b. School of Ecology and Environmental Science & Yunnan Key Laboratory for Plateau Mountain Ecology and Restoration of Degraded Environments, Yunnan University, Kunming 650091, China;
c. CAS Key Laboratory for Plant Diversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650204, China
Over the past three decades, technological and conceptual advances have led to an explosive growth in plant phylogeographic studies (Beheregaray, 2008; Hickerson et al., 2010; Morris and Shaw, 2018). Numerous studies have shown that past geological events and climate fluctuations play a major role in shaping the distributions and genetic structures of extant organisms (Comes and Kadereit, 2003; Qiu et al., 2011; Liu et al., 2012; Tian et al., 2020). Although only a small fraction of the existing taxa have been investigated, phylogeography has revealed various patterns and mechanisms for diversification across a wide range of taxonomic groups (Zamudio et al., 2016).
Drylands, which occupy about 40% of the Earth's land surface, are characterized by low and highly variable precipitation, intense solar radiation, extreme air temperatures and seasonally high potential evaporation (Reynolds et al., 2007; Maestre et al., 2012). These climatic characteristics, coupled with the relatively low soil fertility, impose important limitations on the biota of drylands, and make them highly sensitive and vulnerable to global climate change (Le Houérou, 1996; Körner, 2000; Sala et al., 2000; Reynolds et al., 2007; Maestre et al., 2012). Drylands encompass diverse ecosystems and host about 20% of the major centers of plant diversity worldwide (Field et al., 1998; Kier et al., 2005; Maestre et al., 2012). Consequently, researchers have suggested that drylands are ideal natural laboratories for the study of plant evolution and adaptation to extreme conditions (Maestre et al., 2012; Ward, 2016). The arid northwest region of China is one of the largest mid-latitude, temperate, continental interior dryland areas in the world. The disjunctive deserts of this region (i.e., the Taklamakan Desert, Badain Jaran Desert, and Tengger Desert) (Yang and Scuderi, 2010), have attracted increasing interest from researchers (e.g., Meng et al., 2015; Yin et al., 2015; Zhang et al., 2017b; Zeng et al., 2018; Li et al., 2019; Zhao et al., 2019; Yin et al., 2020). These studies have revealed that three factors have played a role in shaping the population genetic structure of plants occurring in this region: increased aridification, subsequent desert expansion, and changes in the East Asian monsoon system (Yin et al., 2015, 2020; Zhang et al., 2017b). Meanwhile, studies have shown that geographical barriers (e.g., mountains, sand dunes and continuous oases) may have led to intraspecific differentiation and even promoted speciation processes (Zeng et al., 2018; Li et al., 2019; Zhao et al., 2019).
Arnebia szechenyi Kanitz (Boraginaceae) is an herbaceous perennial plant inhabiting sunny rocky slopes and sand dune edges in Northwest China (Zhu et al., 2005). This typically distylous species with heteromorphic self-incompatibility has five distinct black nectar guides on the yellow corolla lobes, and indicating that it requires pollinators for reproductive success (Zhang et al., 2014, 2017a; Wang et al., 2015a). Interestingly, it has a nearly circular distribution surrounding the Tengger Desert (Fig. 1), making it a suitable model to study the diverse responses of desert plants to Pleistocene climatic fluctuations, aridification and desert growth as well as geographical barriers. In this study, we used chloroplast DNA (cpDNA) fragments, the nuclear ribosomal region (nrITS) and low-copy nuclear genes (LCGs) and employed ecological niche modelling (ENM) to infer the phylogeography of populations of A. szechenyi surrounding the Tengger Desert. Our specific study aims were to (a) explain range-wide patterns of population genetic diversity of A. szechenyi at cpDNA and nuclear markers; (b) determine the possible intraspecific divergence and gene flow patterns driven by the Tengger Desert and other factors; (c) infer past population and range dynamics and disentangle the underlying environmental causes.
|
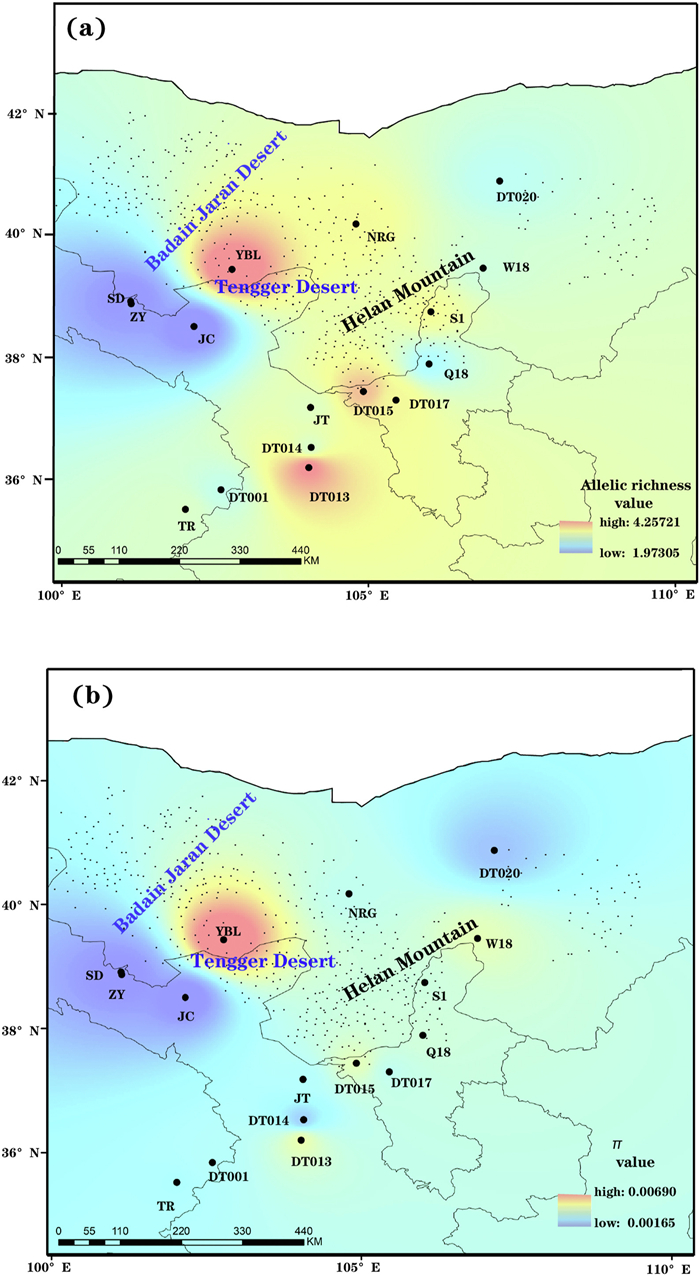
| Fig. 1 Modelled spatial distribution of allelic richness (AR) (a) and nucleotide diversity (π) (b) in Arnebia szechenyi based on seven nDNA loci. |
We collected leaves of Arnebia szechenyi from 16 natural populations covering the whole range of A. szechenyi. We sampled 6–12 individuals per population with sampled individuals located more than 100 m apart in a population. Fresh leaves without disease were collected and dried immediately using silica gel in the field. For each population, one sample was deposited as a voucher specimen for further reference in the Southwest Forestry University Herbarium (SWFC). The latitude, longitude and elevation of each sample site was recorded using a GIS monitor. Detailed information on the distribution of the samples is shown inTable 1 and Fig. 1. Two congeneric species, Arnebia guttata Bge. and Arnebia fimbriata Maxim., located in Zhangye, Gansu (38°52′12″N, 101°7′48.0″E) and Helan Mountain, Ningxia (38°42′35.99″N, 105°59′24″E) were also sampled as outgroups for phylogenetic analysis.
| Region | Pop | Location | Latitude | Longitude | cpDNA | nDNA | ||||||
| n | Hd | π (x 10−4) | Haplotypes | N | AR | π (x 10−3) | ||||||
| eastern | DT001 | Xunhua, Qinghai | 35.84 | 102.6 | 5 | 0.00 | 0.0 | H1(5) | 5 | 2.91 | 3.41 | |
| DT013 | Gaolan, Gansu | 36.20 | 104.03 | 16 | 0.00 | 0.0 | H2(16) | 16 | 4.01 | 4.43 | ||
| DT014 | Baiyinxian, Gansu | 36.53 | 104.07 | 10 | 0.78 | 39.1 | H2(3), H3(1), H4(2), H5(4) | 10 | 3.10 | 2.57 | ||
| DT015 | Zhongweisha, Ningxia | 37.44 | 104.92 | 14 | 0.58 | 72.6 | H2(5), H6(8), H7(1) | 13 | 3.73 | 4.26 | ||
| DT017 | Sizikou, Ningxia | 37.30 | 105.45 | 16 | 0.77 | 13.8 | H2(6), H8(2), H9(4), H10(4) | 15 | 3.44 | 3.60 | ||
| DT020 | Wulate, Neimenggu | 40.87 | 107.14 | 8 | 0.43 | 2.5 | H11(6), H12(2) | 10 | 2.85 | 2.45 | ||
| S1 | Mt.Helan, Ningxia | 38.74 | 106.02 | 9 | 0.50 | 4.8 | H32(6), H33(3) | 10 | 3.37 | 3.81 | ||
| W18 | Mt.Xizhuozi, Ningxia | 39.45 | 106.87 | 10 | 0.00 | 0.0 | H31(10) | 5 | 2.99 | 4.31 | ||
| Q18 | Qingtongxia, Ningxia | 37.89 | 105.99 | 7 | 0.00 | 0.0 | H31(7) | 14 | 2.76 | 3.87 | ||
| JT | Jingtai, Gansu | 37.18 | 104.06 | 9 | 0.95 | 68.0 | H4(1), H18(1), H19(3), H20(2), H21(1), H22(1) | 14 | 3.14 | 3.38 | ||
| TR | Tongrenxian, Gansu | 35.52 | 102.02 | 11 | 0.77 | 52.6 | H35(4), H36(5), H37(2) | 10 | 3.10 | 3.31 | ||
| Total eastern region | 115 | 0.89 | 73.7 | 122 | 23.93 | 5.85 | ||||||
| western | JC | Jinchang, Gansu | 38.50 | 102.16 | 13 | 0.69 | 47.3 | H13(1), H14(3), H15(7), H16(1), H17(1) | 13 | 2.00 | 1.65 | |
| NRG | Nuorigong, Neimenggu | 40.17 | 104.8 | 11 | 0.95 | 47.7 | H23(2), H24(2), H25(1), H26(1), H27(2), H28(1), H29(1), H30 (1) | 14 | 3.34 | 3.59 | ||
| SD | Shandanxian, Gansu | 38.91 | 101.12 | 11 | 0.00 | 0.0 | H34(11) | 16 | 1.97 | 2.01 | ||
| YBL | Yabulai, Neimenggu | 39.43 | 102.78 | 16 | 0.24 | 2.5 | H38(14), H39(1), H40(1) | 11 | 4.26 | 6.90 | ||
| ZY | Zhangye, Gansu | 38.87 | 101.13 | 15 | 0.53 | 3.9 | H34(10), H41(3), H42(2) | 14 | 2.29 | 1.62 | ||
| Total western region | 66 | 0.85 | 53.1 | 68 | 17.57 | 6.28 | ||||||
| Total All regions | 181 | 0.94 | 72.2 | 190 | 17.00 | 7.11 | ||||||
We used DNeasy™ Tissue Kit (Qiagen, Hilden, Germany) to isolate total genomic DNA from dried leaves. We amplified and sequenced six nuclear LCG loci (28490, 33938, 38060, 40789, 40947, 43173), the nuclear ribosomal internal transcribed spacer (nrITS), and three chloroplast DNA (cpDNA) intergenic spacers (psbA-trnH, psbB-psbF and trnL-trnF) for all individuals following previous studies (Rock et al., 1987; White et al., 1990; Taberlet et al., 1991; Sang et al., 1997; Fu et al., 2016; see Appendix S1 for details). MEGA v.7.0 (Kumar et al., 2016) was used to adjust and align generated sequences. DnaSP v.5.0 (Librado and Rozas 2009) was used to phase heterozygous sequences of the nuclear genes. The newly identified sequences were deposited in GenBank under the accession numbers MW229731-MW231889 and MW229277-MW229730. Sequences from the three cpDNA regions were concatenated into a matrix.
2.3. Nuclear DNATo assess selective neutrality of loci from biparentally inherited nuclear data (nrITS and LCGs), we first calculated Tajima's D (Tajima, 1989), Fu and Li's D* (Fu and Li, 1993) and Fu and Li's F* (Fu, 1997) in DnaSP v.5.0 (Librado and Rozas, 2009). Nucleotide diversity (π) and allelic richness (AR) for each population were calculated using SPADS v.1.0 (Dellicour and Mardulyn, 2014). Interpolation surfaces of population genetic diversity were then constructed in the same software. Pearson correlations between latitude/longitude and genetic diversity were performed in R 4.0.1 (R Core Team, 2013). Potential population structure was examined using STRUCTURE v.2.3.4 (Falush et al., 2007) with an admixture model and assuming allele frequencies to be correlated among populations (K = 1 to 10, with 1, 000, 000 Markov chain Monte Carlo (MCMC) steps and a burn-in of the first 10, 000 steps). The most likely number of clusters was determined by combining LnP(D) and ΔK. Additionally, we performed a distance-based clustering method through principal coordinate analysis (PCoA) based on pairwise Fst values among populations using SPADS.
To detect any potential demographic expansions within the identified genetic groups, temporal changes in the effective population size (Ne) were inferred with extended Bayesian skyline plots (EBSP) as implemented in BEAST v.2.4. The best-fit models of nucleotide substitution for each locus were determined using jModelTest. To date, the substitution rates for the markers used in this study have not been estimated in Arnebia szechenyi or other congeners. For most angiosperms, the substitution rates have been estimated to vary between 0.38 and 8.34 × 10−9 s/s/y for nrITS (Kay et al., 2006). To address the uncertainties in these rate values, we used a normal distribution prior with a mean of 4.36 × 10−9 s/s/y and a SD of 1.59 × 10−9 s/s/y for nrITS; this covered the range of rates within 95% of the distribution, in accordance with Jia et al. (2012) and Wang et al. (2016). For the substitution rate of the LCGs, we used a normal distribution prior with a mean of 1.07 × 10−8 s/s/y and a SD of 3.24 × 10−9 s/s/y, assuming a substitution ratio of 3:16 between cpDNA and the LCGs (Drouin et al., 2008). The MCMC was run for 400, 000, 000 steps with sampling once every 400, 000 steps, and the first 20% discarded as burn-in.
To identify periods of long-term isolation among identified genetic groups, we estimated patterns of historical gene flow using the Maximum-Likelihood (ML) algorithm in Migrate v.3.7.2 (Beerli, 2006), we used θ = 4Nμ (N, the effective population size; μ, the mutation rate per generation) and M = m/μ (m, the migration rate per generation) to calculate the effective number of migrants (Nm). Ten short chains (200, 000 trees) and three long chains (1, 000, 000 trees) were run with a burn-in of 10, 000 initial trees. The starting values for θ and M were estimated from Fst values. The analyses were performed three times independently with all individuals included.
2.4. Chloroplast DNAFor each population, we calculated the genetic diversity (nucleotide diversity π and haplotype diversity Hd) of maternally inherited cpDNA using DnaSP v.5.0 (Librado and Rozas, 2009). The total genetic diversity (Ht), within-population genetic diversity (Hs), and between-population genetic differentiation (Nst, Gst) were estimated using the program Permut 1.0 with 1000 permutation tests (Pons and Petit, 1996; available at http://www.pierroton.inra.fr/genetics/labo/Software/Permut/). The partitioning of genetic variation at different levels was estimated by a hierarchical analysis of molecular variance (AMOVA; Excoffier et al., 1992) using 1000 permutations in Arlequin v.3.5 (Excoffier and Lischer, 2010).
To examine the demographic expansions within the genetic groups identified, we conducted a mismatch distribution analysis (MDA; Rogers and Harpending, 1992) and an EBSP analysis as implemented in DnaSP v.5.0 (Librado and Rozas, 2009) and BEAST v.2.4, respectively. The EBSP analysis was run as described above. We also calculated Tajima's D (Tajima, 1989) and Fu & Li's D* test (Fu and Li, 1993) to assess possible expansions. If Arnebia szechenyi genetic groups had undergone expansions, Tajima's D and Fu & Li's D* test should generate large negative values due to excess rare mutations. We calculated significance of the tests with 1000 replicates in DnaSP v.5.0 (Librado and Rozas, 2009).
We analyzed phylogenetic relationships of the different cpDNA and estimated genetic divergence through Bayesian analyses using BEAST v.2.4 (Bouckaert et al., 2014). For most angiosperms, cpDNA substitution rates have been estimated to vary between 1 and 3 × 10−9 substitutions per site per year (s/s/y) (Wolfe et al., 1987). In this study, we used a normal distribution prior with a mean cpDNA substitution rate of 2. 0 × 10−9 s/s/y and a SD of 6.08 × 10−10 s/s/y (Jia et al., 2012; Wang et al., 2016). Best-fit models of nucleotide substitution for cpDNA matrices were determined to be HKY by the AIC using jModelTest v.2.1.10 (Darriba et al., 2012). A strict clock model and Yule process with a piecewise linear and constant population size model were applied. The length of the MCMC algorithm was set to 10, 000, 000 steps. We sampled all parameters once every 1000 steps out of 10, 000, 000 MCMC steps, with the first 10% of samples discarded as burn-in. The convergence of chains to a stationary distribution (effective population sizes > 200) was examined in Trace v.1.7 (Rambaut et al., 2018). Trees were then compiled into a maximum clade credibility (MCC) tree using TreeAnnotator v.2.4 (Bouckaert et al., 2014) to display posterior probabilities (PP) and mean node ages for each node. The program FigTree v.1.7.1 (available at http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the MCC tree.
To detect genealogical relationships among sequences with shallow genetic divergences, we also constructed the most parsimonious cpDNA haplotype networks using Network v.5.0 (available at http://www.fluxus-engineering.com/sharenet.htm).
2.5. Ecological niche modellingWe used a maximum-entropy modelling technique to predict the potential distribution of Arnebia szechenyi during the Last Glacial Maximum (LGM) and at present as implemented in MAXENT v.3.4.1 (Phillips et al., 2006). A total of 34 occurrences, including the 16 sampled locations, were obtained from the Chinese Virtual Herbarium (http://www.cvh.org.cn/), the Global Biodiversity Information Facility (http://data.gbif.org) and records reported in grassland community studies. To reduce the influence of sampling deviation on the accuracy of simulation results, we used a spatial filtering method to randomly reserve one of any two distribution coordinates with a distance less than 1 km, and a total of 34 coordinate points were reserved after filtering. Environmental variables, including 19 bioclimatic and one elevation variable, eight soil variables and six ultraviolet rays, were downloaded from the world biological climate database (https://www.worldclim.org/), HWSD database (http://www.fao.org/) and world soil GIUV database (http://www.ufz.de/gluv/). To avoid model overfitting caused by strong correlations between environmental variables, Pearson correlation coefficients were used to screen variables. When each pair of environmental variables was correlated (|r| > 0.75), the variable that contributed the most to the model was retained Appendix S2). To better project the simulation results to the LGM period, only linear (L) and quadratic (Q) were selected for the eigenfunction, and appropriate regularization parameters were selected for the model using the R package ENMeval (Muscarella et al., 2014). We performed a 10-fold cross-validation procedure to evaluate the models. The accuracy of model predictions was evaluated based on the area under the curve of the receiver operating characteristic (AUC; Fawcett, 2006). Finally, Arcgis v.10.5 was used to reclassify the simulation results and map the suitability of species distribution in each period.
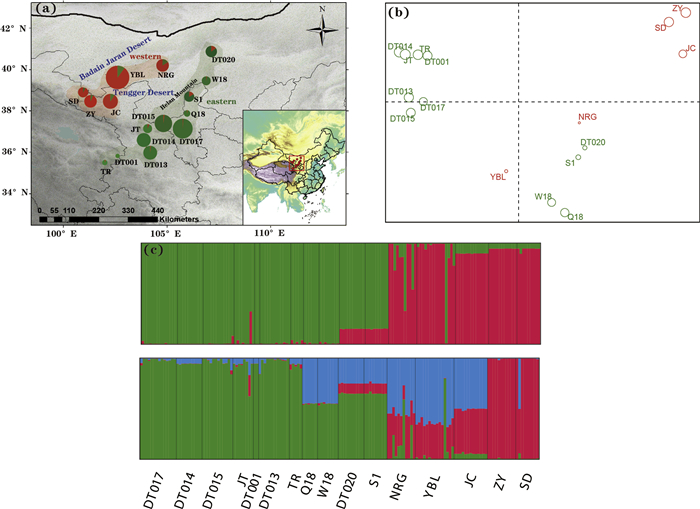
3. Results 3.1. Nuclear DNAMeasures of genetic variation varied across populations, with nucleotide diversity (π) ranging from 1.62 × 10−3 to 6.90 × 10−3, and allele richness (AR) varying from 1.97 to 4.26 (Table 1). The highest levels of genetic diversity were found in population YBL (AR = 4.26 and π = 6.90 × 10−3) (Table 1, Fig. 1). Neutrality tests detected significant negative D and F* values and a large negative D* value for locus 33938 (Appendix S3), indicating a departure from neutral expectations. We therefore excluded this locus from further analyses. Bayesian clustering analysis implemented in STRUCTURE clearly showed two distinct genetic groups (Fig. 2a and c; Appendix S4), corresponding to the two geographic regions separated by the Tengger Desert. The western genetic group was composed of five populations (SD, ZY, JC, YBL and NRG) located between the Badain Jaran Desert and the Tengger Desert, while the eastern genetic group consisted of the remaining eleven populations located southeast of the Tengger Desert and along the Helan Moutains. However, PCoA yielded a different pattern of genetic clustering, with the six northern populations from the western genetic group (populations YBL and NRG) and the eastern genetic group (populations DT020, W18, S1 and Q18) clustered into another genetic group (Fig. 2b). The genetic differentiation between the two genetic groups was supported by a large Fst value (0.71). Migrate analysis indicated that historical migration between eastern and western genetic groups was significantly asymmetrical. Unidirectional gene flow was considerably higher from eastern to western genetic groups (Nm = 32.90; 95% HPD: 26.64–34.41) than from western to eastern groups (Nm = 0.64; 95% HPD: 0.52–0.68). Nevertheless, we did not observe any significant correlation between genetic diversity (AR and π), latitude, or longitude (Appendix S5).
|
| Fig. 2 (a) Frequency distribution of the two genetic clusters as revealed in STRUCTURE analysis with K = 2 in the 16 Arnebia szechenyi populations.The eastern genetic group (EAST) which consists of eleven populations located southeast of the Tengger Desert and along the Helan Moutains; the western genetic group (WEST), which is composed of five populations (SD, ZY, JC, YBL and NRG) located between the Badain Jaran Desert and the Tengger Desert; (b) Principle component analysis based on pairwise genetic differentiation (Fst) of populations. (c) Histogram of the STRUCTURE assignments for all individuals at K = 2 and K = 3. |
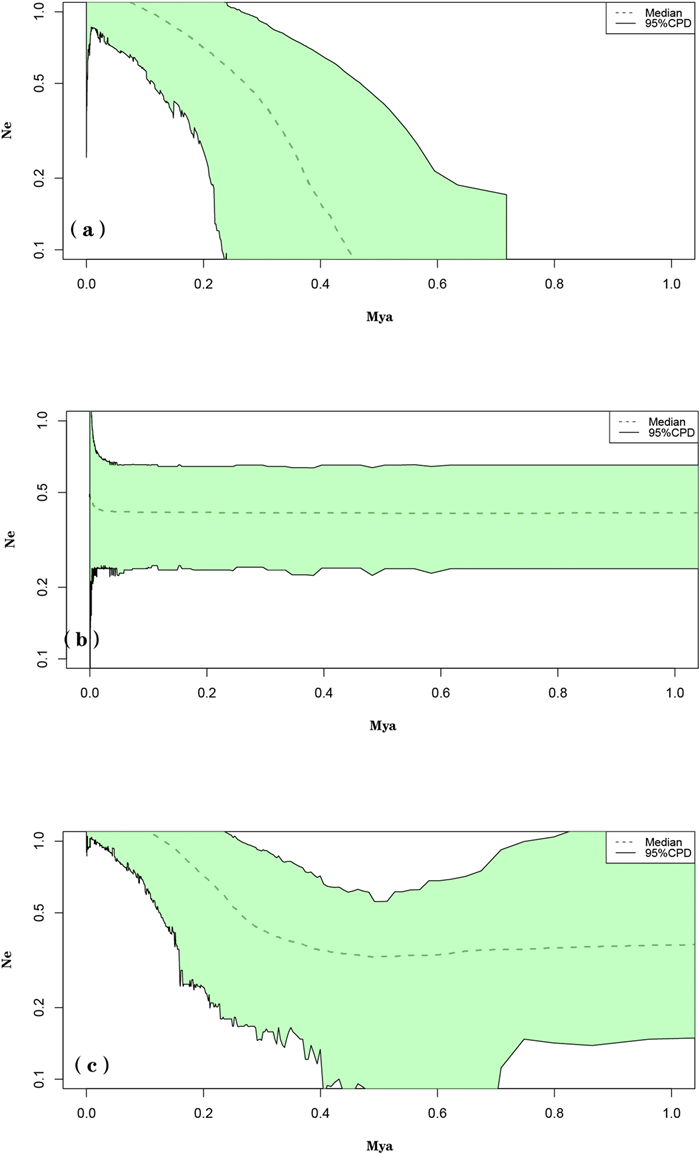
Extended Bayesian skyline plots (EBSP) (see models in Appendix S6) suggest that the eastern genetic group and all populations combined have undergone historical expansion episodes, and that the effective population size has increased over the last 0.4–0.5 Mya (Fig. 3a and c).
|
| Fig. 3 BEAST-derived extended Bayesian skyline plot (EBSP) based on nDNA for (a) the eastern genetic group; (b) the western genetic group and (c) all populations combined. |
After combining the three cpDNA fragments, we identified a total of 42 chlorotypes (H1–H42; Table 1; see details in Appendix S7). Within the 16 populations, nucleotide diversity (π) ranged from 0 to 72.60 × 10−4 and haplotype diversity (Hd) varied from 0 to 0.95 (Table 1). The highest levels of haplotype diversity were found in populations NRG and JT (0.95), while population DT015 showed the highest nucleotide diversity (72.60 × 10−4). The total genetic diversity (Ht) (0.98 ± 0.02) throughout all populations was much higher than the average within-population diversity (Hs) (0.50 ± 0.09), indicating that the majority of the cpDNA diversity was distributed among populations. The difference between the coefficient of genetic differentiation for non-ordered alleles (Gst = 0.49) and that for ordered alleles (Nst = 0.48) was non-significant (P = 0.10), thus revealing the absence of phylogeographical structure in Arnebia szechenyi (Appendix S8). The hierarchical AMOVA attributed most of the total genetic variation (64%) within populations, while a relatively low percentage (7%) to differences among the two genetic groups Appendix S9).
Neither the eastern nor western genetic groups show signs of unimodal mismatch distribution under models of population expansion, nor do all population combined (Appendix S10 and Appendix S11). However, the EBSP analysis inferred that the effective population underwent slight fluctuations in size around 0–0.05 Mya (Appendix S12).
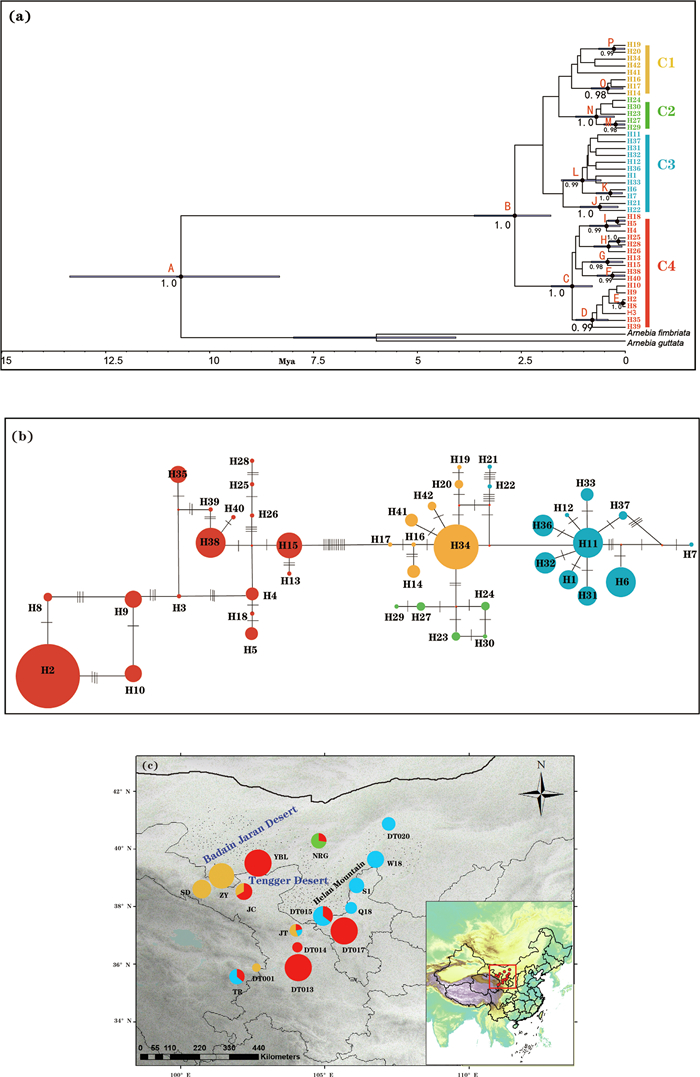
All haplotypes from Arnebia szechenyi formed a monophyletic lineage with four major clades (C1–C4) (Fig. 4a and b). The haplotypes placed in clades C2 and C3 were exclusively distributed in the western genetic groups and the eastern genetic groups, respectively; the haplotypes place in C1 or C4 were mainly distributed in either the western (C1) or eastern (C4) genetic groups (Fig. 4c). The haplotype network was largely congruent with the phylogenetic tree, although it depicted relationships between haplotypes in more detail. The dating analysis estimated that A. szechenyi originated in the Late Miocene (10.69 Mya; 95% HPD: 8.32–13.36 Mya), and started to diversify in the early Pleistocene (2.66 Mya; 95% HPD: 1.79–3.64 Mya) (Fig. 4a).
|
| Fig. 4 (a) BEAST-derived chronogram of divergence times within Arnebia szechenyi based on three chloroplast DNA fragments (psbA-trnH, psbB-psbF, trnL-trnF). Blue bars on each node indicate 95% highest posterior density (HPD) intervals for the TMRCA estimates. Posterior probabilities (PP) > 0.95 are shown above nodes. (b) The most parsimonious network of the haplotypes. Circles sizes are proportional to chlorotype frequencies within the total sample. Small red dots represent hypothetical unsampled or extinct ancestral haplotypes, and lines between circles represent single mutational steps. Pie colors correspond the different clades identified. (c) Geographical distribution of the four cpDNA clades. Pie charts show the relative proportion of each cpDNA clade within each population. |
Ecological niche modelling indicated that the current distribution of Arnebia szechenyi is a good fit to its potential distribution (AUC value = 0.97 ± 0.03). Our analysis also indicated that the potential range of A. szechenyi underwent a considerable contraction during the Last Glacial Maximum (LGM) (Fig. 5). However, compared with the current distribution, the most suitable habitat remained in the areas of the Hexi corridor, south and east of the Tengger Desert and Helan Mountains (Fig. 5).
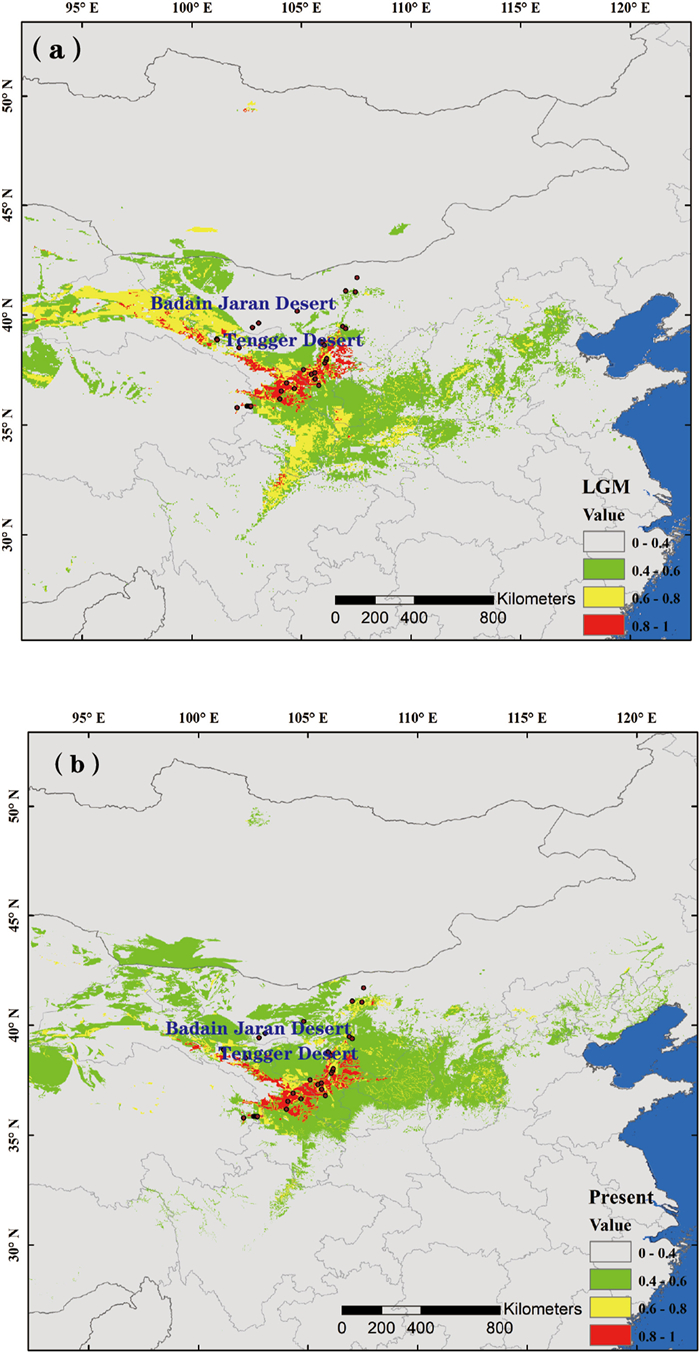
|
| Fig. 5 Potential species distribution of Arnebia szechenyi based on ecological niche model of the Last Glacial Maximum (a), and at present (b). |
Our analyses based on both nuclear and chloroplast data revealed a considerably high level of genetic variation within Arnebia szechenyi. From the 181 individuals sampled, we recovered a total of 42 chlorotypes. The total genetic diversity (Ht = 0.98) across all populations was much higher than the average within-population diversity (Hs = 0.50) indicating that the majority of cpDNA diversity was distributed among populations. This pattern of genetic variation has also been observed in other herbaceous plants occurring in the drylands of Northwest China (e.g. Lagochilus ilicifolius: Meng and Zhang, 2011; Allium mongolicum: Zhang et al., 2017). This genetic variation is probably associated with the particular ecological conditions in the drylands and the long evolutionary histories of desert plants (Meng and Zhang, 2011). The biophysical characteristics and landscapes of drylands likely imposed important limitations on their biota and produced discrete vegetation patches (Maestre et al., 2012). As a consequence, isolated populations may have limited gene flow to maintain genetic similarity among populations. Additionally, the distyly and heteromorphic self-incompatibility ofA. szechenyi, which is considered a mechanism that evolved to promote outcrossing and avoid selfing (Lloyd and Webb 1992; Barrett et al., 2000; Zhou and Wang, 2009), ensures higher levels of genetic diversity within populations, conducive to evolution and adaptation to the harsh environment (Zhang et al., 2014).
4.2. Intraspecific divergence and chloroplast-nuclear discordanceThe chloroplast genome, which is maternally inherited and does not undergo recombination, has lower rates of gene flow and smaller effective population sizes than nuclear DNA (Du et al., 2009). These differences in inheritance underlie how the two genomes have responded to past demographic fluctuations and biogeographic events (Després, 2019; Hinojosa et al., 2019). Our Bayesian clustering analysis based on nDNA showed a clear split between the western and the eastern genetic groups, which corresponds to the two geographic regions separated by the Tengger Desert (Fig. 2a; Appendix S4). However, analyses based on cpDNA revealed a different pattern. Although the haplotypes in clade C2 and in C3 were exclusively distributed in the western and eastern genetic groups, respectively, the haplotypes in C1 or C4 were only mostly distributed in either the western or the eastern genetic groups, with haplotypes from each clade distributed in the eastern and western genetic groups (Fig. 4c).
Cytonuclear discordance can be caused by incomplete lineage sorting, introgression, selection or gene flow (Barrett and Schluter, 2008; Sloan et al., 2017). In this study, incomplete lineage sorting and selection seem unlikely to have caused such a discordance, due to the high sequence variation among the samples and rapid progression of lineage sorting in plastid DNA (Funk and Omland, 2003). Further, because Arnebia szechenyi was recovered as monophyletic with respect to the closely related congeners A. guttata, A. linearifolia and A. fimbriata (Wang et al., 2015), the role of introgression can also be neglected.
Our estimations based on cpDNA suggest that genetic divergence within Arnebia szechenyi occurred 2.66 Mya (95% HPD: 1.79–3.64 Mya), corresponding to the late Pliocene. The estimated timescales for this divergence fit well with the uplift of the Helan Mountains and the Qilian Mountains during the middle-late Pliocene (2.60–3.55 Mya; Li, 2013), which blocked water vapor from the Pacific Ocean and the Arctic Ocean (Xie and Zhang, 2013; Meng et al., 2015). Between 2.85 and 2.60 Mya, the deposition rate of alluvial-diluvial deposits and fluvial rocks in the Tengger desert area increased. At the same time, the high Himalayan Mountains and the Qinghai-Tibet Plateau (QTP) impeded warm and moist air flow from the Indian Ocean (Zhang et al., 2000). Together, these factors intensified the aridification in the Tengger desert region (Li, 2013). The severe drought conditions combined with habitat loss and destruction could have caused local or even regional extinction of A. szechenyi in the hinterland, and/or blocked gene flow between surviving geographically isolated populations and therefore promoted intraspecific divergence. After the Tengger Desert formed during the middle Pleistocene (1.1–0.9 Mya; Li, 2013; Li et al., 2014; Wang et al., 2015), its repeated expansions and contractions further accelerated differentiation within each genetic group (Fig. 4a).
During repeated geographical and demographic expansions and contractions, the nuclear genomes may have come into contact and fully recombined (in absence of reproductive isolation); however, divergent nonrecombinant chlorotypes may have been retained, as drift plays no role in large expanding populations. In addition, we found no phylogeographical structure across all populations of the species (Nst = 0.48 vs. Gst = 0.49; P = 0.10). Such a result might also indicate a complex history of colonization by gene pools that had been genetically distinct for a long period and spread across the whole distribution range. In light of this evidence, the most likely explanation for the chloroplast-nuclear discordance observed in the Arnebia szechenyi populations is that these populations experienced long periods of geographic isolation followed by range expansions precipitated by the formation of the Tengger Desert area, which homogenized the nuclear genome (which evolves rapidly) but not the chloroplast genome.
4.3. Regional expansionIn addition to intraspecific divergence and diversification due to long-term isolation, geological processes and climatic fluctuations may have driven range shifts in regions that were not continuously isolated (Avise, 2004). Generally, range expansions result in a series of founder events leading to loss of genetic diversity and wide fixtures of single chlorotypes (Avise et al., 1987; Hewitt, 1996, 1999, 2000; Comes and Kadereit, 1998). However, genetic signs of expansion were not found in either eastern or western genetic groups, indicating that this species did not undergo a recent rapid range expansion. This result was supported by our ENM analysis, which suggested an overall retention of the species' potential distribution range, with the most suitable habitat remaining relatively stable since the LGM (Fig. 5).
Although neither the neutrality test nor mismatch distribution analysis based on cpDNA indicated that the populations in the western, eastern or combined regions underwent sudden demographic expansions, EBSP analysis based on nDNA suggested historical expansion episodes for the eastern group and all populations combined. According to these analyses, the effective population size of the eastern genetic group and all populations combined have been increasing since around 0.4–0.5 Mya (Fig. 3a and c), which is highly consistent with the period during when the Tengger Desert began to contract dramatically (0.47–0.12 Mya) after a large-scale expansion (Wang et al., 2015). This desert contraction likely provided an opportunity for Arnebia szechenyi to expand northward from south of the Tengger Desert and along the Helan Mountains. Previous studies have also shown that the Helan Mountains acted as a migration corridor for recolonization of desert plants in this area (Meng et al., 2015; Zhang et al., 2017; Zhao et al., 2019).
4.4. Asymmetric gene flowAdditionally, we detected noteworthy asymmetry in historical migration from the east to the west (32.90 > > 0, 64). Historical gene flow mainly occurred among the six northern populations from the western genetic group (populations YBL and NRG) and eastern genetic group (populations DT020, W18, S1 and Q18). This pattern was also supported by PCoA on pairwise Fst, which grouped the six populations into another group (Fig. 2b). This secondary gene exchange between two genetic groups was reflected by the nuclear DNA, implying that pollen-mediated gene flow was limited but not blocked entirely in spite of the presence of the desert. Located at the margin of the East Asian monsoon region, the Tengger Desert area is influenced dramatically by strong anomalous northwesterly and westerly winds. It is likely that the northwestward movement of the East Asian monsoon intensified the spread from the eastern to western genetic groups. The important role of East Asian monsoon system in shaping population genetic structure has also been demonstrated in other plants in this region (Yin et al., 2015, 2020; Zhang et al., 2017b).
5. ConclusionsOur analyses revealed that Arnebia szechenyi have high levels of genetic diversity distributed among its populations. Genetic variation of the species is not geographically structured across its whole distribution, and two nuclear genetic groups of populations were revealed, corresponding to the two geographic regions that are separated by the Tengger Desert and the Helan Mountain. However, the results from cpDNA do not support the boundary of these two geographic groups. We suggest that this chloroplast-nuclear discordance may have resulted from long periods of geographic isolation followed by lineage fusion processes that occurred in the geologic history of the Tengger Desert area. A significant asymmetry in historical migration between the two genetic groups was found, with a considerably higher rate of unidirectional gene flow from eastern to western genetic groups than from the opposite. Additionally, western genetic group showed signs of long-term local persistence and stability, while eastern genetic group showed signs of an expansion process. Our study highlights that cpDNA and nDNA may follow different mechanisms of evolution, and that their respective evolutionary history could reflect complementary aspects of past demographic and biogeographic events.
Author contributionsB.T. designated the study and managed the project. D.R.J. and B.T. collected samplings and completed the species identification. B.T. prepared DNA samples and performed sequencing. M.J.F., H.Y.W. performed the molecular phylogenetic, population structure, demographic history and niche modeling analyses. M.J.F. wrote the manuscript. D.R.J. and B.T. modification and improvement the manuscript. All authors read and approved the final manuscript.
Declaration of competing interestWe believe that the results presented here are of general interests to the readers of Plant Diversity, and we promise that our manuscript has not been published elsewhere nor is under consideration by another journal. All people contributed to this work have been acknowledged or co-authored according to his/her contributions. The authors have no conflict of interest to declare. All the authors agree to the submission of this manuscript.
AcknowledgmentsWe are grateful to Dr. Jun-Wei Ye for assistant on data analysis. We thank Dr. Yuan-Wen Duan for help during the field survey. This study was supported by the National Natural Science Foundation of China (41861008), Science Foundation of Yunnan Education Department (2018JS347), and the Ten-thousand Talents Program of Yunnan Province (YNWR-QNBJ-2020).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.04.002.
Avise, J.C., 2004. Molecular Markers, Natural History, and Evolution. Sunderland, MA: Sinauer Associates.
|
Avise, J.C., Arnold, J., Ball, R.M., et al, 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Systemat., 18: 489-522. DOI:10.1146/annurev.es.18.110187.002421 |
Barrett, R.D.H., Schluter, D., 2008. Adaptation from standing genetic variation. Trends Ecol. Evol., 23: 38-44. DOI:10.1016/j.tree.2007.09.008 |
Barrett, S.C.H., Jesson, L.K., Baker, A.M., 2000. The evolution and function of stylar polymorphisms in flowering plants. Ann. Bot., 85: 253-265. DOI:10.1006/anbo.1999.1067 |
Beerli, P., 2006. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics, 22: 341-345. DOI:10.1093/bioinformatics/bti803 |
Beheregaray, L.B., 2008. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol. Ecol., 17: 3754-3774. |
Bouckaert, R., Heled, J., Kühnert, D., et al, 2014. Beast 2: a software Platform for bayesian evolutionary analysis. PLoS Comput. Biol., 10: e1003537. DOI:10.1371/journal.pcbi.1003537 |
Comes, H.P., Kadereit, J.W., 1998. The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci., 3: 432-438. DOI:10.1016/S1360-1385(98)01327-2 |
Comes, H.P., Kadereit, J.W., 2003. Spatial and temporal patterns in the evolution of the flora of the European Alpine System. Taxon, 52: 451-462. DOI:10.2307/3647382 |
Darriba, D., Taboada, G.L., Doallo, R., et al, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods, 9: 772-772. |
Dellicour, S., Mardulyn, P., 2014. Spads 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Mol. Ecol. Resour., 14: 647-651. DOI:10.1111/1755-0998.12200 |
Després, L., 2019. One, two or more species? Mitonuclear discordance and species delimitation. Mol. Ecol., 28: 3845-3847. DOI:10.1111/mec.15211 |
Drouin, G., Daoud, H., Xia, J.N., 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol., 49: 827-831. DOI:10.1016/j.ympev.2008.09.009 |
Du, F.K., Petit, R.J., Liu, J.Q., 2009. More introgression with less gene flow: chloroplast vs. mitochondrial DNA in the Picea asperata complex in China, and comparison with other Conifers. Mol. Ecol., 18: 1396-1407. DOI:10.1111/j.1365-294X.2009.04107.x |
Excoffier, L., Lischer, H.E.L., 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour., 10: 564-567. DOI:10.1111/j.1755-0998.2010.02847.x |
Excoffier, L., Smouse, P.E., Quattro, J.M., 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479-491. DOI:10.1093/genetics/131.2.479 |
Falush, D., Stephens, M., Pritchard, J.K., 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes, 7: 574-578. DOI:10.1111/j.1471-8286.2007.01758.x |
Fawcett, T., 2006. An introduction to ROC analysis. Pattern Recogn. Lett., 27: 861-874. DOI:10.1016/j.patrec.2005.10.010 |
Field, C.B., Behrenfeld, M.J., Randerson, J.T., et al, 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science, 281: 237-240. DOI:10.1126/science.281.5374.237 |
Fu, Y., He, C.Z., Tian, B., 2016. Development of low copy nuclear gene primers for Arnebia szechenyi based on transcriptome. J. S. W. Forest. Univ., 36: 86-90. |
Fu, Y.X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147: 915-925. DOI:10.1093/genetics/147.2.915 |
Fu, Y.X., Li, W.H., 1993. Statistical tests of neutrality of mutations. Genetics, 133: 693-709. DOI:10.1093/genetics/133.3.693 |
Funk, D.J., Omland, K.E., 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst., 34: 397-423. DOI:10.1146/annurev.ecolsys.34.011802.132421 |
Hewitt, G., 2000. The genetic legacy of the Quaternary ice ages. Nature, 405: 907-913. DOI:10.1038/35016000 |
Hewitt, G.M., 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. Lond, 58: 247-276. DOI:10.1006/bijl.1996.0035 |
Hewitt, G.M., 1999. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. Lond., 68: 87-112. DOI:10.1111/j.1095-8312.1999.tb01160.x |
Hickerson, M.J., Carstens, B.C., Cavender-Bares, J., et al, 2010. Phylogeography's past, present, and future: 10 years after Avise, 2000. Mol. Phylogenet. Evol., 54: 291-301. DOI:10.1016/j.ympev.2009.09.016 |
Hinojosa, J.C., Koubínová, D., Szenteczki, M.A., et al, 2019. A mirage of cryptic species: genomics uncover striking mitonuclear discordance in the butterfly Thymelicus sylvestris. Mol. Ecol., 28: 3857-3868. DOI:10.1111/mec.15153 |
Jia, D., Abbott, R., Liu, T., et al, 2012. Out of the Qinghai-Tibet plateau: evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophae rhamnoides (Elaeagnaceae). New Phytol., 194: 1123-1133. DOI:10.1111/j.1469-8137.2012.04115.x |
Kay, K.M., Whittall, J.B., Hodges, S.A., 2006. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol. Biol., 6: 1-9. DOI:10.1186/1471-2148-6-1 |
Kier, G., Mutke, J., Dinerstein, E., et al, 2005. Global patterns of plant diversity and floristic knowledge. J. Biogeogr., 32: 1107-1116. DOI:10.1111/j.1365-2699.2005.01272.x |
Körner, C., 2000. Biosphere responses to CO2 enrichment. Ecol. Appl., 10: 1590-1619. |
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Le Houérou, H.N., 1996. Climate change, drought and desertification. J. Arid Environ., 34: 133-185. DOI:10.1006/jare.1996.0099 |
Li, Y., Zhang, X.N., Lv, G.H., 2019. Phylogeography of Ixiolirion songaricum, a spring ephemeral species endemic to Northwest China. Plant Systemat. Evol., 305: 205-221. DOI:10.1007/s00606-018-1563-7 |
Li, Z., Sun, D., Chen, F., et al, 2014. Chronology and paleoenvironmental records of a drill core in the central Tengger Desert of China. Quat. Sci. Rev., 85: 85-98. DOI:10.1016/j.quascirev.2013.12.003 |
Li, Z.J., 2013. Formation and Paleo-Environmental Evolution of the Tengger Desert Revealed by Drilling Holes in the Hinterland of the Tengger Desert (Doctoral Dissertation. Lanzhou University.
|
Librado, P., Rozas, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Liu, J.Q., Sun, Y.S., Ge, X.J., et al, 2012. Phylogeographic studies of plants in China: advances in the past and directions in the future. J. Systemat. Evol., 50: 267-275. DOI:10.1111/j.1759-6831.2012.00214.x |
Lloyd, D.G., Webb, C.J., 1992. The Evolution of Heterostyly. Evolution and Function of Heterostyly. Springer, Berlin, Heidelberg, pp. 151-178.
|
Maestre, F.T., Salguero-Gomez, R., Quero, J.L., 2012. It is getting hotter in here: determining and projecting the impacts of global environmental change on drylands. Philos. Trans. R. Soc. Lond. B Biol. Sci., 367: 3062-3075. DOI:10.1098/rstb.2011.0323 |
Meng, H.H., Gao, X.Y., Huang, J.F., et al, 2015. Plant phylogeography in arid Northwest China: retrospectives and perspectives. J. Systemat. Evol., 53: 33-46. DOI:10.1111/jse.12088 |
Meng, H.H., Zhang, M.L., 2011. Phylogeography of Lagochilus ilicifolius (Lamiaceae) in relation to Quaternary climatic oscillation and aridification in northern China. Biochem. Systemat. Ecol., 39: 787-796. DOI:10.1016/j.bse.2011.07.015 |
Morris, A.B., Shaw, J., 2018. Markers in time and space: a review of the last decade of plant phylogeographic approaches. Mol. Ecol., 27: 2317-2333. DOI:10.1111/mec.14695 |
Muscarella, R., Galante, P.J., Soley-Guardia, M., et al, 2014. ENM eval: an R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol., 5: 1198-1205. DOI:10.1111/2041-210X.12261 |
Phillips, S.J., Anderson, R.P., Schapire, R.E., 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model., 190: 231-259. DOI:10.1504/IJGENVI.2006.010156 |
Pons, O., Petit, R.J., 1996. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics, 144: 1237-1245. DOI:10.1093/genetics/144.3.1237 |
Qiu, Y.X., Fu, C.X., Comes, H.P., 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Mol. Phylogenet. Evol., 59: 225-244. DOI:10.1016/j.ympev.2011.01.012 |
R Core Team, 2013. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
|
Rambaut, A., Drummond, A.J., Xie, D., et al, 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol., 67: 901. DOI:10.1093/sysbio/syy032 |
Reynolds, J.F., Maestre, F.T., Kemp, P.R., et al., 2007. Natural and human dimensions of land degradation in drylands: causes and consequences. In: Terrestrial Ecosystems in a Changing World. Springer, Berlin, Heidelberg, pp. 247-257.
|
Rock, C.D., Barkan, A., Taylor, W.C., 1987. The maize plastid psbB-psbF-petB-petD gene cluster: spliced and unspliced petB and petD RNAs encode alternative products. Curr. Genet., 12: 69-77. DOI:10.1007/BF00420729 |
Rogers, A.R., Harpending, H., 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol., 9: 552-569. |
Sala, O.E., Chapin, F.S., Armesto, J.J., et al, 2000. Global biodiversity scenarios for the year 2100. Science, 287: 1770-1774. DOI:10.1126/science.287.5459.1770 |
Sang, T., Crawford, D.J., Stuessy, T.F., 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot., 84: 1120-1136. DOI:10.2307/2446155 |
Sloan, D.B., Havird, J.C., Sharbrough, J., 2017. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol., 26: 2212-2236. DOI:10.1111/mec.13959 |
Taberlet, P., Gielly, L., Pautou, G., et al, 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol., 17: 1105-1109. DOI:10.1007/BF00037152 |
Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123: 585-595. DOI:10.1093/genetics/123.3.585 |
Tian, B., Fu, Y., Milne, R.I., et al, 2020. A complex pattern of post-divergence expansion, contraction, introgression, and asynchronous responses to Pleistocene climate changes in two Dipelta sister species from western China. J. Systemat. Evol., 58: 247-262. DOI:10.1111/jse.12524 |
Wang, F., Sun, D., Chen, F., et al, 2015. Formation and evolution of the Badain Jaran Desert, North China, as revealed by a drill core from the desert centre and by geological survey. Palaeogeogr. Palaeoclimatol. Palaeoecol., 426: 139-158. DOI:10.1016/j.palaeo.2015.03.011 |
Wang, L.L., Zhang, C., Tian, B., et al, 2015. Reproductive isolation is mediated by pollen incompatibility in sympatric populations of two Arnebia species. Ecol. Evol., 5: 5838-5846. DOI:10.1002/ece3.1849 |
Wang, P., Zhang, X., Tang, N., et al, 2016. Phylogeography of Libanotis buchtormensis (Umbelliferae) in disjunct populations along the deserts in Northwest China. PLoS One, 11: e0159790. DOI:10.1371/journal.pone.0159790 |
Ward, D., 2016. The Biology of Deserts. Oxford University Press.
|
White, T.J., Bruns, T., Lee, S.J. W.T., et al., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR Protocols: A Guide to Methods and Applications. Academic Press, Inc., New York, pp. 315-322.
|
Wolfe, K.H., Li, W.H., Sharp, P.M., 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A, 84: 9054-9058. DOI:10.1073/pnas.84.24.9054 |
Xie, K.Q., Zhang, M.L., 2013. The effect of Quaternary climatic oscillations on Ribes meyeri (Saxifragaceae) in northwestern China. Biochem. Systemat. Ecol., 50: 39-47. DOI:10.1016/j.bse.2013.03.031 |
Yang, X., Scuderi, L.A., 2010. Hydrological and climatic changes in deserts of China since the late Pleistocene. Quat. Res., 73: 1-9. DOI:10.1016/j.yqres.2009.10.011 |
Yin, H., Wang, L., Shi, Y., et al, 2020. The East Asian Winter Monsoon acts as a major selective factor in the intraspecific differentiation of drought-tolerant Nitraria tangutorum in Northwest China. Plants, 9: 1100. DOI:10.3390/plants9091100 |
Yin, H.X., Yan, X., Shi, Y., et al, 2015. The role of East Asian monsoon system in shaping population divergence and dynamics of a constructive desert shrub Reaumuria soongarica. Sci. Rep., 5: 1-14. |
Zamudio, K.R., Bell, R.C., Mason, N.A., 2016. Phenotypes in phylogeography: species' traits, environmental variation, and vertebrate diversification. Proc. Natl. Acad. Sci. U.S.A., 113: 8041. DOI:10.1073/pnas.1602237113 |
Zeng, Y.F., Zhang, J.G., Abuduhamiti, B., et al, 2018. Phylogeographic patterns of the desert poplar in Northwest China shaped by both geology and climatic oscillations. BMC Evol. Biol., 18: 1-14. DOI:10.1186/s12862-017-1072-2 |
Zhang, C., Vereecken, N.J., Wang, L., et al, 2017. Are nectar guide colour changes a reliable signal to pollinators that enhances reproductive success?. Plant Ecol. Divers., 10: 89-96. DOI:10.1080/17550874.2017.1350763 |
Zhang, C., Wang, L.L., Lan, D., et al, 2014. Pollination ecology of Arnebia szechenyi (Boraginaceae), a Chinese endemic perennial characterized by distyly and heteromorphic self-incompatibility. Ann. Zool. Fenn., 51: 297-304. DOI:10.5735/085.051.0505 |
Zhang, D.F., L, F.Q., B, J.M., 2000. Eco-environmental effects of the Qinghai-Tibet Plateau uplift during the quaternary in China. Environ. Geol., 39: 1352-1358. DOI:10.1007/s002540000174 |
Zhang, H.X., Zhang, M.L., Sanderson, S.C., 2017. Spatial genetic structure of forest and xerophytic plant species in arid Eastern Central Asia: insights from comparative phylogeography and ecological niche modelling. Biol. J. Linn. Soc. Lond., 120: 612-625. |
Zhang, Y., Yu, Q., Zhang, Q., et al, 2017. Regional-scale differentiation and phylogeography of a desert plant Allium mongolicum (Liliaceae) inferred from chloroplast DNA sequence variation. Plant Systemat. Evol., 303: 451-466. DOI:10.1007/s00606-016-1383-6 |
Zhao, Y.F., Zhang, H.X., Pan, B.R., et al, 2019. Intraspecific divergences and phylogeography of Panzerina lanata (Lamiaceae) in northwest China. PeerJ, 7: e6264. DOI:10.7717/peerj.6264 |
Zhou, W., Wang, H., 2009. Heterostyly in angiosperms and its evolutionary significance. Ann. Bot., 44: 742. |
Zhu, G.L., Riedl, H., Kamelin, R., 2005. Boraginaceae. In: Wu, Z.-Y., Raven, P.H. (Eds.), Flora of China. Science Press and Missouri Botanical Garden, Beijing and St Louis.
|



