b. Sino-Africa Joint Research Center, Chinese Academy of Sciences, Wuhan, 430074, China;
c. University of Chinese Academy of Sciences, Beijing, 100049, China;
d. Jiangxi Provincial Key Laboratory for Bamboo Germplasm Resources and Utilization, Forestry College, Jiangxi Agricultural University, Nanchang, 330045, China;
e. Administration of Siguniang Mountains National Nature Reserve, Xiaojin, 624206, China
Orchidaceae is one of the largest families of flowering plants, comprising about 736 known genera and 28, 000 species worldwide (Chase et al., 2015; Christenhusz and Byng, 2016). During an investigation on wild orchid resources in western Sichuan, China in 2020, a distinctive orchid species was discovered in the Siguniang Mountains National Nature Reserve under shady Populus forests. Some morphological characters such as lack of leaf and coralloid rhizome immediately caught our attention, and it is morphologically similar to the species in the genus Corallorhiza. Corallorhiza Gagnebin is the largest temperate genus of leafless putative heterotrophs in Orchidaceae, comprising 11 species that are restricted to the northern hemisphere of the New World, except for a single circumboreal species, C. trifida Chatelain (Chase et al., 2015; Freudenstein, 1997, 1999; Freudenstein and Senyo, 2008). According to the Flora of China, only C. trifida has been recorded in China, distributed in Gansu, Guizhou, Hebei, Jilin, Nei Mongol, Qinghai, Sichuan, Xinjiang (Chen and Stephan, 2009). We compared the new collection with C. trifida, and discovered that it can be distinguished from the latter by bigger flowers, both sepals and petals with 3 veins, and longer lip lateral lobes. We further compared it with other congeneric species. It is also similar to C. striata Lindley and C. bentleyi Freudenstein in sharing a distinctive morphological character: lack a mentum (i.e. a vestigial nectar spur at the base of the perianth), but can be easily distinguished from these two species by its lip broad oblong and obviously 3-lobed the toward base. After carefully comparing the plant with all previously known congeneric taxa, it could not be matched with any known species. Hence, our collection might be a new species of Corallorhiza.
Chase et al. (2015) presented a revised classification of Orchidaceae and divided the family into five subfamilies, namely Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae. The genus Corallorhiza was placed in the subtribe Calypsoinae (Epidendreae, Epidendroideae) which contains 13 genera, namely Corallorhiza, Aplectrum Nutt., Calypso Salisb., Changnienia S.S. Chien, Coelia Lindl., Cremastra Lindl., Dactylostalix Rchb. f., Danxiaorchis J.W. Zhai, F.W. Xing & Z.J. Liu, Ephippianthus Rchb. f., Govenia Lindl., Oreorchis Lindl., Tipularia Nutt., and Yoania Maxim. Freudenstein et al. (2017) reconstructed phylogenetic relationships among subtribe Calypsoinae. Based on matK marker, results showed Corallorhiza was paraphyletic, but the support for the node was relatively weak. In the results of ITS sequence and combined analyses (matK + ITS sequences), Corallorhiza was shown to be monophyletic with strong support. An earlier study based upon combined ITS, plastid datasets, and morphological character matrix strongly support Oreorchis and Corallorhiza are both monophyletic groups, but the Corallorhiza is far from Oreorchis (Zhai et al., 2013). In recent molecular phylogenetic studies based on the plastomes, however, Corallorhiza is still non-monophyletic, and the Oreorchis and Corallorhiza were nested together and formed a single clade (Kim et al., 2020; Li et al., 2020). In the taxonomic revision of Oreorchis (Pearce and Cribb, 1997), except for Kitigorchis itoana, the remaining species of Kitigorchis were transferred into Oreorchis. Moreover, Pearce and Cribb (1997) remarked that Kitigorchis be a monotypic genus intermediate between the Oreorchis and Corallorhiza, and suggested that K. itoana has coral-like rhizomes quite different from the subterranean organs of Oreorchis. Interestingly, the final study revealed that K. itoana and O. indica were conspecific (Yukawa et al., 2003). In fact, O. indica often possesses coralloid rhizomes (Suetsugu et al., 2020). An earlier study has shown that coralloid rhizomes occur in the Aplectrum hyemale (Muhl. ex Willd.) Nutt. (Macdougal and Dufrenoy, 1944). Thus the habit has happened many times in Calypsoinae. Similarly, in the Calypsoinae, a previous study also demonstrated that leafless habit has happened many times (Freudenstein et al., 2017). Suetsugu et al. (2020) suggested that O. indica is a partially mycoheterotrophic orchid, and the full mycoheterotrophy evolved after the establishment of partial mycoheterotrophy, rather than through direct shifts from autotrophy in the Oreorchis-Corallorhiza clade. In the genus Corallorhiza, it is well known that C. trifida is partial mycoheterotrophy (Zimmer et al., 2008). Due to the phylogenetic relationship of Oreorchis and Corallorhiza remaining uncertain, we hypothesize that our collection might be a new species of Corallorhiza.
Here we describe and illustrate the new species Corallorhiza sinensis. To distinguish the new Corallorhiza species, we sequenced its complete plastome. Its plastome was analyzed in comparison with other members of subtribe Calypsoinae. Finally, we reconstructed its phylogenetic relationship within subtribe Calypsoinae based upon the nrDNA sequence and 79 coding sequences (CDSs).
2. Materials and methods 2.1. Morphological analysesMorphological characters of the new species were based on the study of living plants and herbarium specimens. Measurements of flowers and rhizomes were taken from living plants. Photographs of Corallorhiza sinensis and C. trifida were taken in the field and are included in Fig. 1, Fig. 2, respectively. A total of 11 diagnostic characteristics of these two species are compared in Table 1. Measurements of C. trifida were based on the combination of Flora of China (Chen and Stephan, 2009) and observation on living plants. Given that the phylogenetic relationship of Oreorchis and Corallorhiza remains uncertain, we compared the new collection with the members of Oreorchis as well and found the new species resembles O. patens and O. foliosa var. indica. To better distinguish the new species, we also presented its supplementary photograph (Fig. S1). Furthermore, the photographs of O. patens and O. foliosa var. indica were combined in the Fig. S2, which will provide important morphological evidence for this study. The voucher specimens of the new species were deposited in the Herbarium of Wuhan Botanical Garden, Chinese Academy of Sciences (HIB).
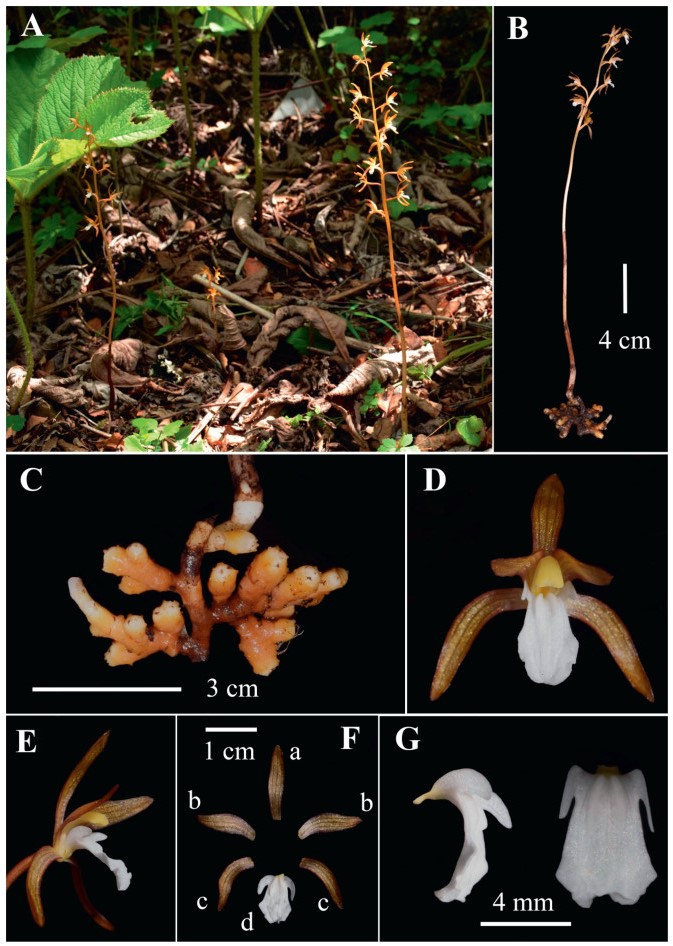
|
| Fig. 1 Photographs of Corallorhiza sinensis. (A) habit; (B) individual; (C) root; (D) front view of flowers; (E) lateral view of flowers; (F) anatomy of flowers (a = dorsal sepal, b = petal, c = lateral sepal, d = lip); (G) lateral and front view of lips. |

|
| Fig. 2 Photographs of Corallorhiza trifida. (A) individual; (B) habit; (C) root; (D) front view of flowers; (E) lateral view of flowers; (F) anatomy of flowers (a = dorsal sepal, b = petal, c = lateral sepal, d = lip); (G) lateral and front view of lips. |
| Characters | Corallorhiza sinensis | C.trifida |
| Plant height | 10-46cm | 10-28cm |
| Inflorescence | 5-13-flowered | 3-7-flowered |
| Flower colour | light reddish brown | pale yellowish green to white |
| Pedicel and ovary | 9-12mm | 3.5-5mm |
| Lateral sepals | ca. 10×2mm, 3-veined | 4-6×1-1.5mm, 1-veined |
| Dorsal sepal | ca. 12×2mm, 3-veined | 4-6×1-1.5mm, 1-veined |
| Petal | long ovate-lanceolate, 11×4mm, 3-veined | lanceolate to oblanceolate, 3-5×ca. 1.5-2mm, 1-veined |
| Lip | ca. 8×6mm, obviously 3-lobed toward base | 2.5-4×ca. 1.5-3mm, obscurely 3-lobed toward base |
| Midlobe of lip | oblong to broadly dilated toward apex, ca. 7×5mm | elliptic to oblong, 1-1.5×ca. 0.8mm |
| Lateral lobe of lip | ca. 3mm long | less than 1mm long |
| Column | yellowish, ca. 8mm long, arcuate | greenish, 2.5-3mm long, erect |
Oreorchis patens Lindl., China: Hunan, Sangzhi county, July-4-1998, Beijingdui, 2362 (PE, 01340734).
Oreorchis foliosa var. indica (Lindl.) N. Pearce & P.J. Cribb, China: Xizang, June-2-1972, Xizang Herbal Medicine Investigation Team 89 (PE, 00850746).
2.3. DNA extraction and sequencingPurified total DNA of Corallorhiza sinensis was fragmented to construct short-insert libraries for sequencing (Illumina HiSeq, 2000 platform from the Biomarker Technologies, Inc.), and 6 GB of reads was obtained.
2.4. Plastome and nrDNA assembly, annotationThe complete plastome of Corallorhiza sinensis was assembled using GetOrganelle v.1.7.1 with appropriate parameters: K-mer is "21, 45, 65, 85, 105", the Word size is 0.6 (Jin et al., 2020). Five key steps were included in the workflow: 1. Mapping reads to seed and assembling seed-mapped reads for parameter estimation; 2. Recruiting more target-associated reads through extending iterations; 3. Conducting de novo assembly; 4. Roughly filtering for target-like contigs; 5. Identifying target contigs and exporting all configurations (Bankevich et al., 2012; Camacho et al., 2009; Jin et al., 2020; Langmead and Salzberg, 2012). Final assembly graphs were visualized by Bandage to assess the completeness of the final graph (Wick et al., 2015). The nrDNA sequence (18S-ITS1-5.8S-ITS2-26S) of C. sinensis was assembled using GetOrganelle v.1.7.1 with default parameters.
Initially, PGA (Qu et al., 2019) was utilized to annotate the complete plastome of Corallorhiza sinensis, with the C. trifida (GenBank: MN990435) selected as the reference, and then manually adjusted protein-coding genes and exons boundaries using software Geneious 11.0.4 (Kearse et al., 2012). The final plastome and nrDNA sequence of C. sinensis have been submitted to GenBank under accession numbers MW191791 and MW413395, respectively. Gene map of the plastome of C. sinensis was drawn online using OGDRAW (Lohse et al., 2007).
2.5. Genome features and comparative plastid genomic analysisLength of the whole plastome of Corallorhiza sinensis, numbers of genes, and categories of Genes were analyzed in Geneious 11.0.4 (Kearse et al., 2012). The software MEGA (Kumar et al., 2016) was used for calculating the guanine-cytosine (GC) content. In order to further identify potential variations between C. sinensis and other Corallorhiza species, the genome rearrangement analyses of ten Corallorhiza sequences were performed using Mauve (Darling et al., 2004) plugin in Geneious 11.0.4 (Kearse et al., 2012). Geneious 11.0.4 (Kearse et al., 2012) was used to detect the contraction/expansion of the inverted repeat regions (IRs), and the graph of expansions/contractions was visualized using Adobe Illustrator.
2.6. Phylogenetic analysisTo estimate the evolutionary history of Corallorhiza sinensis within Calypsoinae, phylogenies were reconstructed by Maximum likelihood (ML) and Bayesian Inference (BI) analyses using the nrDNA sequence and coding sequences (CDSs), the total of 79 CDSs from 27 plastomes were used for constructing the phylogenetic relationship of the members within Calypsoinae (Table S1). Lost genes and pseudogenes were treated as missing data. All nrDNA sequences and plastomes were downloaded from the NCBI database except C. sinensis. In the plastid tree and nrDNA tree, Cattleya crispata and Chysis bractescens are selected as outgroup, respectively. We extracted 79 CDSs using Geneious 11.0.4 (Kearse et al., 2012). Under the codon model, each single CDS matrix was aligned and edited by MAFFT (Katoh and Standley, 2013) and Gblocks (Talavera and Castresana, 2007) plugin in PhyloSuite (Zhang et al., 2020), respectively, and each matrix is eventually concatenated into one supermatrix by PhyloSuite (Zhang et al., 2020). ModelFinder was utilized to select the best-fit model for accurate phylogenetic estimates (Kalyaanamoorthy et al., 2017). ML analysis was performed using IQ-TREE with 1000 bootstrap replicates (Nguyen et al., 2015), BI analysis was performed in MrBayes 3.2.2 (Ronquist et al., 2012) using the Markov chain Monte Carlo (MCMC) method, with two independent runs for 10 million generations (Number of Chains is four). Phylogenetic trees were sampled every one thousand generations, the first 25% of trees generated were discarded as burn-in and the remaining trees were used to construct majority-rule consensus tree (Ronquist et al., 2012). Finally, the phylogenetic trees were edited using FigTree 1.4 (https://github.com/rambaut/figtree).
3. Results 3.1. Taxonomic treatment 3.1.1. Corallorhiza sinensis G.W. Hu & Q.F. Wang (Figs. 1 and 3)
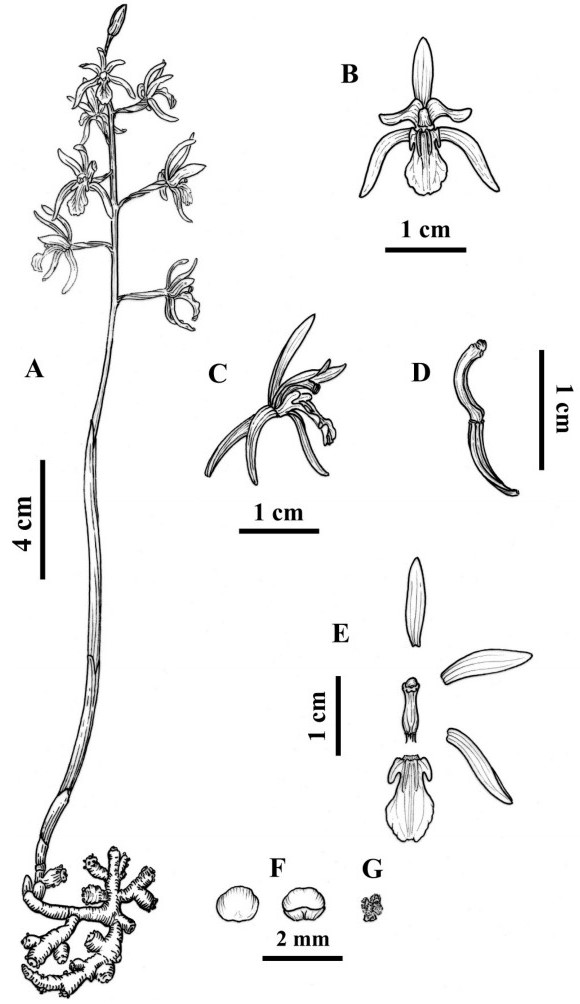
|
| Fig. 3 Illustrations of Corallorhiza sinensis. (A) individual; (B) front view of flowers; (C) lateral view of flowers; (D) column and ovary; (E) anatomy of flowers; (F) anther cap; (G) pollinia. Drawn by Jing Tian. |
Type: China. Sichuan: Xiaojin County, Siguniang Mountains National Nature Reserve, 102°44'31.5"E, 30°58'26.5"N, under Populus forests ca. 2984 m in elevation, 9 Jun 2020, S. Peng, J. X. Yang et J. J. Wang YJX-0060 (holotype: HIB, isotype: HIB).
Diagnosis: Corallorhiza sinensis resembles C. trifida, but prominently differs from the latter in having bigger flowers, both petals and sepals with 3 veins, and longer lateral lobes of lip. It is also similar to C. striata and C. bentleyi by sharing a distinctive morphological character: mentum absent, but can be easily distinguished by its lip broadly oblong and obviously 3-lobed toward base. Comparative analysis of morphology with the Oreorchis species showed that C. sinensis resembles O. patens and O. foliosa var. indica. The labellum of C. sinensis with two lamellae looks almost exactly like that of O. patens (Fig. S2, D-F), but O. patens has smaller flowers than C. sinensis and the subterranean system of O. patens indicated it is similar to normal (Fig. S2, D-F). The O. foliosa var. indica possess coralloid rhizomes and normal roots, which present obvious dual subterranean system, while the pseudobulb is still an important morphological character. Moreover, O. foliosa var. indica has a broadly 3-lobed to unlobed lip (Fig. S2, A-C). By observation of different populations, the C. sinensis has no leaves, not even visible green tissue in all individuals. Hence, our collection most closely resembles the Corallorhiza.
3.1.2. DescriptionPerennial mycoheterotrophic herbs, 10-46 cm tall. Rhizome fleshy, extensively branching, coralloid, nodiferous, internodes 2-4 mm long, often brown at the base of branch and beige at the apex of branch, diameter ca. 5 mm. Stems erect, cylindrical. Inflorescence light reddish brown, glabrous, leafless, covered with 3 or 4 membranous tubular sheaths from base to middle; sheaths amplexicaul, reddish brown, 2-12 cm long; rachis 5-18 cm, laxly 5-13-flowered; floral bracts small, lanceolate, apex acuminate, 1-3.5 mm long. Flowers light reddish brown, pedicel and ovary 9-12 mm long, torsion at base. Dorsal sepal arching forward, narrowly oblong-lanceolate, ca. 12 × 2 mm, 3-veined, apex acuminate; lateral sepals similar to dorsal sepal, slightly oblique, 3-veined, ca. 10 × 2 mm, connate at base, apex obtuse or acute. Petals long ovate-lanceolate, ca. 11 × 4 mm, 3-veined, apex obtuse or acute; lip broadly oblong, pure white with a yellowish claw, ca. 8 × 6 mm, obviously 3-lobed toward base; lateral lobes 2, erect, lanceolate but slightly curved, ca. 3 × 1 mm; middle lobe oblong to broadly dilated toward apex, ca. 7 × 5 mm, margin entire and slightly wavy at apex, with 2 distinct basal lamellae. Column yellowish, ca. 8 mm long, arcuate, basally with ridges or poorly developed auricles; ovary ca. 8.5 mm; mentum absent. Fruit not seen.
3.1.3. EtymologyThe specific epithet "sinensis" refers to China, because the new species is endemic to China.
3.1.4. Distribution, habitat and conservation statusCurrently, Corallorhiza sinensis is known only from Siguniang Mountains National Nature Reserve in Xiaojin County, Sichuan, China. The plants grow in shady Populus forests, flowering from May to June. Only two small populations of the new species were discovered, each population comprises approximately 12 mature individuals. Based on a previous study, Corallorhiza cannot be cultivated reliably because of the require of fungal associations for growth (Freudenstein, 1997). A preliminary assessment of the risk of extinction was conducted according to the International Union for Conservation of Nature (IUCN) Red List Categories and Criteria (https://www.iucn.org/), we thereby consider that C. sinensis should be classified as critically endangered (CR).
3.1.5. Additional specimens examined (paratypes)China. Sichuan: Xiaojin County, Siguniang Mountains National Nature Reserve, grows Populus forests, elev. 2984 m, 102°48'21"E, 30°59'15.6"N, 8 Jun 2020, S. Peng, J. X. Yang et J. J. Wang PS-00166 (HIB).
3.2. Characteristics of the Corallorhiza sinensis plastomeThe complete plastome of Corallorhiza sinensis is 148, 124 bp in length, and exhibits a typical quadripartite structure, consisting of a large single copy (LSC) region of 82, 207 bp and a small single copy (SSC) region of 82, 207 bp, which were separated by a pair of 26, 165 bp inverted repeat regions (IRs). The gene map of C. sinensis plastome is presented in Fig. 4. The gene composition in plastome of C. sinensis could be divided into four categories: genes related to photosynthesis, genes related to self-replication, protein-coding genes with unknown functions, and other genes. A total of 109 unique genes were identified in the plastome, it contains 68 protein-coding genes, 30 tRNAs, and four rRNAs (Table 2). Among these genes, 15 genes are annotated as pseudogenes (ccsA, ndhA, ndhB, ndhC, ndhI, ndhJ, ndhK, petA, petB, psaB, psaC, rbcL, rpoB, ycf1, rpl22). A total of 21 genes were duplicated in the IR regions, including eight tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC), four rRNA genes (rrn4.5, rrn5, rrn16, rrn23), and nine PCGs (ndhB, ndhG, rpl2, rpl22, rpl23, rps7, rps19, rps12, ycf1). A total of seven protein-coding genes and six tRNA genes contain a single intron, only clpP and ycf3 contain two introns.
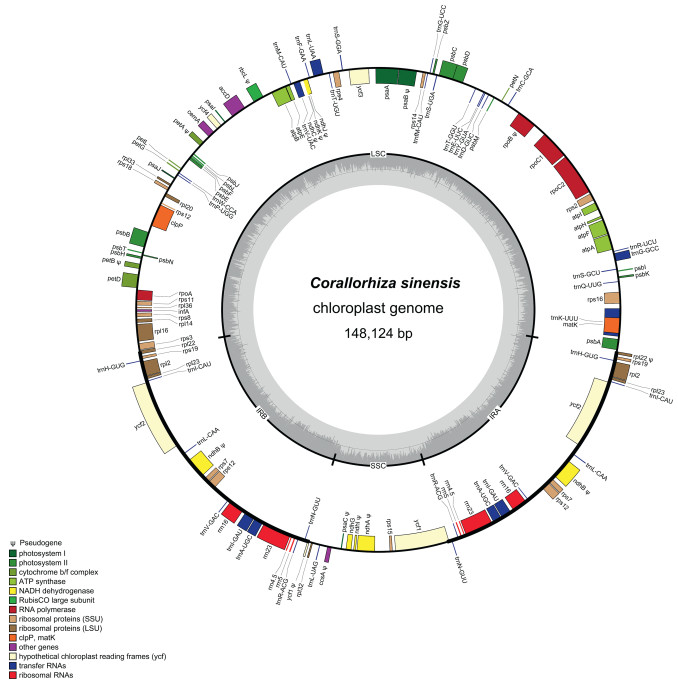
|
| Fig. 4 Circular gene map of the plastid genome of Corallorhiza sinensis. Genes drawn within the circle are transcribed clockwise, while those drawn outside are transcribed counterclockwise. Genes are color-coded according to their functional groups. |
| Category, Group of Genes | Gene Names |
| Photosynthesis: | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI |
| Subunits of NADH dehydrogenase | ndhAa, ndhBa (x2), ndhCa, ndhG(x2), ndhIa, ndhJa, ndhKa |
| Cytochrome b/f complex | petAa, petBa, petD*, petG, petL, petN |
| Subunits of photosystem Ⅰ | psaA, psaBa, psaCa, psaI, psaJ |
| Subunits of photosystem Ⅱ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Large subunit of rubisco | rbcLa |
| Other genes | |
| Subunit of Acetyl-CoA-carboxylase | accD |
| c-type cytochrome synthesis gene | ccsAa |
| Envelope membrane protein | cemA |
| Protease | clpP** |
| Translational initiation | infA |
| Maturase | matK |
| Self-replication | |
| Large subunit of ribosome | rpl14, rpl16*, rpl2*(x2), rpl20, rpl22b (x2), rpl23(x2), rpl32, rpl33, rpl36 |
| DNA dependent RNA polymerase | rpoA, rpoBb, rpoC1*, rpoC2 |
| Small subunit of ribosome | rps2, rps3, rps4, rps7(x2), rps8, rps11, rps12*c (x2), rps14, rps15, rps16*, rps18, rps19(x2) |
| rRNA Genes | rrn4.5(x2), rrn5(x2), rrn16(x2), rrn23(x2) |
| tRNA Genes | trnA-UGC*(x2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC*, trnG-UCC, trnH-GUG(x2), trnI-CAU(x2), trnI-GAU*(x2), trnK-UUU*, trnL-CAA(x2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(x2), trnP-UGG, trnQ-UUG, trnR-ACG(x2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(x2), trnV-UAC*, trnW-CCA, trnY-GUA |
| Unknown function | |
| Conserved open reading frames | ycf1b (x2), ycf2, ycf3**, ycf4 |
| Note: a indicates the pseudogene; c indicates trans-spliced gene; b indicates one of them is a pseudogene; * genes containing a single intron; ** genes containing two introns; (x2) genes present as two copies in the IR regions. | |
A comparison of the Corallorhiza and Oreorchis plastomes was conducted in the Table 3. The full-length plastome of the 10 Corallorhiza species ranged from 124, 482 bp (C. bentleyi) to 151, 031 bp (C. macrantha), and the C. bentleyi has the shortest plastome. The range of length variation among members of the Corallorhiza for the LSC and SSC region is obvious. Further, we also compared them with publicated Oreorchis plastomes (Table 3). The length of the plastome ranges from 158, 256 bp in O. patens to 158, 654 bp in O. angustata, obviously larger than Corallorhiza. In addition, the results indicated that Oreorchis plastome have longer LSC region than Corallorhiza. Comparative analysis of gene content showed that the number of PCGs in Oreorchis is very similar to most angiosperms, but it is very low in some Corallorhiza members. As our results displayed, GC content and the number of tRNA and rRNA genes are highly conserved in all these plastomes.
| Species | Accession no. | Length (bp) | LSC (bp) | SSC (bp) | IR (bp) | GC content (%) | No. of PCGs | No. of tRNA | No. of rRNA |
| Corallorhiza sinensis | MW191791 | 148, 124 | 82, 207 | 13, 587 | 26, 165 | 36.6 | 68 | 38 | 8 |
| C.bulbosa | KM390013 | 148, 643 | 82, 851 | 12, 368 | 27, 712 | 37.1 | 74 | 38 | 8 |
| C.mertensiana | KM390018 | 147, 941 | 81, 109 | 13, 774 | 26, 529 | 36.8 | 60 | 38 | 8 |
| C.wisteriana | KM390020 | 146, 437 | 82, 350 | 11, 743 | 26, 172 | 37.1 | 73 | 38 | 8 |
| C.odontorhiza | KM390021 | 147, 317 | 82, 259 | 13, 508 | 25, 775 | 37.0 | 73 | 38 | 8 |
| C.trifida | MN990435 | 149, 408 | 83, 171 | 14, 397 | 25, 920 | 37.2 | 74 | 38 | 8 |
| C.macrantha | NC_025660 | 151, 031 | 84, 263 | 12, 544 | 27, 112 | 37.2 | 72 | 38 | 8 |
| C.striata var. striata | NC_040978 | 141, 202 | 75, 881 | 12, 999 | 26, 161 | 36.3 | 50 | 37 | 8 |
| C.bentleyi | NC_040979 | 124, 482 | 63, 581 | 7, 781 | 26, 560 | 36.6 | 45 | 39 | 8 |
| C.maculata | NC_046814 | 146, 198 | 79, 612 | 13, 018 | 26, 784 | 36.8 | 55 | 38 | 8 |
| Oreorchis angustata | MN990443 | 158, 654 | 86, 294 | 18, 312 | 27, 024 | 37.0 | 86 | 38 | 8 |
| O.patens | MN990436 | 158, 256 | 86, 253 | 18, 341 | 26, 831 | 37.0 | 86 | 38 | 8 |
| O.foliosa var. indica | MN990440 | 158, 599 | 86, 109 | 17, 616 | 27, 437 | 36.9 | 82 | 38 | 8 |
| O.foliosa | MN990441 | 158, 496 | 86, 061 | 18, 361 | 27, 037 | 36.9 | 82 | 38 | 8 |
Expansion and contraction of the IRs were compared among Calypsoinae (Fig. S3). The LSC/IRb boundary information of Corallorhiza sinensis is similar to that of ten plastomes, including Changnienia amoena, Calypso bulbosa, Cremastra appendiculata, two Corallorhiza species (C. bentleyi and C. striata var. striata), three Oreorchis species (O. angustata, O. foliosa, and O. foliosa var. indica), and two Tipularia species (T. josephi and T. szechuanica). The gene rpl22 spanned the IRb/LSC region and the length of rpl22 ranged from 94 bp to 364 bp in LSC region. For the most species, the rpl22 locates in the LSC region near the LSC/IRb border, and it is separated from the LSC/IRb border by a spacer varying from 17 to 68 bp. Notabley, the LSC/IRb border of D. singchiana is rps3 gene. The SSC/IRa border is crossed by the gene ycf1 in all plastomes, and the fragment located in IRa region ranged from 61 to 1565 bp. Correspondingly, the gene ycf1 in the junction region between SSC and IRb is treated as a pseudogene because of the incomplete duplication of the normal copy. However, for SSC/IRb border, gene ndhF located in SSC has a 71 bp extension to the IRb of four plastomes (O. angustata, O. patens, Ch. amoena, Cr. appendiculata). Similarly, the rpl22 in the junction region between LSC and IRb is annotated as a pseudogene because of the incomplete duplication. The psbA gene locates in the LSC region, 70-147 bp from the LSC/IRa border. These results showed the obvious expansion of the IR region in C. sinensis plastome.
The result of the plastome structure comparison showed that there is one small inversion in Corallorhiza sinensis but none in other members of Corallorhiza and Oreorchis (Fig. S4). The inversion occurred in the LSC region between trnE-UUC and psbD genes, and was approximately 700 bp in length. Gene trnT-GGU was contained in the inversion region. We also detected an obvious inversion in plastome of C. maculata var. maculata and C. maculata. Furthermore, two inversions were detected in D. singchiana.
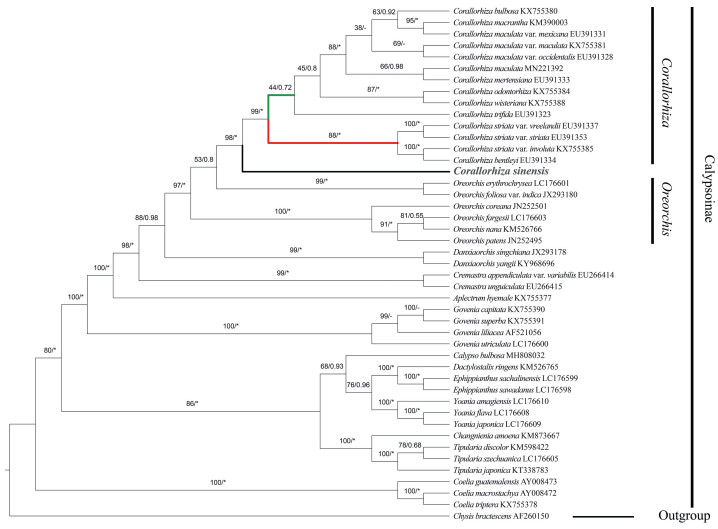
3.4. Phylogenetic analysisPhylogenetic result of CDSs dataset showed Corallorhiza sinensis is sister to the clade comprised of C. striata var. vreelandii + C. striata var. striata + C. cf. striata + C. bentleyi + C. striata var. involuta with strong support in both Maximum likelihood and Bayesian Inference (ML: BS = 100; BI: PP = 1.00) (Fig. 5). In the plastid tree, Corallorhiza did not form a monophyletic clade within Calypsoinae. Both ML and BI phylogenetic trees strongly indicated that the Oreorchis is nested within Corallorhiza (ML: BS = 100; BI: PP = 1.00). However, phylogenetic analysis based on nrDNA sequence suggest that Corallorhiza is a monophyletic group, and strongly support C. sinensis as sister to the rest spcies of Corallorhiza (ML: BS = 98; BI: PP = 1.00) (Fig. 6).
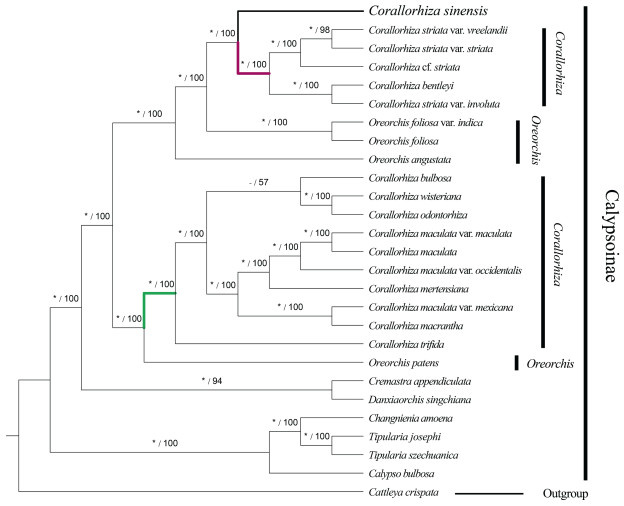
|
| Fig. 5 Phylogenetic tree reconstruction of Calypsoinae using the maximum likelihood (ML) method based on 79 coding sequences. Only the ML tree is shown, because its topology is nearly identical to that of the obtained BI tree. Clade support values in the tree are posterior probabilities/bootstrap supports in percent from Bayesian Inference/Maximum likelihood, "*" indicates that the node is 1.00 supported. "-" indicates that the node is incongruent between the topology of the ML tree and the Bayesian tree. Red and green clade indicates mentum absent and present, respectively. |
|
| Fig. 6 Phylogenetic tree reconstruction of Calypsoinae using the maximum likelihood (ML) method based on nrDNA sequences. Only the ML tree is shown, because its topology is nearly identical to that of the obtained BI tree. Numbers associated with the branches are ML bootstrap value (BS) and BI posterior probabilities (PP). "*" indicates that the node is 1.00 supported. "-" indicates that the node is incongruent between the topology of the ML tree and the Bayesian tree. Red and green clade indicates mentum absence and presence, respectively. The species name is followed by accession number of the nrDNA sequences. |
The study is the first time to report new species of mycoheterotrophic orchid from western Sichuan: Corallorhiza sinensis. Previous studies indicated the members of Oreorchis possess coralloid rhizomes (Pearce and Cribb, 1997; Suetsugu et al., 2020), while the new species exhibits more abundant coralloid rhizomes than Oreorchis. C. sinensis present many synapomorphies that involved in Corallorhiza, such as leafless, no visible green tissue in all individuals. Despite the populations are characteristic of the intermediate morphological traits between Corallorhiza and Oreorchis, it most closely resembles the Corallorhiza. The complete plastome of C. sinensis was sequenced, assembled, and analyzed in comparison with these members of subtribe Calypsoinae. The plastome of C. sinensis shows a typical quadripartite structure, as reported for most angiosperms (Khayi et al., 2020; Wu et al., 2020; Zhu et al., 2020). A total of 109 unique genes were identified in the plastome, which contains 68 protein-coding genes, 30 tRNAs, and four rRNAs. C. sinensis further shows evidence of some photosynthesis-related genes either lost or pseudogenized, which is consistent with some previous studies of Corallorhiza (Barrett and Davis, 2012; Kim et al., 2020; Li et al., 2020). In the current study, a common feature of the plastome of Corallorhiza and Oreorchis species was that IR regions were more conserved than the LSC and SSC regions. C. sinensis is closely related to Corallorhiza in terms of the length of whole plastome. Comparative analysis of plastomes showed that C. sinensis has one small inversion occurred in the LSC region. However, the larger inversions appear in Danxiaorchis singchiana (Lee et al., 2020), C. maculata var. maculata (Barrett et al., 2014) and C. maculata (Kim et al., 2020), which had been reported. IR boundaries of C. sinensis present obvious expansion in these members of Calypsoinae.
Phylogenetic analysis based on coding sequences (CDS) from 26 Calypsoinae members strongly supports Corallorhiza sinensis to be sister to the clade consisting of C. striata var. vreelandii + C. striata var. striata + C. cf. striata + C. bentleyi + C. striata var. involuta. All members of the sister clade of C. sinensis are distributed in North America (Freudenstein, 1997, 1999). C. sinensis is extremely similar to these members in sepals and petals morphology, but can be easily distinguished by its lip broad oblong and obviously 3-lobed toward base. In the clade consisting of C. sinensis, C. striata complex, and C. bentleyi, their mentum is absent. According to a monograph of Corallorhiza (Freudenstein, 1997), Corallorhiza was divided into two clades by the mentum absent or present. In current phylogenetic trees, red and green clade indicates mentum absent and present, respectively. The monograph indicated that the mentum is one of the characters to distinguish C. striata and other congeneric species (Freudenstein, 1997), thus the character presents a possible synapomorphy for the clade. Our phylogenetic result is consistent with the previous studies (Kim et al., 2020; Li et al., 2020), that Corallorhiza is not a monophyletic group, because the genus Oreorchis is nested within the clade of Corallorhiza with strong bootstrap support (BS = 100) and posterior probability (PP = 1.00). In the Calypsoinae, however, phylogenetic analysis based on the nrDNA sequence suggests that Corallorhiza is a monophyletic group and strongly supports C. sinensis as sister to Corallorhiza (ML: BS = 98; BI: PP = 1.00). Although the topological conflicts are presented between plastome and nrDNA phylogenies of C. sinensis, it is still the most closely related to Corallorhiza. Both the plastid tree and nrDNA tree indicate Oreorchis is paraphyletic. The finding of this new species from China shed new light on the phylogenetic relationship of Oreorchis and Corallorhiza. Finally, we treat it here as a new species of Corallorhiza based upon morphological and molecular evidence.
Author ContributionsJXY, SP, JJW and HY collected these materials; GWH and QFW identified species; SX D, YW, JT, and HY designed and performed the experiments; JXY, SP, and JJW performed the analyses; JT drew the illustrations of the species; JY and SP wrote the manuscript; GWH and QFW revised the paper. All authors read and approved the final manuscript.
Declaration of competing interestThe author declares no conflict of interest.
AcknowledgementsThe authors are grateful to the Project of Orchid Biodiversity Survey of China from National Forestry and Grassland Administration. We also thank professor Xiao-Hua Jin for confirming the identification and encouraging us to publish this new species, and staffs from Siguniangshan National Nature Reserves for their help during the field work. We acknowledge the National Wild Plant Germplasm Resource Center for all kinds of support. This work was supported by the grants from the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), the National Natural Science Foundation of China (31970211).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.01.002.
Bankevich A., Nurk S., Antipov D., et al, 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol, 19: 455-477. DOI:10.1089/cmb.2012.0021 |
Barrett C.F., Davis J.I., 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot, 99: 1513-1523. DOI:10.3732/ajb.1200256 |
Barrett C.F., Freudenstein J.V., Jeff L., et al, 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol, 31: 3095-3112. DOI:10.1093/molbev/msu252 |
Camacho C., Coulouris G., Avagyan V., et al, 2009. BLAST+: architecture and applications. BMC Bioinf, 10: 421. DOI:10.1186/1471-2105-10-421 |
Chase M.W., Cameron K.M., Freudenstein J.V., et al, 2015. An updated classification of Orchidaceae. Bot. J. Linn. Soc, 177: 151-174. DOI:10.1111/boj.12234 |
Chen, X.Q., Gale, S.W., Cribb, P.J., 2009. Corallorhiza. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds. ), Flora of China, vol. 25. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press, pp. 252-253.
|
Christenhusz M.J.M., Byng J.W., 2016. The number of known plants species in the world and its annual increase. Phytotaxa, 261: 201-217. DOI:10.11646/phytotaxa.261.3.1 |
Darling A.C.E., Mau B., Blattner F.R., et al, 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res, 14: 1394-1403. DOI:10.1101/gr.2289704 |
Freudenstein J.V., 1997. A monograph of Corallorhiza (Orchidaceae). Harv. Pap. Bot, 1: 5-51. DOI:10.2307/41761525 |
Freudenstein J.V., 1999. A new species of Corallorhiza (Orchidaceae) from West Virginia. U.S.A. Novon, 9: 511-513. DOI:10.2307/3392151 |
Freudenstein J.V., Senyo D.M., 2008. Relationships and evolution of matK in a group of leafless orchids (Corallorhiza and Corallorhizinae; Orchidaceae: Epidendroideae). Am. J. Bot, 95: 498-505. DOI:10.3732/ajb.95.4.498 |
Freudenstein J.V., Yukawa T., Luo Y.B., 2017. A reanalysis of relationships among Calypsoinae (Orchidaceae: Epidendroideae): floral and vegetative evolution and the placement of Yoania. Syst. Bot, 42: 17-25. DOI:10.1600/036364417x694944 |
Jin J.J., Yu W.B., Yang J.B., et al, 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol, 21: 241. DOI:10.1186/s13059-020-02154-5 |
Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., et al, 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 : 587-589. https://doi. org/10, 14: 587-589. DOI:10.1038/nmeth.4285 |
Katoh K., Standley D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol, 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse M., Moir R., Wilson A., et al, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Khayi S., Gaboun F., Pirro S., et al, 2020. Complete chloroplast genome of Argania spinosa: structural organization and phylogenetic relationships in sapotaceae. Plants: 9. DOI:10.3390/plants9101354 |
Kim Y.K., Jo S., Cheon S.H., et al, 2020. Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Fron. Plant Sci, 11: 22. DOI:10.3389/fpls.2020.00022 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol. Biol. Evol, 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Langmead B., Salzberg S.L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9: 357-359. DOI:10.1038/nmeth.1923 |
Lee S.Y., Meng K., Wang H., et al, 2020. Severe plastid genome size reduction in a mycoheterotrophic orchid, Danxiaorchis singchiana, reveals heavy gene loss and gene relocations. Plants, 9: 521. DOI:10.3390/plants9040521 |
Li Z.H., Jiang Y., Ma X., et al, 2020. Plastid genome evolution in the subtribe Calypsoinae (Epidendroideae, Orchidaceae). Genome Biol. Evol, 12: 867-870. DOI:10.1093/gbe/evaa091 |
Lohse M., Drechsel O., Bock R., 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet, 52: 267-274. DOI:10.1007/s00294-007-0161-y |
Macdougal D.T., Dufrenoy J., 1944. Mycorrhizalsymbiosis in Aplectrum, Corallorhiza and Pinus. Plant Physiol. (Wash. D C), 19: 440-465. DOI:10.2307/4257795 |
Nguyen L.T., Schmidt H.A., von Haeseler A., et al, 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol, 32(1): 268-274. DOI:10.1093/molbev/msu300 |
Pearce N., Cribb P., 1997. A revision of the genus Oreorchis (Orchidaceae). Edinb. J. Bot, 54: 289-328. DOI:10.1017/S0960428600004145 |
Qu X.J., Moore M.J., Li D.Z., et al, 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 50: 50. DOI:10.1186/s13007-019-0435-7 |
Ronquist F., Teslenko M., van der Mark P., et al, 2012. MrBayes 3. 2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys029 |
Suetsugu K., Haraguchi T.F., Tanabe A.S., et al, 2020. Specialized mycorrhizal association between a partially mycoheterotrophic orchid Oreorchis indica and a Tomentella taxon. Mycorrhiza. DOI:10.1007/s00572-020-00999-z |
Talavera G., Castresana J., 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol, 56: 564-577. DOI:10.1080/10635150701472164 |
Wick R.R., Schultz M.B., Zobel J., et al, 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics, 31: 3350-3352. DOI:10.1093/bioinformatics/btv383 |
Wu L.W., Nie L.P., Xu Z.C., et al, 2020. Comparative and phylogenetic analysis of the complete chloroplast genomes of three Paeonia section Moutan species(Paeoniaceae). Front. Genet: 11. DOI:10.3389/fgene.2020.00980 |
Yukawa T., Chung S.W., Luo Y.B., et al, 2003. Reappraisal of Kitigorchis (Orchidaceae). Bot. Bull. Acad. Sin. (Taipei), 44: 345-351. |
Zhai J.W., Zhang G.Q., Chen L.J., et al, 2013. A new orchid genus, Danxiaorchis, and phylogenetic analysis of the tribe Calypsoeae. PLoS One, 8: e60371. DOI:10.1371/journal.pone.0060371 |
Zhang D., Gao F.L., Jakovlic I., et al, 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour, 20: 348-355. DOI:10.1111/1755-0998.13096 |
Zhu B., Feng Q., Yu J., et al, 2020. Chloroplast genome features of an important medicinal and edible plant: Houttuynia cordata (Saururaceae). PLoS One, 15: e0239823. DOI:10.1371/journal.pone.0239823 |
Zimmer K., Meyer C., Gebauer G., 2008. The ectomycorrhizal specialist orchid Corallorhiza trifida is a partial myco-heterotroph. New Phytol, 178: 395-400. DOI:10.1111/j.1469-8137.2007.02362.x |




