on both Delta K and mean LnP (K) (Fig. 2a and b). In the case of K = 3, three wild populations of R. meddianum were clearly distinguished, forming three clusters. When it was assumed K = 2, the CJ and PM populations were grouped into the same cluster, indicating that they share a close genetic composition. The results of K = 4 was consistent with K = 3, i.e., the populations were clearly distinguished in accord with geographical distribution (Fig. 2c). Consistent with STRUCTURE results, both the PCA and NJ trees of R. meddianum samples revealed three major clusters corresponding to the CJ, PM and HQ populations (Fig. 2d and e).
3.3. Population demographic historyTo reconstruct the demographic history of Rhododendron meddianum, we used a stairway plot to assess population size changes and obtained a demographic history from 150 to 100, 000 years ago (Fig. 3). The results revealed that a population bottleneck occurred about 70, 000 years ago, followed by a rapid expansion 60, 000–70, 000 years ago, and then the populations maintained a stable Ne from 150 to 60, 000 years ago, including the Last Glacial Maximum (LGM: 21, 000 years ago). However, the Ne declined 300 years ago (Fig. 3).
3.4. Phylogeographic patterns of Rhododendron meddianum and R. cyanocarpumA total of 1495 loci with 207, 927 variant sites were retained after sequence data processing and quality control. After retaining one SNP per locus, 1495 SNPs were ultimately selected. Our BayeScan analysis did not identify any outlier loci with significantly elevated FST values (q < 0.05), which indicates that 1495 loci of the two species were neutral (Fig. S2b).
STRUCTURE analysis showed that the optimal K value was K = 2 with Delta K of 235.302 (Figs. S3a, b, c), which suggests that the populations of R. meddianum and R. cyanocarpum are grouped into different clusters. This result is accordance with the conclusions of PCA and NJ trees analysis (Figs. S3d and e).
3.5. Evolutionary history of Rhododendron meddianum and R. cyanocarpumModel 4 (AIC weight = 0.9999; Table 4) estimated that the divergence time between Rhododendron meddianum and R. cyanocarpum from a common ancestor occurred in the late Pliocene, ~3, 048, 100 years ago (TDIV, 95%CI: 2, 213, 628–5, 026, 950) (Table 5, Fig. S4). The Ne of the two species ranged from 33, 827 to 35, 113, which is higher than that of the common ancestor (25, 546, 95%CI: 15, 435–37, 242). The gene flow/migration (Mmc) between the two species was extremely small (1.2477e-5, 95%CI: 1.1301e-5–1.7042e-5).
| Parameter | Point estimation | 95% CI lower bound | 95% CI upper bound |
| Ne_NANC | 25, 546 | 15, 435 | 37, 242 |
| TDIV | 3, 048, 100 | 2, 213, 628 | 5, 026, 950 |
| Ne_R.meddianum | 35, 113 | 23, 552 | 38, 231 |
| Ne_R.cyanocarpum | 33, 827 | 20, 661 | 37, 853 |
| Mmc | 1.2477e-5 | 1.1301e-5 | 1.7042e-5 |
Compared to widespread plant species, the genetic diversity of narrowly distributed species is likely to be lower because of inbreeding depression and genetic drift (Godt et al., 1997; Setoguchi et al., 2011). Unexpectedly, ddRAD sequencing indicated that nucleotide genetic diversity (0.0671–0.0813) of Rhododendron meddianum is higher than that of other endangered plant species, such as Viola uliginosa Muhl. (0.013–0.023), Cymbidium aloifolium (Linn.) Sw. (0.0136), and Geodorum densiflorum (Lam.) Schltr. (0.0359) (Roy et al., 2016; Lee et al., 2020). In addition, R. meddianum has higher genetic diversity than other widespread species, e.g. Taxus wallichiana Zucc. (0.0281–0.0433) (Qin, 2017). Yet, a similar level of genetic diversity was detected in R. cyanocarpum (0.0702) (Liu et al., 2020a). These results generally support the view that some rare and endangered species can maintain high-level genetic diversity even at small population sizes (Li et al., 2015; Stone et al., 2019; Liu et al., 2020a).
The genetic diversity of species is generally influenced by their distribution ranges, life span, breeding system, seed dispersal mechanism, and evolutionary history. In addition, genetic diversity is usually higher in outcrossing species than in selfing species (Hamrick, 1990; Nybom, 2004). Previous studies found that Rhododendron have adhesive pollen and require pollinators, indicating that Rhododendron may primarily rely on outcrossing (Ng and Corlett, 2000; Ma et al., 2015; Huang et al., 2017; Li et al., 2018). Our field surveys indicate that the primary pollination mode of R. meddianum is cross-pollination by insect (mainly Bombus spp.) and birds. The high genetic diversity of the three remaining populations of R. meddianum might also be derived from their ancestral populations, which has been the case in other endangered plants, e.g. Tricyrtis ishiiana (Kitag. et T. Koyama) Ohwi et Okuyama and Rhododendron protistum var. gigantum (Forrest ex Tagg) D.F. Chamb (Setoguchi et al., 2011; Wu et al., 2015).
4.2. Genetic differentiation and genetic structureSTRUCTURE analyses, PCA and NJ trees demonstrated that the three populations of Rhododendron meddianum (CJ, PM and HQ) are genetically separated in accordance with their geographical distributions. The FST of R. meddianum (0.1582–0.2388) revealed that a moderate genetic differentiation among the populations occurred (Wright, 1922). Furthermore, AMOVA showed that 19.88% of the genetic variation occurred amongR. meddianum populations. For rare and endangered plants, population genetic differentiation is usually affected by large geographical distances between sampled populations (Hamrick, 1990; Nybom, 2004). The distance among the populations in this study was more than 50 km, which hardly allows gene flow (pollen or seeds) between populations. Previous studies have indicated that the seed dispersal distance of Rhododendron species ranges from approximately 30–80 m (Ng and Corlett, 2000), and its pollen can be transmitted by bees and birds with dispersal distance of 3–10 km of (Ng and Corlett, 2000; Huang et al., 2017; Li et al., 2018). In addition, in line with our observation, seedlings grow sporadically around the parent tree. Hence, the differentiation among the populations could be attributed to distance-limited pollen flow and short-distance seed dispersal.
4.3. Population dynamic historyWe used stairway plot to explore the demographic history of Rhododendron meddianum between 150 and 100, 000 years ago (Fig. 3). Our analyses showed that a population bottleneck occurred approximately 70, 000 years ago. This bottleneck event, which is consistent with that of other plant species (e.g. Cycas balansae Warb., see Feng et al., 2014; Rhododendron rex Lévl., see Zhang et al., 2020), may have been caused by climate oscillations. This result also indicates a population expansion around 0.06–0.07 Ma, which is supported by the significantly negative value of Tajima'D (−0.6740) (Tajima, 1989). However, the Ne of R. meddianum experienced a contraction 300 years ago, which may have been the result of human activity and climate change. According to our survey, the CJ population habitat was severely damaged and fragmented by the construction of wind power stations and roads. Moreover, the habitat of the other two populations are all located near farmlands and threatened by grazing. Endangered plants usually experience decreases in its genetic diversity after bottleneck events (Wu et al., 2017). In our study, genetic diversity of R. meddianum was higher than that of other species, suggesting that a historical bottleneck did not reduce its genetic diversity, similar to in other Rhododendron species, such as R. cyanocarpum, R. protistum and R. rex (Wu et al., 2017; Liu et al., 2020a; Zhang et al., 2020). Adaptive radiations often occur when a species expands its ecological niche to adapt to diverse habitats (Cronk, 1995). Thus, one explanation for the high-level genetic diversity of R. meddianum is that when climate oscillations led to a population bottleneck, this event may have initially reduced the genetic diversity of the population, but then generated new genetic variation. In addition, a rapid population expansion took place after the population bottleneck, which might have contributed to the accumulation of mutations.
4.4. Evolutionary history of Rhododendron meddianum and R. cyanocarpumSTRUCTRUE, PCA and NJ trees analysis indicated that genetic structure ofRhododendron meddianum and R. cyanocarpum populations differed clearly. These two species, which are distributed in Western Yunnan, are divided by rivers and mountains (elev. > 2500 m) (Fig. S1). Topographical uplift may have isolated populations of R. meddianum and R. cyanocarpum, shaping their patterns of genetic diversity.
We estimated that Rhododendron meddianum and R. cyanocarpum diverged around 3.05 (2.21–5.03) Ma. This date is consistent with the conclusion of Mline et al. (2004), who estimated that most species of the Rhododenron subgenus Hymenanthes (mainly distribute in Himalayas and southern China) diverged from one another around 3–5 Ma. Neogene climate reconstructions of Western Yunnan indicate that the climate during the late Pliocene was slightly warmer and much wetter than the present climate (Su et al., 2013a, b). The Asian winter monsoon became significantly stronger after the late Pliocene, which was characterized by drier winters in southwestern China (Su et al., 2013a; Wang, 2006). Our field surveys indicated that the climate conditions of R. meddianum populations are drier than those of R. cyanocarpum populations. This ecological difference may have promoted genetic differentiation and led to speciation (Spicer et al., 2020). Therefore, we hypothesize that the intensification of the Asian winter monsoon after the late Pliocene may have contributed to genetic divergence of R. meddianum and R. cyanocarpum.
4.5. Conservation implicationsOur study on the genetic structure of Rhododendron meddianum provide important conservation implications for this narrowly distributed species. The most effective approach to conserving endangered species is in situ conservation (Wu et al., 2015). In wild populations of R. meddianum, seedlings and saplings are rarely found, and its habitat is disturbed and destroyed by various factors, including wind power and grazing. Therefore, it is imperative to establish conservation plots to protect its natural habitat. Meanwhile, seed collection from the all three R. meddianum populations (CJ, PM and HQ) should be conducted as soon as possible so they can be used for germplasm storage and ex situ conservation. Moreover, because moderate genetic differentiation and strong genetic structure were revealed among populations, germplasm resources from each population should be not mixed and instead should be used separately to avoid the risk of outbreeding depression (Liu et al., 2020b).
Author contributionsXJZ, ZHL, and DTL collected plant samples. XJZ, XFL and YRC performed the experiments. XJZ analyzed the data and wrote the manuscript. HM and YPM designed and supervised the study, and also wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interestThe authors declare that there are no conflict of interest.
AcknowledgementsThis work was supported by the Fundamental Research Funds for the Central Non-profit Research Institution of the Chinese Academy of Forestry (Grant No. CAFYBB2019ZB007); the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China (Grant No. 2019HJ2096001006); the Ten Thousand Talent Program of Yunnan Province (Grant No. YNWR-QNBJ-2019-010; YNWR-QNBJ-2018-174); and the Young and Middle-aged Academic and Technical Leader Reserve Talent Project of Yunnan Province (Grant No. 2018HB066).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.05.005.
Amor, M.D., Johnson, J.C., James, E.A., 2020. Identification of clonemates and genetic lineages using next-generation sequencing (ddRADseq) guides conservation of a rare species, Bossiaea vombata (Fabaceae). Perspect. Plant Ecol. Evol. Systemat., 45: 125544. DOI:10.1016/j.ppees.2020.125544 |
Catchen, J., Hohenlohe, P.A., Bassham, S., et al, 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol., 22: 3124-3140. DOI:10.1111/mec.12354 |
Catchen, J.M., Amores, A., Hohenlohe, P., et al, 2011. Stacks: building and genotyping Loci de novo from short-read sequences. G3-Genes Genome. Genet., 1: 171-182. DOI:10.1534/g3.111.000240 |
Cronk, Q., 1995. Images of nature. Nature, 376: 652-653. DOI:10.1038/376652a0 |
Cross, J.R., 1975. Biological flora of the British isles. Rhododendron ponticum L. J. Ecol., 63: 345-364. DOI:10.2307/2258859 |
Danecek, P., Auton, A., Abecasis, G., et al, 2011. The variant call format and VCFtools. Bioinformatics, 27: 2156-2158. DOI:10.1093/bioinformatics/btr330 |
Deschamps, S., Llaca, V., May, G.D., 2012. Genotyping-by-sequencing in plants. Biology, 1: 460-483. DOI:10.3390/biology1030460 |
Doyle, J., Doyle, J., 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull., 19: 11-15. |
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Everaert, H., De Wever, J., Tang, T.K.H., et al, 2020. Genetic classification of Vietnamese cacao cultivars assessed by SNP and SSR markers. Tree Genet. Genomes, 16: 43. DOI:10.1007/s11295-020-01439-x |
Excoffier, L., Dupanloup, I., Huerta-Sánchez, E., et al, 2013. Robust demographic inference from genomic and SNP data. PLoS Genet., 9: e1003905. DOI:10.1371/journal.pgen.1003905 |
Excoffier, L., Laval, G., Schneider, S., 2007. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinf. Online, 1: 47-50. |
Falush, D., Stephens, M., Pritchard, J.K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164: 1567-1587. DOI:10.1093/genetics/164.4.1567 |
Fang, M.Y., Fang, R.C., He, M.Y., et al., 2005. Rhododendron Linnaeus. In: Wu, Z.Y., Raven, P., Hong, D.Y. (Eds.), Flora of China, vol. 14. Science Press, Bejing. China and Missouri Botanical Garden Press, St. Louis Missouri USA, pp. 260-455.
|
Feng, X.Y., Wang, Y.H., Gong, X., 2014. Genetic diversity, genetic structure and demographic history of Cycas simplicipinna (Cycadaceae) assessed by DNA sequences and SSR markers. BMC Plant Biol., 14: 187. DOI:10.1186/1471-2229-14-187 |
Foll, M., Gaggiotti, O., 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics, 180: 977-993. DOI:10.1534/genetics.108.092221 |
Gibbs, D., Chamberlain, D., Argent, G., 2011. The red list of rhododendrons. Botanic Gardens Conservation International, Richmond, UK, p. 57.
|
Godt, M.J.W., Walker, J., Hamrick, J.L., 1997. Genetic diversity in the endangered lily Harperocallis flava and a close relative, Tofieldia racemosa. Conserv. Biol., 11: 361-366. DOI:10.1046/j.1523-1739.1997.95439.x |
Goetsch, L., Eckert, A.J., Hall, B.D., 2005. The molecular systematics of Rhododendron (Ericaceae): a phylogeny based upon RPB2 gene sequences. Syst. Bot., 30: 616-626. DOI:10.1600/0363644054782170 |
Groom, M.J., Chyi, Y. S., Carroll C.R., 2006. Principles of Conservation Biology. Sunderland, MA: Sinauer.
|
Hamrick, J.L, Godt, M.J.W., 1990. Plant Population Genetics, Breeding and Genetic Resources. Sunderland, MA: Sinauer.
|
Huang, C.L., Chen, J.H., Chang, C.T., et al, 2016. Disentangling the effects of isolation-by-distance and isolation-by-environment on genetic differentiation among Rhododendron lineages in the subgenus Tsutsusi. Tree Genet. Genomes, 12: 1-22. DOI:10.1097/NCC.0000000000000420 |
Huang, Z.H., Song, Y.P., Huang, S.Q., 2017. Evidence for passerine bird pollination in Rhododendron species. AoB Plants, 9: x62. |
Jiang, X.L., Gardner, E.M., Meng, H.H., et al, 2019. Land bridges in the Pleistocene contributed to flora assembly on the continental islands of South China: insights from the evolutionary history of Quercus championii. Mol. Phylogenet. Evol., 132: 36-45. DOI:10.1016/j.ympev.2018.11.021 |
Kaeuffer, R., Réale, D., Coltman, D.W., et al, 2007. Detecting population structure using STRUCTURE software: effect of background linkage disequilibrium. Heredity, 99: 374-380. DOI:10.1038/sj.hdy.6801010 |
Kumar, S., Stecher, G., Li, M., et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol., 35: 1547-1549. DOI:10.1093/molbev/msy096 |
Lee, K.M., Ranta, P., Saarikivi, J., et al, 2020. Using genomic information for management planning of an endangered perennial, Viola uliginosa. Ecol. Evol., 10: 2638-2649. DOI:10.1002/ece3.6093 |
Li, M.W., Chen, S.F., Shi, S., et al, 2015. High genetic diversity and weak population structure of Rhododendron jinggangshanicum, a threatened endemic species in Mount Jinggangshan of China. Biochem. Systemat. Ecol., 58: 178-186. DOI:10.3901/JME.2015.05.178 |
Li, T.Q., Liu, X.F., Li, Z.H., et al, 2018. Study on reproductive biology of Rhododendron longipedicellatum: a newly discovered and special threatened plant surviving in limestone habitat in Southeast Yunnan, China. Front. Plant Sci., 9: 33. DOI:10.3389/fpls.2018.00033 |
Lischer, H.E.L., Excoffier, L., 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 28: 298-299. DOI:10.1093/bioinformatics/btr642 |
Liu, D.T., Zhang, L., Wang, J.H., et al, 2020. Conservation genomics of a threatened Rhododendron: contrasting patterns of population structure revealed from neutral and selected SNPs. Front. Genet., 11: 757. DOI:10.1201/9781351187435-95 |
Liu, X.F., Ma, Y.P., Wan, Y.M., et al, 2020. Genetic diversity of Phyllanthus emblica from two different climate type areas. Front. Plant Sci., 11: 580812. DOI:10.3389/fpls.2020.580812 |
Liu, X.M., Fu, Y.X., 2015. Exploring population size changes using SNP frequency spectra. Nat. Genet., 47: 555-559. DOI:10.1038/ng.3254 |
Ma, Y.P., Wu, Z.K., Dong, K., et al, 2015. Pollination biology of Rhododendron cyanocarpum (Ericaceae): an alpine species endemic to NW Yunnan, China. J. Systemat. Evol., 53: 63-71. DOI:10.1111/jse.12114 |
Meffe, G.K., Carroll, C.R., 1997. Principles of Conservation Biology. Sunderland, MA: Sinauer.
|
Milne, R.I., 2004. Phylogeny and biogeography of Rhododendron subsection Pontica, a group with a Tertiary relict distribution. Mol. Phylogenet. Evol., 33: 389-401. DOI:10.1016/j.ympev.2004.06.009 |
Morimoto, J., Shibata, S., Hasegawa, S., 2003. Habitat requirement of Rhododendron reticulatum and Rhododendron macrosepalum in germination and seedling stages-filed experiment for restoration of native Rhododendron by seedling. J. Jpn. Soc. Reveg. Technol., 29: 15-140. DOI:10.7211/jjsrt.29.15 |
Ng, S., Corlett, R.T., 2000. Comparative reproductive biology of the six species of Rhododendron (Ericaceae) in Hong Kong, South China. Can. J. Bot., 78: 221-229. |
Nybom, H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol., 13: 1143-1155. DOI:10.1111/j.1365-294X.2004.02141.x |
Peterson, B.K., Weber, J.N., Kay, E.H., et al, 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One, 7: e37135. DOI:10.1371/journal.pone.0037135 |
Pritchard, J.K., Stephens, M., Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1093/genetics/155.2.945 |
Purcell, S., Neale, B., Todd-Brown, K., et al, 2007. Plink: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81: 559-575. DOI:10.1086/519795 |
Qin, H.N., Yang, Y., Dong, S.Y., 2017. Threatened species list of China's higher plants. Biodivers. Sci., 25: 696-744. DOI:10.17520/biods.2017144 |
Qin, H.T., 2017b. Population Genomics of Natural Hybrid Species of Taxus in Hengduan Mountains region. University of Chinese Academy of Sciences.
|
R Core Team, 2016. R: a Language and Environment for Statistical Computing. Available from:. R foundation for statistical computing, Vienna, Austria. Available from https://www.R-project.org/.
|
Roy, S.C., Moitra, K., De Sarker, D., 2016. Assessment of genetic diversity among four orchids based on ddRAD sequencing data for conservation purposes. Physiol. Mol. Biol. Plants, 23: 169-183. DOI:10.1515/ijame-2016-0011 |
Setoguchi, H., Mitsui, Y., Ikeda, H., et al, 2011. Genetic structure of the critically endangered plant Tricyrtis ishiiana (Convallariaceae) in relict populations of Japan. Conserv. Genet., 12: 491-501. DOI:10.1007/s10592-010-0156-y |
Spicer, R.A., Farnsworth, A., Su, T., 2020. Cenozoic topography, monsoons and biodiversity conservation within the Tibetan region: An evolving story. Plant Divers., 42: 229-254. DOI:10.1016/j.pld.2020.06.011 |
Stone, B.W., Ward, A., Farenwald, M., et al, 2019. Genetic diversity and population structure in Cary's Beardtongue Penstemon caryi (Plantaginaceae), a rare plant endemic to the eastern Rocky Mountains of Wyoming and Montana. Conserv. Genet., 20: 1149-1161. DOI:10.1007/s10592-019-01204-1 |
Su, T., Jacques, F., Spicer, R.A., et al, 2013. Post-Pliocene establishment of the present monsoonal climate in SW China: evidence from the late Pliocene Longmen megaflora. Clim. Past, 9: 1911-1920. DOI:10.5194/cp-9-1911-2013 |
Su, T., Liu, Y.S., Jacques, F., et al, 2013. The intensification of the East Asian winter monsoon contributed to the disappearance of Cedrus (Pinaceae) in southwestern China. Quat. Res. (Tokyo), 80: 316-325. DOI:10.1016/j.yqres.2013.07.001 |
Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123: 585-595. DOI:10.1093/genetics/123.3.585 |
Wagutu, G.K., Fan, X., Njeri, H.K., et al, 2020. Development and characterization of EST-SSR markers for the endangered tree Magnolia patungensis (Magnoliaceae). Ann. Bot. Fenn., 57: 97-107. DOI:10.5735/085.057.0114 |
Wang, B., 2006. The Asian Monsoon. Berlin, Heidelberg: Springer Praxis Books, Springer.
|
Wright, S., 1922. Coefficients of inbreeding and relationship. Am. Nat., 56: 330-338. DOI:10.1086/279872 |
Wu, F.Q., Shen, S.K., Zhang, X., et al, 2017. Inferences of genetic structure and demographic history of Rhododendron protistum var. giganteum-The world's largest Rhododendron using microsatellite markers. Flora, 233: 1-6. DOI:10.1109/ATNAC.2017.8215386 |
Wu, F.Q., Shen, S.K., Zhang, X.J., et al, 2015. Genetic diversity and population structure of an extremely endangered species: the world's largest Rhododendron. AoB Plants, 7: plu082. DOI:10.1093/aobpla/plv082 |
Xiao, J.H., Ding, X., Li, L., et al, 2020. Miocene diversification of a golden-thread nanmu tree species (Phoebe zhennan, Lauraceae) around the Sichuan Basin shaped by the East Asian monsoon. Ecol. Evol., 10: 10543-10557. DOI:10.1002/ece3.6710 |
Yamashita, H., Katai, H., Kawaguchi, L., et al, 2019. Analyses of single nucleotide polymorphisms identified by ddRAD-seq reveal genetic structure of tea germplasm and Japanese landraces for tea breeding. PLoS One, 14: e220981. |
Yilmaz, A., 2016. Analysis of genetic variation among some Turkish oaks using random amplified polymorphic DNA (RAPD) method. Afr. J. Biotechnol., 231: 17-18. |
Yoichi, W., Tamaki, I., Sakaguchi, S., et al, 2016. Population demographic history of a temperate shrub, Rhododendron weyrichii (Ericaceae), on continental islands of Japan and South Korea. Ecol. Evol., 6: 8800-8810. DOI:10.1002/ece3.2576 |
Zhang, X., Liu, Y.H., Wang, Y.H., et al, 2020. Genetic diversity and population structure of Rhododendron rex subsp. rex inferred from microsatellite markers and chloroplast DNA sequences. Plants, 9: 338. DOI:10.3390/plants9030338 |



