b. University of Chinese Academy of Sciences, Beijing, 100049, China
Natural hybridization has been regarded as a crucial pathway of speciation and provides the raw materials for the evolution of biodiversity (Harrison, 1990; Arnold, 1997). To date, more and more botanists have turned their attention to natural hybridization due to the importance of hybridization in plant evolution (Rieseberg, 1997; Rieseberg et al., 2000; Abbott et al., 2008, 2010; Soltis and Soltis, 2009; Chase et al., 2010). Hybrids can occupy new ecological niches that could not be occupied by the parental species and form new species eventually by accumulation of genetic variation and/or ecological factors (Arnold, 1997; Burke and Arnold, 2001). Homoploid hybrid speciation (Mallet, 2007; Abbott et al., 2010) and allopolyploid speciation (Tate et al., 2005; Arnold, 2006) are two forms of hybrid speciation. Allopolyploid speciation can lead to rapidly reproductive isolation from parental lineages by doubled chromosome and produce new species (Rieseberg and Willis, 2007; Soltis and Soltis, 1999), whereas homoploid hybrid speciation occurs without changes in chromosome numbers (Mallet, 2007; Abbott et al., 2010). Thus, allopolyploid speciation was considered more common than homoploid hybrid speciation in flowering plant (Arnold, 1992; Abbott et al., 2010). However, hybridization events do not always lead to the origin of new species (Arnold, 1997). Oppositely, some hybridization events may form hybrid swarms or hybrid zones depending on the extent of introgression (Nolte and Tautz, 2010). Introgression is a fairly widespread and significant consequence of hybridization (Buerkle et al., 2000; Rieseberg et al., 2003; Martin et al., 2006). If introgression is strong enough, gene flow will occur across two parental species through hybrids and hybrid populations as bridges, thus introgression can play a key role in adaptive evolution (Arnold, 1992; Rieseberg and Wendel, 1993).
Natural hybridization is common in Ligularia species and has accelerated the evolutionary process of Ligularia species (Pan et al., 2008; Saito et al., 2011; Yu et al., 2011, 2014a, b). Pan et al. (2008) confirmed a hybrid species between L. paradoxa Hand.-Mazz. and L. duciformis (C. Winkl.) Hand.-Mazz. based on a comprehensive study. Moreover, Saito et al. (2011) presumed that L. lamarum (Diels) C.C. Chang and L. subspicata (Bur. and Franch.) Hand.-Mazz. hybridize with other Ligularia species in certain populations according to the examination of chemical similarities. Yu et al.(2011, 2014a, b) demonstrated the occurrence of natural hybridization between sympatric distributions of L. vellerea (Franch.) Hand.-Mazz. and L. subspicata, between sympatric distributions of L. subspicata and L. nelumbifolia (Bur. and Franch.) Hand.-Mazz., and between sympatric distributions of L. cymbulifera (W.W. Smith) Hand.-Mazz. and L. tongolensis (Franch.) Hand.-Mazz. based on both morphological traits and molecular data (e.g. chloroplast DNA (cpDNA) and internal transcribed spacer (ITS) sequence, ISSR markers, SSR markers). Particularly, most of the current studies have demonstrated that the directions of natural hybridization among most Ligularia species are bidirectional (Yu et al., 2011, 2014b). Although the natural hybridization between L. vellerea and L. subspicata was preliminarily confirmed based on both morphological traits and molecular data, the direction of natural hybridization was uncertain due to the limitation of sampling (only 5 putative hybrids were sampled) (Yu et al., 2014a). In the present study, we sampled 16 putative hybrids from the same mixed population for confirming natural hybridization again and elucidating the pattern of hybridization with the hypothesis of bidirectional hybridization. Moreover, L. tongolensis also grows in the mixed population and has overlapped flowering period with the two putative parental species. In order to investigate whether L. tongolensis participated in the process of hybridization, the individuals of L. tongolensis were also sampled and studied.
Molecular markers have been widely applied for natural hybridization studies (Rieseberg and Ellstrand, 1993). Although multiple molecular markers have been used for identifying plant hybridization since last century (Heiser, 1947; Anderson, 1949; Rieseberg and Brunsfeld, 1992), direct sequencing of DNA have been proven to be the most direct method to detect plant hybridization (Rieseberg and Ellstrand, 1993; Pan et al., 2008; Yu et al., 2011, 2014a, b; Liao et al., 2015). Numerous hybridization events have been verified based on sequences of biparentally inherited nuclear ribosomal DNA (nrDNA), and it has been feasible to confirm the paternal and maternal parents of hybrids according to uniparentally inherited DNA (e.g. Yu et al., 2011, 2014a, b; Yan et al., 2013; Zhang et al., 2014). Here, we investigated the natural hybridization between two morphologically distinct species of L. vellerea and L. subspicata using ITS region of nrDNA and three cpDNA fragments (trnL-rpl32, trnQ-5'rps16 and atpB-rbcL).
2. Materials and methods 2.1. Experimental materialsPlant materials were collected from Daxue Mountain, Shangri-La County, Yunnan, China, at 99°49.818′E and 28°33.722′N. Leaves of 85 individuals were collected and dried in silica gel in field for DNA extraction and the number of all samples was listed in Table 1. Leaf shape, stem and inflorescence type were primarily diagnostic characters in the field and the main morphological differences among L. vellerea, L. subspicata, L. tongolensis and putative hybrids were listed in Table 2. The voucher specimens were deposited in Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN), with accession numbers PG14081401-PG14081404.
| Taxa | No. |
| Ligularia vellerea | V1, V2, V3, V4, V5, V6, V7, V8, V9, V10, V11, V12, V13, V14, V15, V16, V17, V18, V19, V20, V21, V22, V23, V24, V25 |
| Putative hybrids | H1, H2, H3, H4, H5, H6, H7, H8, H9, H10, H11, H12, H13, H14, H15, H16 |
| L. subspicata | S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19, S20, S21, S22, S23, S24 |
| L. tongolensis | T1, T2, T3, T4, T5, T6, T7, T8, T9, T10, T11, T12, T13, T14, T15, T16, T17, T18, T19, T20 |
| Taxa | Morphological traits | |||
| Inflorescence | Stem | Leaf shape | Florets | |
| Ligularia vellerea | Racemiform | Densely long white puberulent | Lanceolate | Numerous tubular florets, with several ray florets |
| L. subspicata | Racemiform | Proximally glabrous | Ovate-cordate hastate arrow-shaped | Tubular florets |
| Putative hybrids | Racemiform | Some glabrous Some sparsely Some densely |
Between L. vellerea and L. subspicata | Numerous tubular florets, some several ray florets, some without ray florets |
| L. tongolensis | Corymbose | Spider filiform pilose | Ovate-cordate ovate-oblong | Numerous tubular florets, with several ray florets |
Total genomic DNA was extracted from the silica-dried leaf tissues using the CTAB method (Doyle and Doyle, 1987) with minor modifications. Because we excluded the probability that L. tongolensis participated in the hybridization in the preliminary experiment (i.e., L. tongolensis was completely separated with other two species and putative hybrids based on the ITS sequences), we only selected 8 individuals (Table 1) of L. tongolensis in subsequent sequencing experiment. The ITS region of all sampled individuals was amplified using primers ITS4 and ITS5 (White et al., 1990). PCR was conducted in a total reaction volume of 20 μL, containing 12.4 μL ultrapure water, 2.0 μL 10 × PCR Buffer (Mg2+ free), 1.0 μL MgCl2 (25 mmol/L), 1.0 μL dNTP (2.5 mmol/L), 0.8 μL BSA (20 mg/mL), 0.3 μL Taq polymerase (5 units/μL), 0.5 μL each primer (10 μmol/L) and 1.5 μL template DNA (20–60 ng). The amplification was performed as following conditions: 1 cycle, 94 ℃, 5 min; 36 cycles, 94 ℃, 45 s; 56 ℃, 45 s; 72 ℃, 50 s, and a final extension of 1 cycle, 72 ℃, 8 min. The purified PCR products were sequenced by sequencing company with an ABI 3730 automated sequencer. Direct sequencing was successful for L. vellerea and L. tongolensis, but it produced chimeric or unreadable peaks in the chromatograms for putative hybrids and L. subspicata. Therefore, cloning was carried out for all putative hybrids and all L. subspicata individuals. Purified PCR products were cloned into plasmids using the pUM-T vector system (Bioteke Corporation, Beijing, China). Four to ten positive clones were selected for each amplification product and cultured to isolate plasmids. Positive clones with inserts of the correct size were confirmed by colony PCR. Plasmids with correct inserts were sequenced using universal M13F/M13R primers. Two cpDNA fragments were amplified using the following universal primers: trnL-rpl32, trnQ-5'rps16 (Shaw et al., 2007). PCR was conducted in a reaction volume of 20 μL containing 12.5 μL ultrapure water, 2.0 μL 10 × PCR Buffer (Mg2+ free), 1.0 μL MgCl2 (25 mmol/L), 1.0 μL dNTP (2.5 mmol/L), 1.0 μL DMSO (20 mg/mL), 0.3 μL Taq polymerase (5 units/μL), 0.35 μL each primer (10 μmol/L) and 1.5 μL template DNA (20–60 ng). The amplification was performed as following conditions: 1 cycle, 80 ℃, 5 min; 30 cycles, 94 ℃, 45 s; 53 ℃, 45 s; 65 ℃, 50 s, and a final extension of 1 cycle, 65 ℃, 7 min. The third cpDNA fragment was amplified using universal primers atpB-rbcL (Chiang et al., 1998). PCR was conducted in a reaction volume of 20 μL, containing 10.8 μL ultrapure water, 2.0 μL 10 × PCR Buffer (Mg2+ free), 2.0 μL MgCl2(25 mmol/L), 1.6 μL dNTP (2.5 mmol/L), 1 μL DMSO (20 mg/mL), 0.3 μL Taq polymerase (5 units/μL), 0.4 μL each primer (10 μmol/L) and 1.5 μL template DNA (20–60 ng). The amplification was performed as following conditions: 1 cycle, 94 ℃, 3 min; 36 cycles, 94 ℃, 45 s; 53 ℃, 1 min, 65 ℃, 90 s, and a final extension of 1 cycle, 65 ℃, 7 min. The PCR products of trnL-rpl32, trnQ-5'rps16 and atpB-rbcL fragments were purified and then directly sequenced using an ABI 3770 automated sequencer.
2.3. Data analysisThe sequences were aligned and compared in SeqMan (DNA Star package; DNAStar Inc., Madison, WI, USA, Burland, 1999), then sequences were edited using BioEdit V.7 (Hall, 1999) and adjusted manually. Three fragments of cpDNA were combined using PAUP*version 4.0b (Swofford, 2002). The base additivity of L. vellerea and L. tongolensis were disposed using DnaSP5.0 (Rozas et al., 2003). Haplotypes for all sampled individuals were obtained using DnaSP5.0. The all haplotypes of three cpDNA fragments and ITS region examined in this study were archived in NCBI GenBank with accession numbers KY788676 to KY788755. The obtained haplotypes were used to infer the relationships for all sampled individuals using the criterion of neighbor-joining (NJ) tree, which was implemented in MEGA 7.0 (Tamura et al., 2007) and tested by bootstrap method. Bootstrap values were calculated with 1000 replications. It is generally believed that when some haplotypes get together into a cluster with the supporting rate ≥50%, these haplotypes have closer relationships (Tripathi et al., 2013).
3. Results 3.1. ITS analysisThe aligned length of all the ITS sequences was 612 bp. Clearly, there were twelve nucleotide substitutions and one insertion/deletion (indel) were detected between two putative parents (Table 3). For putative hybrids, all individuals showed twelve chromatogram additivity sites between L. vellerea and L. subspicata and one deletion. Although at these fixed sites, sympatric L. tongolensis showed identical informative sites with either L. vellerea or L. subspicata, Ligularia tongolensis had twenty-three fixed informative sites separately. For L. subspicata and putative hybrids, direct sequencing of the nrITS region generated many chimeric and unreadable peaks in the chromatogram. Thus, in the subsequent cloning sequencing, we obtained more haplotypes than L. vellerea. Interestingly, one exception was detected among four L. subspicata individuals (S1, S2, S3 and S15). The four individuals had the accordant variable sites with putative hybrids. Namely, the four individuals also showed chromatogram additivity at fixed sites between L. vellerea and L. subspicata.
| Taxa | Polymorphic sites | |||||||||||||||||||||||||
| 21 | 25 | 47 | 62 | 71 | 72 | 80 | 91 | 95 | 99 | 104–106 | 123 | 127 | 133 | 175 | 188 | 192 | 195 | 201–202 | 204 | 211 | 214 | 219 | 221 | 260 | 266 | |
| V1–V4, V6–V7, V13–V25 | C | T | C | G | G | C | C | A | C | A | CTA | G | T | A | G | G | – | C | AC | G | C | G | G | A | T | C |
| V5, V8–V12 | C | T | C | K | K | C | C | A | C | A | CTA | G | T | A | G | G | – | C | AC | G | C | G | G | A | T | C |
| S6–1, S10–1, S12–1, S17–1, S21–1, S21–3, S22–3 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | G | G | G | T | C |
| S7–7, S17–5 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | G | G | G | T | C |
| S13–1, S13–2, S14–5, S18–2, S23–2 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | G | G | G | T | T |
| S14–4, S22–2 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | G | G | G | T | C |
| S4–3, S6–2, S8–6, S8–7, S9–3, S10–4, S16–2, S18–3, S19–1, S19–2, S21–2, S22–6, S24–1, S24–2 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | G | G | G | T | C |
| S5–1, S7–5, S7–6, S8–3, S11–2, S11–3, S16–5, S17–2, S20–4, S20–6 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | T | C | GT | T | C | G | G | G | T | C |
| S4–1, S4–2 | C | T | T | G | G | C | T | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | C | G | G | T | C |
| S10–2 | C | T | C | G | G | C | C | A | C | G | TCC | G | C | A | G | G | C | C | GT | G | C | G | G | G | T | C |
| S20–5 | C | T | C | G | G | T | C | A | C | G | TCC | G | C | A | G | G | – | C | GT | G | C | G | G | G | A | C |
| S1 | C | T | C | G | G | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | G | G | R | T | C |
| S2, S3, S15 | C | T | C | G | G | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | C | C | G | G | R | T | C |
| H6, H10, H13 | C | T | C | K | K | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | C | G | R | T | C |
| H4, H8, H14 | C | T | C | K | K | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | G | G | R | T | C |
| H3, H16 | C | T | C | G | G | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | G | G | R | T | C |
| H1, H12, H15 | C | T | C | G | G | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | G | G | R | T | C |
| H7, H11 | C | T | C | G | G | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | G | G | R | T | C |
| H2, H5, H9 | C | T | C | G | G | C | C | A | C | R | YYM | G | Y | A | G | G | – | C | RY | G | C | G | G | R | T | C |
| T1–T8 | T | C | C | G | G | C | C | G | T | G | CTA | T | C | C | A | T | – | T | GC | G | T | G | A | G | T | C |
| Taxa | Polymorphic sites | Haplotype | ||||||||||||||||||||||||
| 369 | 373 | 403 | 411 | 418 | 419 | 421–422 | 427 | 430 | 432 | 436 | 468 | 504 | 511 | 515 | 532 | 556 | 574 | 599 | 603 | 609 | ||||||
| V1–V4, V6–V7, V13–V25 | C | C | C | C | A | C | CC | C | T | A | T | T | T | T | C | C | G | T | C | T | T | Hap3 | ||||
| V5, V8–V12 | C | C | C | C | A | C | CC | C | T | A | T | T | T | T | C | C | G | T | C | T | T | Hap3, Hap15 | ||||
| S6–1, S10–1, S12–1, S17–1, S21–1, S21–3, S22–3 | C | C | C | T | A | C | CC | C | C | G | T | T | C | C | C | C | G | T | C | C | T | Hap1 | ||||
| S7–7, S17–5 | C | C | C | T | A | C | CC | C | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap2 | ||||
| S13–1, S13–2, S14–5, S18–2, S23–2 | C | C | C | T | G | C | – | C | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap4 | ||||
| S14–4, S22–2 | C | C | C | T | G | C | – | C | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap8 | ||||
| S4–3, S6–2, S8–6, S8–7, S9–3, S10–4, S16–2, S18–3, S19–1, S19–2, S21–2, S22–6, S24–1, S24–2 | C | C | C | T | A | C | CC | T | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap9 | ||||
| S5–1, S7–5, S7–6, S8–3, S11–2, S11–3, S16–5, S17–2, S20–4, S20–6 | C | C | C | T | A | C | CC | C | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap10 | ||||
| S4–1, S4–2 | C | C | C | T | A | C | CC | C | C | G | T | T | C | C | C | C | G | T | C | C | T | Hap19 | ||||
| S10–2 | C | C | C | T | A | C | CC | C | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap28 | ||||
| S20–5 | C | C | C | T | G | C | – | C | T | G | T | T | C | C | C | C | G | T | C | C | T | Hap30 | ||||
| S1 | C | C | C | Y | A | C | CC | C | T | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap3, Hap10, Hap12 | ||||
| S2, S3, S15 | C | C | C | Y | A | C | CC | Y | Y | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap10, Hap11, Hap26, Hap27, Hap33 | ||||
| H6, H10, H13 | C | C | C | Y | A | C | CC | C | T | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap3, Hap10, Hap15, Hap20–Hap21, Hap31, Hap32 | ||||
| H4, H8, H14 | C | C | C | Y | A | C | CC | Y | T | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap1, Hap12–Hap13, Hap15, Hap22–Hap24, Hap29 | ||||
| H3, H16 | C | C | C | Y | A | C | CC | C | Y | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap1, Hap3, Hap14, Hap19 | ||||
| H1, H12, H15 | C | C | C | Y | A | C | CC | Y | Y | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap1–Hap3, Hap7–Hap8, Hap15–Hap16 | ||||
| H7, H11 | C | C | C | Y | A | C | CC | C | T | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap3, Hap15, Hap25, Hap28, Hap34 | ||||
| H2, H5, H9 | C | C | C | Y | A | C | CC | Y | T | R | T | T | Y | Y | C | C | G | T | C | Y | T | Hap3–Hap6, Hap9, Hap17–Hap18, Hap35 | ||||
| T1–T8 | T | T | T | C | A | T | CC | C | T | G | A | A | C | C | Y | A | A | G | T | T | C | Hap36–Hap37 | ||||
| –, deletions; K = G + T; Y = C + T; R = A + G; M = A + C. | ||||||||||||||||||||||||||
A strict consensus tree from nrDNA haplotypes was generated to show the relationships among three species and putative hybrids. The NJ tree from the nrDNA suggested that the haplotypes of putative parental species and putative hybrids formed two respective branches with high supporting values (92 and 88). However, the haplotypes of sympatric L. tongolensis formed the third branch (Fig. 1). Hence, all putative hybrids were products of hybridization between L. vellerea and L. subspicata (Koch et al., 2003) and sympatric L. tongolensis was not apparently involved in the hybridization between L. vellerea and L. subspicata.

|
| Fig. 1 Phylogenetic relationships of the nrITS haplotypes of all Ligularia species distributed within the mixed population. Supporting rate > 50% is shown above branches. |
The aligned sequences of atpB-rbcL, trnL-rpl32 and trnQ-5'rps16 fragments were 746 bp, 866 bp and 945 bp, respectively. The combined length was 2557 bp. Ligularia vellerea and L. subspicata differed in twenty nucleotide substitutions and four fixed indels (Table 4). All individuals of putative hybrids had the same cpDNA sequences with either L. vellerea or L. subspicata. Ligularia vellerea and L. subspicata both had two haplotypes respectively. Sympatric L. tongolensis had one haplotype and differed from the putative parents by two nucleotide substitutions and two fixed indels in the chloroplast sequences. Similarly, one exception was also detected among three L. subspicata individuals (S1, S8 and S10). Namely, the three individuals all showed the identical cpDNA sequences with L. vellerea.
| Taxa | Polymorphic sites | |||||||||||||||||||||
| atpB-rbcL | trnQ-5'rps16 | trnL-rpl32 | ||||||||||||||||||||
| 43 | 314 | 379 | 695 | 161 | 213 | 263–265 | 293 | 404–405 | 512 | 580 | 127 | 188 | 189–195 | 249 | 332 | 363 | 459–461 | 496 | 511–515 | |||
| V1, V19–V20, V22–V25 | T | T | A | T | T | A | – | A | TT | C | C | T | T | ▼ | T | T | G | – | T | – | ||
| V2–V18, V21 | G | T | A | C | T | A | – | A | AG | C | C | T | – | – | T | T | G | ◆ | T | – | ||
| H6, H11 | T | T | A | T | T | A | – | A | TT | C | C | T | T | ▼ | T | T | G | – | T | – | ||
| H5 | G | T | C | T | T | – | – | A | TT | C | C | T | G | ▼ | T | A | G | – | T | – | ||
| H10, H13 | G | T | A | C | T | A | – | A | AG | C | C | T | – | – | T | T | G | ◆ | T | – | ||
| H2 | G | A | A | C | G | A | – | A | TT | – | A | T | T | ▼ | A | T | T | – | – | – | ||
| H1, H3–H4, H7–H9, H12, H14–H16 | G | A | A | C | G | A | – | A | TT | – | A | T | T | ▼ | T | T | G | – | T | – | ||
| S1 | G | T | A | C | T | A | – | A | AG | C | C | T | – | – | T | T | G | ◆ | T | – | ||
| S8, S10 | G | T | C | T | T | – | – | A | TT | C | C | T | G | ▼ | T | A | G | – | T | – | ||
| S3, S5, S6, S9, S11, S14–S18, S23–S24 | G | A | A | C | G | A | – | A | TT | – | A | T | T | ▼ | T | T | G | – | T | – | ||
| S2, S4, S7, S12–S13, S19–S22 | G | A | A | C | G | A | – | A | TT | – | A | T | T | ▼ | A | T | T | – | – | – | ||
| T1–T8 | G | T | A | C | T | A | ▲ | G | TT | C | C | G | T | ▼ | T | T | G | – | T | ■ | ||
| Taxa | Polymorphic sites | Haplotype | ||||||||||||||||||||
| trnL-rpl32 | ||||||||||||||||||||||
| 774 | 776–777 | 779 | 781–786 | 787–791 | 793 | 797 | 802 | 804–805 | 807–810 | 811–866 | ||||||||||||
| V1, V19–V20, V22–V25 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap3 | ||||||||||
| V2–V18, V21 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap2 | ||||||||||
| H6, H11 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap3 | ||||||||||
| H5 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap5 | ||||||||||
| H10, H13 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap2 | ||||||||||
| H2 | T | GA | C | ★ | TTATC | A | T | C | TT | CTAA | ▶ | Hap4 | ||||||||||
| H1, H3–H4, H7–H9, H12, H14–H16 | T | GA | C | ★ | TTATC | A | T | C | TT | CTAA | ▶ | Hap1 | ||||||||||
| S1 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap2 | ||||||||||
| S8, S10 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap5 | ||||||||||
| S3, S5, S6, S9, S11, S14–S18, S23–S24 | T | GA | C | ★ | TTATC | A | T | C | TT | CTAA | ▶ | Hap1 | ||||||||||
| S2, S4, S7, S12–S13, S19–S22 | T | GA | C | ★ | TTATC | A | T | C | TT | CTAA | ▶ | Hap4 | ||||||||||
| T1–T8 | G | TC | A | – | CTCGA | G | – | A | AA | TAGG | – | Hap6 | ||||||||||
| –, deletions; ▲▼◆■★▶, presence of insertion; ▲, TAA(3bp); ▼, AAGATTA(7bp); ◆, ACT(3bp); ■, TTATA(5bp); ★, AGTTTT(6bp); ▶, AAAAACTTATTTGATTGAATTAACTTGTTCAATCTCGACGATTGAA TATAAATAGG(56bp). | ||||||||||||||||||||||
The NJ tree from combined three cpDNA fragments showed clearly relationships of all samples. The haplotypes of two putative parental species and putative hybrids formed two respective branches with high supporting values (100 and 99), and the haplotypes of sympatric L. tongolensis shared a branch with the L. vellerea (Fig. 2). Because chloroplast DNA is maternally inherited in Ligularia species (Zhang et al., 2003), the hybridization between L. vellerea and L. subspicata is bidirectional. Namely, one of the two parents of L. vellerea and L. subspicata can not only act the maternal parent, but also act the paternal parent.
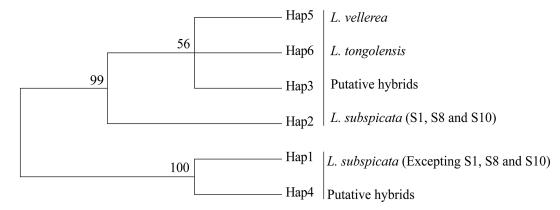
|
| Fig. 2 Phylogenetic relationships of the three cpDNA intergenic spacer regions haplotypes of all Ligularia species distributed within the mixed population. Supporting rate > 50% is shown above branches. |
In sympatric two closely related species, interspecific natural hybridization may arise if the flowering periods overlap partly or totally (Nishiwaki et al., 2011). In the case of our study system, the flowering periods of two putative parents are somewhat overlap because they all bloom in late June to August (Liu, 1989). Moreover, pollinator observations in the highlands have shown that pollinators do not strictly discriminate among Ligularia species (Liu, 2002; Cao et al., 2008). Thus, when different species are flowering at the same place and time, nectar-collecting insects may transfer pollen from one Ligularia species to another Ligularia species by chance. Hence, these conditions provide the probability for the natural hybridization between L. vellerea and L. subspicata. Based on the morphological comparisons, we preliminary judged that morphologically intermediate individuals are produced by the hybridization between L. vellerea and L. subspicata in the field. The leaf shape, stem and inflorescence type are easily distinguished and the putative hybrids have the intermediate morphology of these characteristics between L. vellerea and L. subspicata (Table 2). In other words, the possibility of hybridization between them has been implied by the morphological characteristics, sympatric distribution and overlapped flowering periods. Furthermore, the results of ITS data provided another evidence for the occurrence of hybridization between L. vellerea and L. subspicata, which is exactly consistent with that of Yu et al. (2014a). In addition, the results of cpDNA data clearly show that the direction of natural hybridization between L. vellerea and L. subspicata is bidirectional, which supports our previous hypothesis. Interestingly, the result is inconsistent with that of Yu et al. (2014a), which shows the evidence of unidirectional hybridization between the two species. And notably, we notice that the flowering periods of L. tongolensis also partly overlap with the L. vellerea and L. subspicata, which implies L. tongolensis may participate in natural hybridization. However, this can be strongly excluded by the results of ITS data analyses, because L. tongolensis is completely separated from other two species and putative hybrids based on ITS data (see Table 3 and Fig. 1). Although L. vellerea, L. subspicata and L. tongolensis grow together, L. vellerea and L. subspicata have the identical inflorescences, which may make pollinating insects transfer pollen between them other than L. tongolensis. On the other hand, in Ligularia phylogeny, the relationship between L. vellerea and L. subspicata is closer (He and Pan, 2015) than L. tongolensis, which may suggests that, in comparison with L. tongolensis, the reproductive isolation between them may be easier to be broken and the natural hybridization can be promoted.
4.2. The inconsistency between cpDNA and nrDNA for sympatric L. tongolensisFor sympatric L. tongolensis, the results of cpDNA and nrDNA are inconsistent. For example, the haplotypes of cpDNA of L. tongolensis formed a cluster with L. vellerea (Fig. 2), whereas the nrDNA results suggested L. tongolensis was separated completely from L. vellerea (Fig. 1). The inconsistency between results of cpDNA and nrDNA may be caused by the heterogeneity of evolution rate (Coyne and Orr, 2004), incomplete lineage sorting (Comes and Abbott, 2001), recurrent hybridization (Tsukaya et al., 2003; Yatabe et al., 2009), and convergent evolution (Davis, 1998; Desplanque et al., 2000). Here, we argue that this discordance may be explained by the heterogeneity of evolution rate. The evolutionary rate of nrDNA is faster than cpDNA (Wolfe et al., 1987). Moreover, the ITS region of nrDNA can better distinguish the species of Ligularia than cpDNA (He and Pan, 2015). Hence, in the case of our study, the slower evolution rate of cpDNA results in that the combined cpDNA fragments can not distinguish the L. tongolensis from L. vellerea explicitly.
4.3. The confirmation of some individuals (S1, S2, S3, S8, S10 and S15) of putative parent L. subspicataIn this study, we notice that some individuals of L. subspicata are peculiar. For example, individual S1 had the identical nrITS sequence with putative hybrids and identical cpDNA sequences with L. vellerea. Furthermore, individual S2, S3 and S15 had the identical nrITS sequences with putative hybrids and identical cpDNA sequences with L. subspicata. Individual S8 and S10 had the identical nrITS sequences with L. subspicata and identical cpDNA sequences with L. vellerea (Tables 3 and 4). This phenomenon may be attributed to introgression (Harrison, 1986). Introgression can produce hybrids that are similar to one parent and have some particular characters from another parent. Further, introgression can provide the opportunity for gene flow between the putative parents, resulting in more complex relationships among these taxa (Harrison, 1986). In addition, introgressive hybridization can produce considerable numbers of new genotypes and lead to the establishment of new ecotypes which can adapt to particular environments (Arnold and Hodges, 1995; Arnold, 1997). Consequently, the occurrence of introgressive hybridization may increase the difficulties for confirming these individuals of L. subspicata. Namely, these individuals may be the progeny which are produced by the backcrossing between hybrids and putative parents. The occurrence of backcrosses can provide the opportunities for gene flow between two putative species and may indicate that introgression occurs between L. vellerea and L. subspicata.
Declaration of authorship
Xun Gong conceived and designed the research. Jiaojun Yu collected the experimental materials and analyzed the data. Huai Ning conducted the experiment, analyzed the data and wrote the manuscript. And all authors contributed to reviewing the manuscript.
AcknowledgmentsWe would like to thank Yuezhi Pan, Yujuan Zhao, Ningning Zhang, Rong Zhang, and Weiying He for their contributions to the experiments and the revision of the manuscript. The work is supported by the National Science Foundation of China (31470336 to XG).
Aanderson, E., 1949. Introgressive Hybridization. Wiley, New York.
|
Arnold M.L., 1992. Natural hybridization as an evolutionary process. Annu. Rev.Ecol. Syst: 237-261. |
Arnold M.L., Hodges S.A., 1995. Are natural hybrids fit or unfit relative to their parents?. Trends Ecol. Evol. , 10: 67-71. |
Arnold M.L., 1997. Natural Hybridization and Evolution. New York: Oxford University Press.
|
Arnold M.L., 2006. Evolution through Genetic Exchange. New York: Oxford University Press.
|
Abbott R.J., Ritchie M.G., Hollingsworth P.M., 2008. Introduction. Speciation in plants and animals:pattern and process. Philos. Trans. R. Soc. B Biol. Sci, 363: 2965-2969. |
Abbott R.J., Hegarty M.J., Hiscock S.J., et al, 2010. Homoploid hybrid speciation in action. Taxon, 59: 1375-1386. |
Burland T.G., 1999. DNASTAR's Lasergene sequence analysis software. Methods Mol.Biol, 132: 71-91. |
Buerkle C.A., Morris R.J., Asmussen M.A., et al, 2000. The likelihood of homoploid hybrid speciation. Heredity, 84: 441-451. DOI:10.1046/j.1365-2540.2000.00680.x |
Burke J.M., Arnold M.L., 2001. Genetics and the fitness of hybrids. Annu. Rev. Genet, 35: 31-52. DOI:10.1146/annurev.genet.35.102401.085719 |
Chiang T.Y., Schaal B.A., Peng C.I., 1998. Universal primers for amplification and sequencing a non-coding spacer between the atpB and rbcL genes of chloroplast DNA. Botanical Bull. Botanical Bull. Acad. Sinica, 39: 245-250. |
Comes H.P., Abbott R.J., 2001. Molecular phylogeography, reticulation, and lineage sorting in Mediterranean Senecio sect. Senecio (Asteraceae).. Evolution, 55: 1943-1962. DOI:10.1111/evo.2001.55.issue-10 |
Coyne, J. A., Orr, H. A., 2004. Speciation. Sinauer Associates Sunderland, M. A.
|
Cao Y., Ma R.J., Wang G.X., 2008. he breeding system of three species of genus Ligularia in the east of Qinghai-Tibet Plateau. Guihaia, 28: 302-306. |
Chase M.W., Paun O., Fay M.F., 2010. Hybridization and speciation in angiosperms:a role for pollinator shifts?. BMC Biol, 8: 45. DOI:10.1186/1741-7007-8-45 |
Doyle J.J., Doyle J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19: 11-15. |
Davis J.I., Simmons M.P., Stevenson D.W., et al, 1998. Data decisiveness, data quality, and incongruence in phylogenetic analysis:an example from the monocotyledons using mitochondrial atpA sequences. Syst. Biol, 47: 282-310. DOI:10.1080/106351598260923 |
Desplanque B.F., Viard F., Bernard J., et al, 2000. The linkage disequilibrium between chloroplast DNA and mitochondrial DNA haplotypes in Beta vulgaris ssp. maritima (L.):the usefulness of both genomes for population genetic studies. Mol. Ecol, 9: 141-154. DOI:10.1046/j.1365-294x.2000.00843.x |
Heiser C.B., 1947. Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution, 1: 249-262. DOI:10.1111/evo.1947.1.issue-4 |
Harrison R.G., 1986. Pattern and process in a narrow hybrid zone. Heredity, 56: 337-349. DOI:10.1038/hdy.1986.55 |
Harrison R.G., 1990. Hybrid zones:windows on evolutionary process. Oxf. Surv.Evol. Biol, 7: 69-128. |
Hall T.A., 1999. Bioedit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp, 41: 95-98. |
He W.Y., Pan Y.Z., 2015. Study on DNA barcoding of genus Ligularia Cass. (Asteraceae). Plant Divers. Resour, 37: 693-703. |
Koch M.A., Dobes C., Mitchell-Olds T., 2003. Multiple hybrid formation in natural populations:concerted evolution of the internal transcribed spacer of nuclear ribosomal DNA (ITS) in North American Arabis divaricarpa (Brassicaceae). Mol.Biol. Evol, 20: 338-350. DOI:10.1093/molbev/msg046 |
Liu, S. W., 1989. Compositae-senecioneae. In: Ling, Y., Liu, S. W. (Eds. ), Flora Reipublicae Popularis Sinicae. Science Press, Beijing, pp. 4-115.
|
Liu, Z. J., 2002. Life History Strategies of Ligularia virgaurea, an Advantage Toxic Forb in Degradative Rangland of Alpine Meadow (Doctor Degree). Lanzhou University China, Lanzhou.
|
Liao R.L., Ma Y.P., Gong W.C., et al, 2015. Natural hybridization and asymmetric introgression at the distribution margin of two Buddleja species with a large overlap. BMC Plant Biol, 15(1): 146. DOI:10.1186/s12870-015-0539-9 |
Martin N.H., Bouck A.C., Arnold M.L., 2006. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics, 172: 2481-2489. |
Mallet J., 2007. Hybrid speciation. Nature, 446: 279-283. DOI:10.1038/nature05706 |
Nolte A.W., Tautz D., 2010. Understanding the onset of hybrid speciation. Trends Genet, 26: 54-58. DOI:10.1016/j.tig.2009.12.001 |
Nishiwaki A., Mizuguti A., Kuwabara S., et al, 2011. Discovery of natural Miscanthus (Poaceae) triploid plants in sympatric populations of Miscanthus sacchariflorus and Miscanthus sinensis in southern Japan. Am. J. Bot, 98: 154-159. DOI:10.3732/ajb.1000258 |
Pan Y.Z., Shi S.H., Gong X., et al, 2008. A natural hybrid between Ligularia paradoxa and L. duciformis (Asteraceae, Senecioneae) from Yunnan, China. Ann. Missouri Botanical Gard, 95: 487-494. DOI:10.3417/2006034 |
Rieseberg, L. H., Brunsfeld, S. J., 1992. Molecular Evidence and Plant Introgression. Molecular Systematics of plants, Springer US, pp. 151-176. http://link.springer.com/10.1007/978-1-4615-3276-7_7
|
Rieseberg L.H., Ellstrand N.C., 1993. What can molecular and morphological markers tell us about plant hybridization? Crit. Rev. Plant Sci, 12: 213-241. |
Rieseberg, L. H., Wendel, J. F., 1993. Introgression and its consequences in plants. In: Harrison, R. G. (Ed. ), Hybrid Zones and the Evolutionary Process. Oxford University Press, New York, pp. 70-109.
|
Rieseberg L.H., 1997. Hybrid origins of plant species. Annu. Rev. Ecol. Syst, 28: 359-389. DOI:10.1146/annurev.ecolsys.28.1.359 |
Rieseberg L.H., Baird S.J.E., Gardner K.A., 2000. Hybridization, introgression, and linkage evolution. Plant Mol. Biol, 42: 205-224. DOI:10.1023/A:1006340407546 |
Rieseberg L.H., Raymond O., Rosenthal D.M., et al, 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science, 301: 1211-1216. DOI:10.1126/science.1086949 |
Rozas J., Sánchez-DelBarrio J.C., Messeguer X., et al, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19: 2496-2497. DOI:10.1093/bioinformatics/btg359 |
Rieseberg L.H., Willis J.H., 2007. Plant speciation. Science, 317: 910-914. DOI:10.1126/science.1137729 |
Soltis D.E., Soltis P.S., 1999. Polyploidy:recurrent formation and genome evolution. Trends Ecol. Evol, 14: 348-352. DOI:10.1016/S0169-5347(99)01638-9 |
Swofford, D. L., 2002. PAUP: Phylogenetic Analysis Using Parsimony version 4. 0 b10.
|
Sinauer Associates Sunderland., Shaw J., Lickey E.B., Schilling E.E., et al, 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms:the tortoise and the hare III. Am. J. Bot, 94: 275. DOI:10.3732/ajb.94.3.275 |
Soltis P.S., Soltis D.E., 2009. The role of hybridization in plant speciation. Annu. Rev.Plant Biol, 60: 561-588. DOI:10.1146/annurev.arplant.043008.092039 |
Saito Y., Hattori M., Iwamoto Y., et al, 2011. Overlapping chemical and genetic diversity in Ligularia lamarum and Ligularia subspicata. Isolation of ten new eremophilanes and a new seco-bakkane compound. Tetrahedron, 67: 2220-2231. DOI:10.1016/j.tet.2011.01.082 |
Tsukaya H., Fukuda T., Yokoyama J., 2003. Hybridization and introgression between Callicarpa japonica and C. mollis (Verbenaceae) in central Japan, as inferred from nuclear and chloroplast DNA sequences. Mol. Ecol, 12: 3003-3011. DOI:10.1046/j.1365-294X.2003.01961.x |
Tate, J.A., Soltis, D.E., Soltis, P.S., 2005. Polyploidy in plants. In:Gregory, T.R. (Ed.), The Evolution of the Genome. Elsevier Science and Technology, Academic Press, San Diego, CA, pp. 371-426.
|
Tamura K., Dudley J., Nei M., et al, 2007. MEGA:molecular evolutionary genetics analysis (MEGA) software version 4. 0. Mol. Biol. Evol, 24: 1596-1599. DOI:10.1093/molbev/msm092 |
Tripathi A.M., Tyagi A., Kumar A., et al, 2013. The internal transcribed spacer (ITS)region and trnH-psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS One, 8: e57934. DOI:10.1371/journal.pone.0057934 |
Wolfe K.H., Li W.H., Sharp P.M., 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci.U. S. A, 84: 9054-9058. DOI:10.1073/pnas.84.24.9054 |
White T.J., Bruns T., Lee S., et al, 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl, 18: 315-322. |
Yatabe Y., Tsutsumi C., et al, 2009. Genetic population structure of Osmunda japonica, rheophilous Osmunda lancea and their hybrids. J. Plant Res, 122: 585-595. DOI:10.1007/s10265-009-0254-4 |
Yu J.J., Kuroda C., Gong X., 2011. Natural hybridization and introgression in sympatric Ligularia species (Asteraceae, Senecioneae). J. Syst. Evol, 49: 438-448. DOI:10.1111/jse.2011.49.issue-5 |
Yan L.J., Gao L.M., Li D.Z., 2013. Molecular evidence for natural hybridization between Rhododendron spiciferum and R. spinuliferum (Ericaceae). J. Syst. Evol, 51: 426-434. DOI:10.1111/j.1759-6831.2012.00243.x |
Yu J.J., Pan L., Pan Y.Z., Gong X., 2014a. Natural hybrids between Ligularia vellerea and L. subspicata (Asteraceae:Senecioneae). Plant Biodivers. Resour, 36: 219-226. |
Yu J.J., Kuroda C., Gong X., 2014b. Natural hybridization and introgression between Ligularia cymbulifera and L. tongolensis (Asteraceae, Senecioneae) in four different locations. PLoS One, 9(12): e115167. DOI:10.1371/journal.pone.0115167 |
Zhang Q., Liu Y., Sodmergen, 2003. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol, 44: 941-951. DOI:10.1093/pcp/pcg121 |
Zhang W.Y., Kuo L.Y., Li F.W., et al, 2014. The hybrid origin of Adiantum meishanianum (Pteridaceae):a rare and endemic species in Taiwan. Syst. Bot, 39: 1034-1041. DOI:10.1600/036364414X682616 |



