b. Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China;
c. University of Chinese Academy of Sciences, Beijing, 100049, China
The Hengduan Mountains region (HDM), which is located in southwest China on the southeast portions of the Qinghai-Tibet Plateau (25°-34°N, 96°-105°E), covers an area of 3.64 × 105 km2 (Li, 1987). This region is a very interesting floristic area in view of the fact that Quaternary glaciations covered only a small part of the Hengduan Mountains and the region was likely a core refugium for temperate species of East Asia during glacial periods (Li et al., 1991). Perhaps partly because of these glacial episodes, the HDM became a center for the rapid evolution of new species (Liu et al., 2006; Sun, 2002; Wang and Liu, 2004; Wu, 1988). It is also one of the major centers of endemism in China (Li, 1994). The flora of HDM comprises more than 8590 species belonging to 1348 genera, 2783 of which are endemic (32.4% of the native flora) (Li and Li, 1993; Wu, 1988; Zhang et al., 2009b). Moreover, it is known as one of the 25 most significant biodiversity hotspots in the world (Myers et al., 2000; Olson and Dinerstein, 2002). However, reports of chromosome number and karyotype are relatively few for the HDM flora.
Polyploidy has long been recognized as an important evolutionary force in plants (Grant, 1981; Jiao et al., 2011; Levin, 2002; Stebbins, 1940, 1950, 1971), however the role of ploidy level on rapid speciation is complex, with different lines of evidence often yielding conflicting inferences. Statistical analyses of chromosome numbers of 552 taxa from the HDM show that polyploidy may have played only a minor role in the evolutionary diversification of plants in this region (Nie et al., 2005). Although this point of view is somewhat supported by some highly diversified genera in the region, such as Cremanthodium (Liu et al., 2001), Cyananthus (Chen et al., 2014), Dolomiaea (Wang et al., 2013), Delphinium (Yuan and Yang, 2008) and Ligularia (Liu, 2004), investigations on other groups have drawn different conclusions. Anaphalis and Leontopodium from the Hengduan Mountains suggest that polyploidization has played a relatively important role in the chromosome evolution (Meng et al., 2012, 2014). Research on Aconitum subgenus Lycoctonum (Yuan and Yang, 2006) and Buddleja (Chen and Sun, 2006) from HDM show similar results. All reported chromosome numbers from HDM have been thus far scattered among different families and genera. Therefore, selecting an appropriate group to evaluate the role of polyploidy in speciation is important.
Asteraceae has a series of ploidy levels from di-, tri-, tetra-, hexa-, to octaploid. Therefore, we chose this family to study the role of polyploidy in the evolution of plants in the HDM region. Asteraceae is one of the largest families in the world. It contains approximately 1700 genera and 24, 000 species distributed all over the world (Shi et al., 2011). There are about 833 taxa of Asteraceae which belong to 137 genera in the HDM (Shi et al., 2011; Wang, 1994; Zhang et al., 2009a, 2009b). They show extreme diversification in morphology, and their geographical distribution extends from dry-hot valley to the alpine subnival belt (from 700 to 5500 m) (Shi et al., 2011; Wang, 1994; Xu et al., 2014a, 2014b).
In this study we report the chromosome numbers and chromosome morphology of 19 Asteraceae species, and calculate the frequency of polyploidy of these genera in the Hengduan Mountains region.
1. Material and methods 1.1. Plant materials and cytological studiesAll of the nineteen species (21 populations) were collected from the Hengduan Mountains (HDM) region of SW China (Table 1). Voucher specimens have been stored in the Herbarium of the Kunming Institute of Botany (KUN).
| Taxon | Origin | Location | Altitude/m | Voucher number |
| Cremanthodium lineare var. eligulatuma | Daocheng, Sichuan | 100°02'36″E; 29°08'16″N | 4715 | SunH-07zx-3332 |
| C. angustifoliuma | Daocheng, Sichuan | 100°02'36″E; 29°08'16″N | 4715 | SunH-07zx-3333 |
| C. principisa | Shangri-La, Yunnan | 99°57'23″E; 28°30'05″N | 4600 | PengDL-15 |
| C. helianthusa | Shangri-La, Yunnan | 99°57'23″E; 28°31'05″N | 4500 | PengDL-20 |
| C. campanulatum var. campanulatuma | Shangri-La, Yunnan | 99°55'40″E; 28°30'39″N | 4542 | PengDL-17 |
| Leontopodium soulieia | Mangkang, Tibet | 98°40'17″E; 29°44'37″N | 3590 | SunH-07zx-3383 |
| Aster pekinensis | Batang, Sichuan | 99°12'10″E; 30°34'13″N | 3831 | SunH-07zx-3389 |
| A. gouldii (CY)a | Chayu, Tibet | 97°10'34″E; 29°19'14″N | 3640 | SunH-07zx-2496 |
| A. gouldii (BM)a | Bomi, Tibet | 96°22'57″E; 29°36'46″N | 3370 | SunH-07zx-2544 |
| Anaphalis xylorhizaa | Ganzi, Sichuan | 100°02'36″E; 29°08'16″N | 4715 | SunH-07zx-3336 |
| A. spodiophylla | Ganzi, Sichuan | 99°37'50″E; 31°42'23″N | 3739 | SunH-07zx-3923 |
| Myriactis nepalensis | Mianning, Sichuan | 101°57'12″E; 28°20'17″N | 2486 | SunH-07zx-3788 |
| M. wightii | Mianning, Sichuan | 101°57'12″E; 28°20'17″N | 2486 | SunH-07zx-3789 |
| Syncalathium roseuma | Zhanang, Tibet | 91°25'0.5″E; 29°10'02.6″N | 3750 | ZhangJW-1054 |
| Pertya phylicoides3 | Deqin, Yunnan | 99°15'26″E; 28°15'17″N | 2686 | PengDL-031 |
| Dubyaea tsarongensisa | Gongshan, Yunnan | 98°28'33″E; 27°46'56″N | 3275 | MS-025 |
| Carpesium cernuum (MN) | Mianning, Sichuan | 101°57'19″E; 28°20'00″N | 2221 | SunH-07zx-3781 |
| C. cernuum (LJ) | Lijiang, Yunnan | 99°38'27″E; 26°47'34″N | 2550 | SunH-07zx-3132 |
| C. lipskyia | Mianning, Sichuan | 101°57'12″E; 28°20'17″N | 2486 | SunH-07zx-3790 |
| C. scapiforme | Batang, Sichuan | 99°12'10″E; 30°34'13″N | 3831 | SunH-07zx-3391 |
| C. velutinum | Jiulong, Sichuan | 101°26'31″E; 29°02'33″N | 3125 | SunH-07zx-3774 |
| a Endemic to the HimalayaneHengduan Mountains region. | ||||
Root tips from the germination of seeds were used to examine somatic chromosomes. Seeds were stored at 4 ℃ for one month. Seeds were then germinated using wet double filter paper in glass culture dishes at 24 ℃. Exuberant root tips (1-2 cm in length) were excised from the seedlings and pretreated in 0.002 or 0.003 mol/L 8-hydroxyquinoline solution at 24 ℃ in darkness for 1-3 h, and then immobilized with Carnoy's solution (1 glacial acetic acid: 3 absolute alcohol, v/v) for at least 24 h at 4 ℃. The fixed roots were hydrolyzed in 1 mol/L HCl at 60 ℃ for 10-15 min, and then washed with distilled water, dyed with carbolfuchsin and squashed for observation.
1.2. Karyotype analysisKaryotype formulae were based on measurements of mitotic-metaphase chromosomes taken from photographs. To standardize the procedure, chromosome metaphase plates from at least six cells were measured. The terminology used for the karyotype position of chromosome composition followed Levan et al. (1964). The degree of karyotype asymmetry was calculated following Arano (1963) "As.K%". The equation: index of Karyotypic Asymmetry (As.K%) = the total of the longest chromosome length/the total of the all chromosome length × 100. Stebbin's asymmetry category was described by karyotypic symmetry division according to Stebbins (1971) and marked as KA. To evaluate karyotype asymmetry, we calculated two parameters suggested by Paszko (2006), Peruzzi et al. (2009), Peruzzi and Eroğlu (2013) and Peruzzi and Altınordu (2014): MCA (Mean Centromeric Asymmetry) and CVCL (Coefficient of Variation of Chromosome Length). Karyotype asymmetry was compared to the CVCI (Coefficient of Variation of Chromosome Index) and karyotype total haploid length (THL) was calculated. To account for these parameters, we used Karyo-Type software (Altınordu et al., 2016), a cytogenetic tool which can measure karyotype asymmetry indices automatically and efficiently.
1.3. Source of karyotype dataWe surveyed the literature to collect available chromosome number data for Asteraceae species distributed in the Hengduan Mountains region. The online database from Index to Plant Chromosome Numbers (IPCN, http://www.tropicos.org/NameSearch.aspx?projectid=9) was also consulted to confirm the diversity of chromosome number and basic number.
To confirm the distribution of a species in the HDM, we consulted the The Vascular Plants of the Hengduan Mountains (Wang, 1994), Flora of China, local flora and an online database of plants and fungi in south-central China (http://hengduan.huh.harvard.edu/fieldnotes). For a standardized database, we used the Taxonomic Name Resolution Service v.4.0 (TNRS: http://tnrs.iplantcollaborative.org/index.html) to examine our checklist for synonyms and illegitimate names.
2. Results 2.1. Karyotype of 19 species from AsteraceaeChromosome numbers and karyotypes for 19 Asteraceae species are briefly described below. These species, which represent nine genera, include 12 species endemic to the HimalayaneHengduan Mountains.
2.1.1. Cremanthodium lineare var. eligulatum Y. Ling & S. W. LiuC. lineare var. eligulatum was collected from Daocheng (Sichuan, China). The karyotype was formulated as 2n = 2x = 58 = 44m + 14sm (2SAT) (Table 2; Fig. 1: A, a). The chromosome length varied from 2.68 to 4.64 μm. The ratio of the longest to the shortest chromosome was 1.72, and As.K% = 59.12. KA belongs to Stebbins's-2A (Table 2). This is the first report of karyotype and chromosome numbers for C. lineare var. eligulatum.
| Taxon | Chromosome length range (μm) | Ratio LC/SC | < 2:1 | As.K% | MCA | CVCL | CVCI | THL | 2n/x/ploidy level | Karyotype formula | KA Type | Figure |
| Cremanthodium lineare var. eligulatuma | 2.68-4.64 | 1.72 | 0.14 | 59.12 | 18.26 | 14.45 | 10.80 | 100.03 | 58/29/2x | 44m + 14sm(2sat) | 2A | A, a |
| C. angustifolium3 | 3.43-4.84 | 1.55 | 0.24 | 61.30 | 22.60 | 7.73 | 18.30 | 116.08 | 58/29/2x | 34m + 22sm + 2st | 2A | B, b |
| C. principis3 | 2.69-4.56 | 1.69 | 0.21 | 61.76 | 23.38 | 14.33 | 13.95 | 99.63 | 58/29/2x | 32m + 26sm(2sat) | 2A | C, c |
| C. helianthusa | 2.65-4.30 | 1.62 | 0.17 | 61.58 | 22.94 | 13.34 | 12.40 | 99.74 | 58/29/2x | 36m + 22sm(2sat) | 2A | D, d |
| C. campanulatum var. campanulatuma | 2.96-4.96 | 1.68 | 0.10 | 59.24 | 18.73 | 13.26 | 11.19 | 101.21 | 58/29/2x | 44m(2sat) + 14sm | 2A | E, e |
| Leontopodium souliei | 2.72-4.21 | 1.51 | 0.13 | 59.09 | 10.16 | 11.23 | 5.50 | 25.02 | 48/12/4x | 36m + 12sm + 0/2/3/4B | 2B | F, f |
| Aster pekinensis | 4.68-6.62 | 1.41 | 0 | 55.10 | 11.62 | 11.91 | 3.90 | 31.06 | 18/9/2x | 18m | 1A | G, g |
| A. gouldii (BM)a | 2.96-3.34 | 1.13 | 0 | 55.92 | 12.86 | 10.48 | 5.24 | 48.57 | 18/9/2x | 18m(2sat) | 1A | H, h |
| A. gouldii (CY)a | 2.84-3.45 | 1.21 | 0 | 55.82 | 18.18 | 15.34 | 12.45 | 58.79 | 18/9/2x | 18m(2sat) | 1A | I, i |
| Anaphalis xylorhiza | 2.25-3.16 | 1.40 | 0.36 | 62.20 | 25.21 | 17.48 | 14.55 | 37.98 | 28/14/2x | 16m + 12sm | 2A | J.j |
| A. spodiophyllaa | 2.68-4.05 | 1.51 | 0.43 | 62.42 | 24.80 | 18.25 | 16.58 | 36.56 | 28/14/2x | 16m + 12sm | 2A | K, k |
| Myriactis nepalensis | 4.56-6.59 | 1.45 | 0.11 | 56.36 | 13.04 | 18.27 | 9.43 | 76.91 | 36/18/2x | 32m + 4sm | 2A | L, l |
| M. wightiia | 3.84-5.34 | 1.39 | 0.06 | 56.08 | 12.13 | 17.4 | 9.29 | 79.27 | 36/18/2x | 32m + 4sm | 2A | M, m |
| Syncalathium roseuma | 4.00-4.76 | 1.50 | 0 | 55.75 | 11.46 | 6.21 | 7.65 | 35.78 | 16/8/2x | 14m + 2sm(2sat) | 1A | N, n |
| Pertya phylicoides3 | 1.48-4.36 | 1.49 | 0 | 58.55 | 17.15 | 31.00 | 5.36 | 42.73 | 34/17/2x | 30m + 4sm | 1B | O, o |
| Dubyaea tsarongensisa | 2.97-4.71 | 1.48 | 0 | 59.72 | 7.37 | 13.42 | 19.03 | 31.89 | 16/8/2x | 14m + 2sm | 1A | P.p |
| Carpesium cernuum (MN) | 1.81-7.07 | 1.89 | 0.4 | 63.19 | 24.76 | 39.95 | 7.69 | 64.17 | 40/20/2x | 26m + 10sm(2sat) + 4st | 2B | Q.q |
| C. cernuum (LJ) | 2.00-6.17 | 1.62 | 0.15 | 60.15 | 23.99 | 31.90 | 12.00 | 64.20 | 40/20/2x | 26m + 10sm(2sat) + 4st | 2B | R, r |
| C. lipskyia | 1.64-4.93 | 1.39 | 0.1 | 57.88 | 13.45 | 35.20 | 5.44 | 47.05 | 40/20/2x | 8M + 26m + 6sm | 2B | S, s |
| C. scapiformea | 2.03-6.05 | 1.75 | 0.35 | 62.14 | 21.26 | 32.30 | 7.46 | 61.83 | 40/20/2x | 14sm + 26m(2sat) | 2B | T, t |
| C. velutinuma | 1.43-5.17 | 1.66 | 0.2 | 61.32 | 22.09 | 35.15 | 9.18 | 49.84 | 40/20/2x | 2M + 22m + 16sm | 2B | U, u |
| LC/SC: the proportion of the longest chromosome length to the shortest chromosome length; As.K%: index of karyotypic asymmetry; MCA: Mean Centromeric Asymmetry; CVCL: the relative variation in chromosome length; CVCI: the relative variation in centromeric index; THL: karyotype total haploid length; M: median point; m: median; sm: submedian; st: subterminal region; sat: satellite chromosome; KA: karyotype asymmetry. a Chromosome number and karyomorphology investigated for the first time. | ||||||||||||
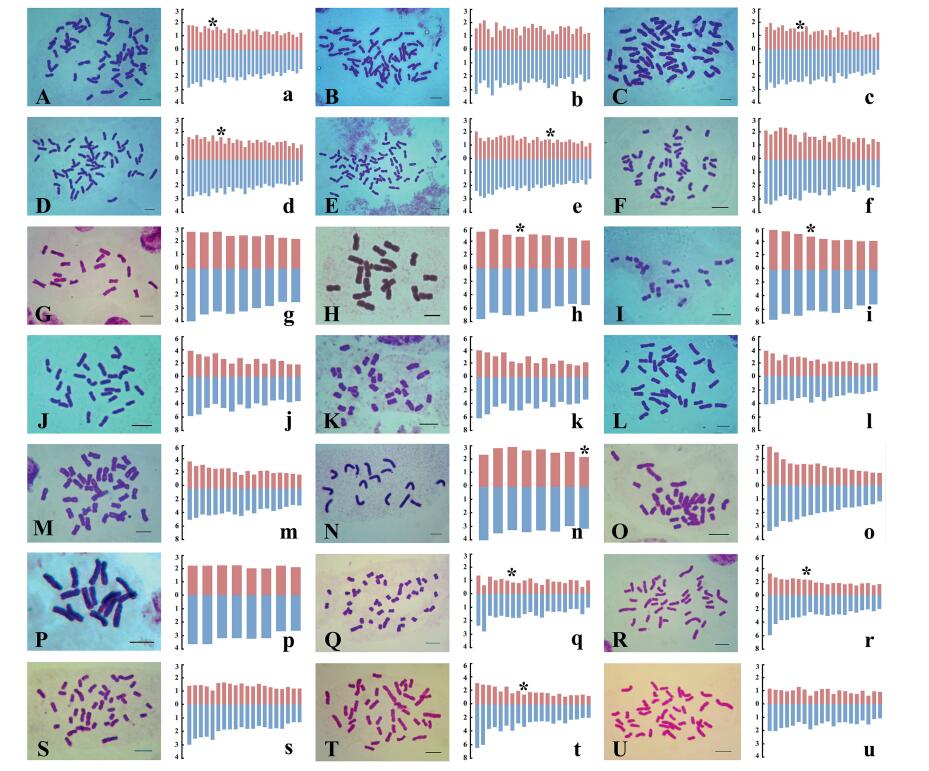
|
| Fig. 1 Mitotic metaphase chromosomes and ideograms of 19 species from the HDM. A, a: Cremanthodium lineare var. eligulatum; B, b: C. angustifolium; C, c: C. principis; D, d: C. helianthus; E, e: C. campanulatum var. campanulatum; F, f: Leontopodium souliei; G, g: Aster pekinensis; H, h: A. gouldii (BM); I, i: A. gouldii (CY); J, j: Anaphalis xylorhiza; K, k: A. spodiophylla; L, l: Myriactis nepalensis; M, m: M. wightii; N, n: Syncalathium roseum; O, o: Pertya phylicoides; P, p: Dubyaea tsarongensis; Q, q: Carpesium cernuum (MN); R, r: C. cernuum (LJ); S, s: C. lipskyi; T, t: C. scapiforme; U, u: C. velutinum (Scale bar = 5 μm; Red: relative length of short arm; Blue: relative length of long arm; *: satellite chromosome). |
C. angustifolium was collected from Daocheng (Sichuan, China). The karyotype was formulated as 2n = 2x = 58 = 34m + 22sm + 2st (Table 2; Fig. 1: B, b). The chromosome length varied from 3.43 to 4.84 μm. The ratio of the longest to the shortest chromosome was 1.55, and As.K% = 61.30. KA belongs to Stebbins's-2A (Table 2). This is the first report of karyotype and chromosome numbers of C. angustifolium.
2.1.3. Cremanthodium principis (Franch.) R. D. GoodC. principis was collected from Shangri-La (Yunnan, China). The karyotype was formulated as 2n = 2x = 58 = 32m + 26sm (2SAT) (Table 2; Fig. 1: C, c). The chromosome length varied from 2.69 to 4.56 μm. The ratio of the longest to the shortest chromosome was 1.69, and As.K% = 61.76. KA belongs to Stebbins's-2A (Table 2). This is the first report of karyotype and chromosome numbers of C. principis.
2.1.4. Cremanthodium helianthus (Franch.) W. W. SmithC. helianthus was collected from Shangri-La (Yunnan, China). The karyotype was formulated as 2n = 2x = 58 = 36m + 22sm (2SAT) (Table 2; Fig. 1: D, d). The chromosome length varied from 2.65 to 4.30 μm. The ratio of the longest to the shortest chromosome was 1.62, and As.K% = 61.58. KA belongs to Stebbins's-2A (Table 2). This is the first report of karyotype and chromosome numbers of C. helianthus.
2.1.5. Cremanthodium campanulatum (Franch.) Diels var. campanulatumC. campanulatum var. campanulatum was collected from Shangri-La (Yunnan, China). The karyotypes were formulated as 2n = 2x = 58 = 44m (2SAT) + 14sm (Table 2; Fig. 1: E, e). The chromosome length varied from 2.96 to 4.96 μm. The ratio of the longest to the shortest chromosome was 1.68, and As.K% = 59.24. KA belongs to Stebbins's-2A (Table 2). This is the first report of karyotype and chromosome numbers of C. campanulatum var. campanulatum.
2.1.6. Leontopodium souliei BeauverdL. souliei was collected from Mangkang (Tibet, China). The karyotype was formulated as 2n = 4x = 48 = 36m + 12sm + 0/2/3/4B (Table 2; Fig. 1: F, f). The chromosome length varied from 2.72 to 4.21 μm. The ratio of the longest to the shortest chromosome was 1.51 and As.K% = 59.09. KA belongs to Stebbins's-2B (Table 2).
2.1.7. Aster pekinensis (Hance) F. H. ChenWe formulated the karyotype of Aster pekinensis as 2n = 2x = 18 = 18m (Table 2; Fig. 1: G, g). The chromosome length varied from 4.68 to 6.62 μm. The ratio of the longest to the shortest chromosome length was 1.41, and the KA belongs to Stebbins's-1A (Table 2).
2.1.8. Aster gouldii C. E. C. FischerIn this study, two populations of Aster gouldii (from Chayu and Bomi) were sampled. The karyotype of the Bomi population was formulated as 2n = 2x = 18 = 18m (2SAT) (Table 2; Fig. 1: H, h). The chromosome length varied from 2.96 to 3.34 μm. The ratio of the longest to the shortest chromosome was 1.13, and the asymmetry of the karyotype belongs to Stebbins's-1A (Table 2). The karyotype of the Chayu population was also formulated as 2n = 2x = 18 = 18m (2SAT) (Table 2; Fig. 1: I, i). The chromosome length varied from 2.84 to 3.45 μm. The ratio of the longest to the shortest chromosome was 1.21, and KA belongs to Stebbins's-1A (Table 2).
2.1.9. Anaphalis xylorhiza Schultz Bipontinus ex J. D. HookerIn this study, the sample from Ganzi (Sichuan) has a formula of 2n = 2x = 28 = 16m + 12sm (Table 2; Fig. 1: J, j). The chromosome length varied from 2.25 to 3.16 μm. The ratio of the longest to the shortest chromosome was 1.40, the As.K% = 62.20. KA belongs to Stebbins's-2A (Table 2).
2.1.10. Anaphalis spodiophylla Y. Ling & Y. L. ChenThe karyotype of Anaphalis spodiophylla was formulated as 2n = 2x = 28 = 16m + 12sm (Table 2; Fig. 1: K, k). The chromosome length varied from 2.68 to 4.05 μm. The ratio of the longest to the shortest chromosome length was 1.51 and the AsK. % = 62.42. KA belongs to Stebbins's-2A (Table 2).
2.1.11. Myriactis nepalensis LessingThe karyotype of Myriactis nepalensis was formulated as 2n = 2x = 36 = 32m + 4sm (Table 2; Fig. 1: L, l). The chromosome length varied from 4.56 to 6.59 μm. The ratio of the longest to the shortest chromosome length was 1.45 and the AsK. % = 56.36. KA belongs to Stebbins's-2A (Table 2).
2.1.12. Myriactis wightii CandolleWe formulated the karyotype of Myriactis wightii as 2n = 2x = 36 = 32m + 4sm (Table 2; Fig. 1: M, m). The chromosome length varied from 3.84 to 5.34 μm. The ratio of the longest to the shortest chromosome length was 1.39 and the AsK. % = 56.08. KA belongs to Stebbins's-2A (Table 2).
2.1.13. Syncalathium roseum Y. LingWe collected seeds of Syncalathium roseum from Zhanang at the type locality. The karyotype of S. roseum was formulated as 2n = 2x = 16 = 14m + 2sm(2sat) (Table 2; Fig. 1: N, n). The chromosome length varied from 4.00 to 4.76 μm. The proportion of the longest to the shortest chromosome length was 1.50 and the AsK. % = 55.75. KA belongs to Stebbins's-1A (Table 2).
2.1.14. Pertya phylicoides JeffreyThe karyotype of Pertya phylicoides was formulated as 2n = 2x = 34 = 30m + 4sm (Table 2; Fig. 1: O, o) in the present study. The chromosome length varied from 1.48 to 4.36 μm. The ratio of the longest to the shortest chromosome length was 1.49 and the AsK. % = 58.55. KA belongs to Stebbins's-1B (Table 2).
2.1.15. Dubyaea tsarongensis (W. W. Smith) StebbinsWe formulated the karyotype of Dubyaea tsarongensis as 2n = 2x = 16 = 14m + 2sm (Table 2; Fig. 1: P, p). The chromosome length varied from 2.97 to 4.71 μm. The ratio of the longest to the shortest chromosome length was 1.48 and the AsK. % = 59.72. KA belongs to Stebbins's-1A (Table 2).
2.1.16. Carpesium cernuum LinnaeusTwo populations of Carpesium cernuum (from Mianning and Lijiang) were sampled. The karyotype of the Mianning population was formulated as 2n = 2x = 40 = 26m + 10sm(2sat) + 4st (Table 2; Fig. 1: Q, q). The chromosome length varied from 1.81 to 7.07 μm. The ratio of the longest to the shortest chromosome was 1.89, and the asymmetry of the karyotype belongs to Stebbins's-2B (Table 2). The karyotype of the Lijiang population was also formulated as 2n = 2x = 40 = 26m + 10sm(2sat) + 4st (Table 2; Fig. 1: R, r). The chromosome length ranged from 2.00 to 6.17 μm. The ratio of the longest to the shortest chromosome was 1.62, and KA belongs to Stebbins's-2B (Table 2).
2.1.17. Carpesium lipskyi C. WinklerC. lipskyi was collected from Mianning (Sichuan, China). The karyotype was formulated as 2n = 2x = 40 = 8M + 26m + 6sm (Table 2; Fig. 1: S, s). The chromosome length varied from 1.64 to 4.93 μm. The ratio of the longest to the shortest chromosome was 1.39, and As.K% = 57.88. KA belongs to Stebbins's-2B (Table 2).
2.1.18. Carpesium scapiforme F. H. Chen & C. M. HuC. scapiforme was collected from Batang (Sichuan, China). The karyotype was formulated as 2n = 2x = 40 = 14sm + 26m(2sat) (Table 2; Fig. 1: T, t) in this study. The chromosome length varied from 2.03 to 6.05 μm. The ratio of the longest to the shortest chromosome was 1.75, and As.K% = 62.14. KA belongs to Stebbins's-2B (Table 2).
2.1.19. Carpesium velutinum C. WinklerC. velutinum was collected from Jiulong (Sichuan, China). The karyotype was formulated as 2n = 2x = 40 = 2M + 22m + 16sm (Table 2; Fig. 1: U. u). The chromosome length varied from 1.43 to 5.17 μm. The ratio of the longest to the shortest chromosome was 1.66, and As.K% = 61.32. KA belongs to Stebbins's-2B (Table 2). This is the first report of karyotype and chromosome numbers of C. velutinum.
2.2. The recorded data of chromosome numbersWe surveyed published papers for the recorded chromosome numbers of species that are both distributed in the Hengduan Mountains and are congeners of the species we sampled (Table 3). By comprehensively surveying these data, the chromosome number or cytogenetics of 69 taxa (include 78 populations) were recorded. We calculated the frequency of polyploidy in these genera in the Hengduan Mountains region, and found that the recorded frequency of Asteraceae polyploidy was only 26.92% (21/78).
| Original taxon | 2n | x | Ploidy | KA | Data source |
| Anaphalis aureopunctata | 28, 56 | 14 | 2x, 4x | 2A, 2B | Meng et al. (2014) |
| A. bicolor | 28, 56 | 14 | 2x, 4x | 2A, 1B, 2B | Meng et al. (2014) |
| A. deserti | 56 | 14 | 4x | 1B | Meng et al. (2010) |
| A. flavescens | 28 | 14 | 2x | 2A | Meng et al. (2014) |
| A. latialata | 56 | 14 | 4x | 2B | Meng et al. (2014) |
| A. likiangensis | 56 | 14 | 4x | 2B | Meng et al. (2014) |
| A. margaritacea | 28, 56 | 14 | 2x, 4x | 1B, 2B | Meng et al. (2010) |
| A. nepalensis var. corymbosa | 28, 84 | 14 | 2x, 6x | 2A, 2B | Meng et al. (2014) |
| A. nepalensis var. nepalensis | 28, 56 | 14 | 2x, 4x | 2A, 2B | Meng et al. (2014) |
| A. pachylaena | 28 | 14 | 2x | 2A | Meng et al. (2014) |
| A. pannosa | 56 | 14 | 4x | 2A | Meng et al. (2014) |
| A. plicata | 56 | 14 | 4x | 2B | Meng et al. (2010) |
| A. rhododactyla | 56 | 14 | 4x | 2B | Meng et al. (2010) |
| A. royleana | 28 | 14 | 2x | 2B | Meng et al. (2010) |
| A. spodiophylla | 28 | 14 | 2x | 2A | Present study |
| A. stenocephala | 56 | 14 | 4x | 2A, 2B | Meng et al. (2014) |
| A. surculosa | 56 | 14 | 4x | 2A | Meng et al. (2014) |
| A. virens | 56 | 14 | 4x | 2B | Meng et al. (2014) |
| A. xylorhiza | 28 | 14 | 2x | 2A | Present study |
| A. yunnanensis | 56 | 14 | 4x | 2B | Meng et al. (2014) |
| Aster albescens var. glabratus | 18 | 9 | 2x | 2A | Meng et al. (2016) |
| A. albescens var. gracilior | 18 | 9 | 2x | 2A | Meng et al. (2016) |
| A. altaicus var. hirsutus | 18 | 9 | 2x | 1A | Meng et al. (2016) |
| A. auriculatus | 18 | 9 | 2x | 1A | Meng et al. (2016) |
| A. crenatifolius | 18 | 9 | 2x | 1A | Chen et al. (2010) and Meng et al. (2016) |
| A. diplostephioides | 18 | 9 | 2x | Liu (1999) | |
| A. gouldii | 18 | 9 | 2x | 1A | Present study |
| A. oreophilus | 18 | 9 | 2x | 1A | Meng et al. (2016) |
| A. pekinensis | 18 | 9 | 2x | 1A | Present study |
| A. poliothamnus | 18 | 9 | 2x | 1A | Meng et al. (2016) |
| A. pycnophyllus | 18, 36 | 9 | 2x, 4x | 1A, 2A | Meng et al. (2016) |
| A. salwinensis | 18 | 9 | 2x | 1A | Meng et al. (2016) |
| A. souliei | 18, 54 | 9 | 2x, 6x | 2A | Liu (1999) and Meng et al. (2016) |
| A. techinensis | 18 | 9 | 2x | 1A | Meng et al. (2016) |
| A. trinervius subsp. ageratoides | 72 | 9 | 8x | 2A | Meng et al. (2016) |
| A. yunnanensis var. labrangensis | 18 | 9 | 2x | Liu (1999) | |
| A. yunnanensis var. yunnanensis | 18 | 9 | 2x | 2A | Meng et al. (2016) |
| Carpesium cernuum | 40 | 20 | 2x | 2B | Present study |
| C. lipskyi | 40 | 20 | 2x | 2B | Present study |
| C. scapiforme | 40 | 20 | 2x | 2B | Present study |
| C. velutinum | 40 | 20 | 2x | 2B | Present study |
| Cremanthodium angustifolium | 58 | 29 | 2x | 2A | Present study |
| C. brunneo-pilosum | 58 | 29 | 2x | 2A | Liu et al. (2001) |
| C. campanulatum | 58 | 29 | 2x | 2A | Present study |
| C. discoideum | 58 | 29 | 2x | 2A | Liu et al. (2001) |
| C. ellisii | 58 | 29 | 2x | 2A | Liu et al. (2001) |
| C. helianthus | 58 | 29 | 2x | 2A | Present study |
| C. humile | 58, 60 | 29, 30 | 2x | 2A | Liu et al. (2001) |
| C. lineare | 58 | 29 | 2x | 2A | Liu et al. (2001) |
| C. lineare var. eligulatum | 58 | 29 | 2x | 2A | Present study |
| C. microglossum | 58 | 29 | 2x | 2A | Liu et al. (2001) |
| C. principis | 58 | 29 | 2x | 2A | Present study |
| C. stenoglossum | 58 | 29 | 2x | 2A | Liu et al. (2001) |
| Dubyaea glaucescens | 34, 51 | 17 | 2x, 3x | Liu et al. (2014) | |
| D. tsarongensis | 16 | 8 | 2x | 1A | Present study |
| Leontopodium artemisiifolium | 26 | 13 | 2x | Russell et al. (2013) | |
| L. dedeckensii | 26 | 13 | 2x | Russell et al. (2013) | |
| L. himalayanum | 24 | 12 | 2x | Russell et al. (2013) | |
| L. sinense | 48 | 12 | 4x | Russell et al. (2013) | |
| L. souliei | 48 | 12 | 4x | 2B | Present study |
| L. stracheyi | 26 | 13 | 2x | Russell et al. (2013) | |
| Myriactis nepalensis | 36 | 18 | 2x | 2A | Present study |
| M. wightii | 36 | 18 | 2x | 2A | Present study |
| Pertya berberidoides | 32 | 16 | 2x | 1B | Chen et al. (2013) |
| P. phylicoides | 34 | 17 | 2x | 1B | Present study |
| Syncalathium chrysocephalum | 16 | 8 | 2x | 1A | Zhang et al. (2009c) |
| S. disciforme | 16 | 8 | 2x | 1A | Zhang et al. (2009c) |
| S. kawaguchii | 16 | 8 | 2x | 1A | Zhang et al. (2009c) |
| S. roseum | 16 | 8 | 2x | 1A | Present study |
From the survey data of Asteraceae species from the Hengduan Mountains, patterns and trends in ploidy level can be seen: (1) Five ploidy levels (x) have been recorded, including 2x (n = 57, 73.08%), 3x (n = 1, 1.28%), 4x (n = 17, 21.79%), 6x (n = 2, 2.56%), and 8x (n = 1, 1.28%). (2) The majority of genera were diploid, while a notable number were tetraploid. A number of Asteraceae species in the HDM had multiple haploid chromosome numbers (n). About four types of n with the largest numbers as 9 (16 species, 20.51%), 14 (10 species, 12.82%), 28 (14 species, 17.95%) and 29 (12 species, 15.38%), respectively; (3) Following Lewis (1980), which defines paleopolyploidy as n ≥ 11, we found that 46.15% (36/78) of Asteraceae species in the HDM could be classified as paleopolyploids. Somatic chromosome number (2n) of the recorded Asteraceae species showed more than 17 patterns of 2n types in HDM. The major patterns of 2n included 18 (16 species, 20.51%), 28 (10 species, 12.82%), 56 (14 species, 17.95%) and 58 (12 species, 15.38%).
3. DiscussionIn this study, we examined the karyotypes of 19 species of Asteraceae from the Hengduan Mountains. We also surveyed previously reported chromosome numbers of congeneric species from the HDM and adjacent regions.
The genus Cremanthodium consists of 69 identified species occurring primarily in the Himalayas-Hengduan Mountains region and all species (46 endemics) can be found in China (Shi et al., 2011). This genus is the largest endemic genus of the Himalayas (Zhang et al., 2009b). All the species of this genus grow almost exclusively on alpine scree or meadow areas at an altitude ranging from 3000 to 5500 m (Shi et al., 2011). Previous studies have shown that Cremanthodium species have one of two chromosome numbers, the most common being 2n = 58, with some species 2n = 60 (Liu et al., 2001). In this study, we examined the karyotypes of five species of Cremanthodium and all of which were examined for the first time. Our results indicate that the basic chromosome numbers are x = 29 and 2n = 58, which is similar to Liu et al. (2004). Combined with the molecular evidence, our chromosome numbers support the treatment of Ligularia-Cremanthodium-Parasenecio as a complex (Liu et al., 2006). Chromosome numbers varied from 2n = 48, 58, 60 in this complex of species (Liu, 2004). The evolution of chromosome number in this genus may represent a good case study to explore the radiation and diversification of species within the uplift of the QinghaiTibet Plateau (Liu et al., 2006).
The genus Leontopodium has about 58 taxa in Eurasia, and 37 species (17 endemics) in China (Shi et al., 2011). Leontopodium souliei is endemic to the Himalayas-Hengduan Mountains region and it grows naturally in grasslands or thickets at altitudes mostly ranging from 2700 to 4500 m (Shi et al., 2011). The previously reported chromosome number of L. souliei (Zuogong, Tibet) is 2n = 4x = 52 (Meng et al., 2012). In our study, L. souliei was collected from Mangkang (Tibet, China) and the karyotype was formulated as 2n = 4x = 48 = 36m + 12sm + 0/2/3/4B. Meng et al.(2012)suggested that the basic chromosome number of Leontopodium is x = 12, 13, 14. The unstable B chromosome range from 0 to 4 differs from Meng et al. (2012). This phenomenon may be due to the different basic chromosome number of Leontopodium with different ploidy in different populations.
The genus Aster is one of the largest genera of Asteraceae. This genus includes about 152 species in the Northern Hemisphere, 123 species (82 endemics) are distributed in China (Shi et al., 2011). A. pekinensis is widely distributed in China and mainly grows in forest margins, thickets, mountain slopes, riverbanks or roadsides from sea level to 1600 m (Shi et al., 2011). Previous studies have shown that A. pekinensis has two chromosome numbers, 2n = 2x = 16 and 2n = 4x = 32 (Gu, 1989). Additional studies have reported that the most common chromosome number in the genus is x = 9 (Meng et al., 2016; Liu, 1999). Our results show that A. pekinensis has the karyotype formula 2n = 2x = 18 = 18m. The two populations of A. gouldii had similar karyotypes, 2n = 2x = 18 = 18m. This is the first report of the karyotype morphology for this species.
Anaphalis is one of the largest genera of Asteraceae and is the most diverse in the Himalayas-Hengduan Mountains region. It comprises about 110 species with 54 species (40 endemics) in China (Shi et al., 2011). More than half of the species occur in the Hengduan Mountains. Anaphalis xylorhiza is widely distributed in alpine grasslands and lichen-covered areas at altitudes from 3800 to 4000 m (Shi et al., 2011). Previous cytology of this species collected from Rikaze (Tibet) showed the formula as 2n = 2x = 28 = 2M + 14m + 12sm (Meng et al., 2010). In this study, the sample from Ganzi (Sichuan) was formulated as 2n = 2x = 28 = 16m + 12sm. A. spodiophylla is endemic to the Hengduan Mountains region and restricted mainly to sunny roadsides at altitudes mostly ranging from 3000 to 3800 m (Shi et al., 2011). Here, we show that the karyotype formula of A. spodiophylla is 2n = 2x = 28 = 16m + 12sm. This is the first report of the karyotype and chromosome number of A. spodiophylla and it is similar to A. xylorhiza.
The genus Myriactis comprises 12 to 16 species, five species (one endemic) in China (Shi et al., 2011). The basic chromosome number of the genus Myriactis has been reported x = 18 and 2n = 36 (Razaq et al., 1994). M. nepalensis is widely distributed in moist or humid areas at altitudes from 700 to 3700 m (Shi et al., 2011). Our results show that the karyotype formula of the M. nepalensis is 2n = 2x = 36 = 32m + 4sm. Previous chromosome number data for M. nepalensis was reported as 2n = 2x = 36 (Gupta et al., 1989). We found a diploid population of M. nepalensis. M. wightii is endemic to the Hengduan Mountains region and is distributed on slopes, mixed forests, grasslands or stream sides at 1900-3600 m (Shi et al., 2011). Our results show that the karyotype formula of M. wightii is 2n = 2x = 36 = 32m + 4sm. The karyotype and chromosome numbers of M. wightii are reported here for the first time and are similar to M. nepalensis.
The genus Syncalathium is a small genus with five identified species endemic to alpine scree of the Sino-Himalayan region, all found in China (Shi et al., 2011; Zhang et al., 2011). All species are restricted mainly to altitudes ranging from 3700 to 5400 m (Shi et al., 2011). It is most likely that 2n = 16 was a common chromosome number in the genus (Zhang et al., 2007, 2009c). To our knowledge, this study is the first to examine karyotype and chromosome number data for S. roseum, a species restricted to sandy riverbanks at altitudes from 3700 to 3800 m (Xizang, Zhanang) (Shi et al., 2011). We collected seeds of S. roseum from Zhanang at the type locality. The karyotype formula of S. roseum is 2n = 2x = 16 = 14m + 2sm(2sat).
The genus Pertya has about 25 species in Eurasia, and 17 species (16 endemics) in China (Shi et al., 2011). P. phylicoides is endemic to the Hengduan Mountains regions and is distributed on dry-hot valley at 2400-3100 m (Shi et al., 2011). In this study, the karyotype of P. phylicoides was formulated as 2n = 2x = 34 = 30m + 4sm. A previous study reported that the karyotype formula for Pertya berberidoides in this area was 2n = 2x = 32 = 28m + 4sm (Chen et al., 2013). These two competing results suggest that Pertya may be an aneuploid or experience unstable structural changes in chromosomes.
The genus Dubyaea has about 15 species, 12 species (8 endemics) in China, with the endemics distributed to the QinghaiTibetan Plateau and adjacent regions (Shi et al., 2011). Our results show that the karyotype formula of the D. tsarongensis is 2n = 2x = 16 = 14m + 2sm. Liu and Yang (2014) reported the cytology of Dubyaea glaucescens with two chromosome numbers, 2n = 34 (diploid) and 2n = 51 (triploid), based on x = 17. The same study transferred D. glaucescens to Faberia after karyological data corroborated evidence from morphology, habitat preference and geographic distribution transferred of D. glaucescens to Faberia from strongly corroborated with karyological characters (). It is most likely that 2n = 16 was the basic chromosome number in this genus (Babcock et al., 1937). Here we show the karyotype and chromosome number of D. tsarongensis for the first time.
The genus Carpesium contains about 20 species in Asia and Europe, 16 (6 endemics) in China (Shi et al., 2011). In this study, we report the karyotype of four species of Carpesium, three for the first time (C. lipskyi, C. scapiforme, C. velutinum). Two populations of C. cernuum (from Mianning and Lijiang) were sampled and the results show that they have the same formula 2n = 2x = 40 = 26m + 10sm(2sat) + 4st. The chromosome number of C. cernuum was previously reported as 2n = 2x = 40 = 24m + 12sm + 2T (Wang et al., 1999). The two populations sampled in our study had similar karyotypes and karyotype morphology as previous reports of C. cernuum.
Polyploidy has long been recognized as an important evolutionary force in plants (Grant, 1981; Jiao et al., 2011; Levin, 2002; Stebbins, 1940, 1950, 1971), especially in Asteraceae, one of the largest angiosperm families (Semple and Watanabe, 2009). Polyploidy has been reported in ca. 570 genera of the family (58.3% of the 978 genera with chromosome counts), and ploidy levels have been found to vary from 2x to 48x (Semple and Watanabe, 2009). Recently, Huang et al. (2016) found that whole-genome duplications (WGDs) in Asteraceae were related to environmental changes and species radiations, providing a possible scenario for polyploids to overcome the disadvantages of WGDs and to evolve into lineages with high biodiversity. The diversity of chromosome evolution patterns in Asteraceae from the HDM may be related to the high level of species richness and endemism. Our findings suggest that polyploidy may not play an important role in how plants from this region have adapted to the environment, although statistical analysis of chromosome number in Asteraceae species indicates that the number of paleopolyploids is large (46.15%). Further studies are needed to discover the relationship between genome polyploidy and genome evolution in Asteraceae and more detailed research in some groups may provide more information about the mechanisms of polyploidization and speciation. Undertaking a full-scale investigation of chromosome databases may potentially reveal the relationship between ploidy level and the evolution of species diversity in the Hengduan Mountains, SW China.
Author contributionsZML conceived and designed the experiments. WGS and FMS collected the data and performed the experiments. WGS and XGM analyzed the data. WGS and XGM wrote the manuscript. JWZ, YHZ and ZML revised the manuscript.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (31670206, 31360049) to Zhi-Min Li, and major Program of NSFC (grant 31590823, 31590820) to Hang Sun, and NSFC (31370004, 31570213) to Jian-Wen Zhang. We gratitude to the Alpine Plant Diversity Group of KIB for help during field trips, we also thank Hui-Xian Yang, Xu-Rui Zhao, Yun Xu, Hui-Ying Li, Ping-Fu Yang and Yan Xu for their help with the experiments.
Altınordu F., Peruzzi L., Yu Y., He X.J., 2016. A tool for the analysis of chromosomes:KaryoType. Taxon, 65: 586-592. DOI:10.12705/653.9 |
Arano H., 1963. Cytological studies in subfamily Carduoideae (Compositae) of Japan Ⅸ. Bot. Mag., 76: 32-39. DOI:10.15281/jplantres1887.76.32 |
Babcock E.B., Stebbins G.L., Jenkins J.A., 1937. Chromosomes and phylogeny in some genera of the Crepidinae. Cytologia: 188-210. |
Chen G., Sun W.B., 2006. Karyotype of four species in Buddleja (Loganiaceae). Acta Bot. Yunnan, 28: 37-40. |
Chen G.F., Ba L.J., Sun W.G., Lou X., Zhang J.W., Li Z.M., 2013. Karyotypes and chromosome numbers of eight species from the family Asteraceae in the Hengduan Mountains and the adjacent regions. Plant Divers. Resour., 35: 367-374. |
Chen G.F., Sun W.G., Hong D.Y., Zhou Z., Niu Y., Nie Z.L., Sun H., Zhang J.W., Li Z.M., 2014. Systematic significance of cytology in Cyananthus (Campanulaceae) endemic to the Sino-Himalayan region. J. Syst. Evol., 52: 260-270. DOI:10.1111/jse.v52.3 |
Chen J.G., Xu B., Li Z.M., Sun H., 2010. Karyological Studies on Two Species of Compositae from the Hengduan Mountains, SW China. Guihaia, 1: 009. |
Grant, V., 1981. Plant Speciation. Columbia University Press, New York xii, 563pp. -illus., maps, chrom. nos. En 2nd edition. Maps, Chromosome numbers. General(KR, 198300748).
|
Gu H.Y., 1989. On chromosome numbers of Kalimeris (Astereae, Asteraceae) and some related taxa. Cathaya, 1: 1-16. |
Gupta, R. C., Gill, B. S., Garg, R. K., 1989. Chromosomal Conspectus of Western Himalayan Compositae. Plant Science Research in India: Present Status and Future Challenges. Part 1.
|
Huang C.H., Zhang C.F., Liu M., Hu Y., Gao T.G., Qi J., Ma H., 2016. Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Mol. Biol. Evol., 33: 2820-2835. DOI:10.1093/molbev/msw157 |
Jiao Y.N., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., 2011. Ancestral polyploidy in seed plants and angiosperms. Nature, 473: 97-100. DOI:10.1038/nature09916 |
Levan A., Fredga K., Sandberg A.A., 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52: 201-220. |
Levin, D. A., 2002. The Role of Chromosomal Change in Plant Evolution. Oxford University Press on Demand.
|
Lewis, W. H., 1980. Polyploidy in Angiosperms: Dicotyledons. Springer, Polyploidy, pp. 241-268.
|
Li B.Y., 1987. On the boundaries of the Hengduan mountains. Mt. Res., 5: 74-82. |
Li X.W., 1994. Two big biodiversity centres of Chinese endemic genera of seed plants and their characteristics in Yunnan Province. Acta Bot. Yunnan, 16: 221-227. |
Li X.W., Li J., 1993. A preliminary floristic study on the seed plants from the region of Hengduan Mountain. Acta Bot. Yunnan, 15: 217-231. |
Li Z.W., Chen Z.R., Wang M.L., 1991. Classification and correlation of the quaternary glacial epoch in the Hengduan (Transverse) mountains. Geol. Rev., 37: 125-132. |
Liu J.Q., 1999. Karomorphological characteristics of three Aster species from southern Qing Hai. Bull. Bot. Res., 19(4): 392-396. |
Liu J.Q., 2004. Uniformity of karyotypes in Ligularia (Asteraceae:Senecioneae), a highly diversified genus of the eastern QinghaieTibet Plateau highlands and adjacent areas. Bot. J. Linn. Soc., 144: 329-342. DOI:10.1111/j.1095-8339.2003.00225.x |
Liu J.Q., Liu S.W., Ho T.N., Lu A.M., 2001. Karyological studies on the SinoHimalayan genus, Cremanthodium (Asteraceae:Senecioneae). Bot. J. Linn. Soc., 135: 107-112. DOI:10.1111/boj.2001.135.issue-2 |
Liu J.Q., Wang Y.J., Wang A.L., Hideaki O., Abbott R.J., 2006. Radiation and diversification within the Ligularia-Cremanthodium-Parasenecio complex(Asteraceae) triggered by uplift of the Qinghai-Tibetan plateau. Mol. Phylogen.Evol., 38: 31-49. DOI:10.1016/j.ympev.2005.09.010 |
Liu Y., Yang Q.E., 2014. Cytotaxonomy of Dubyaea glaucescens (Composita-Cichorieae). Nord. J. Bot., 32: 871-874. DOI:10.1111/njb.2014.v37.i6 |
Meng Y., Nie Z.L., Sun H., Yang Y.P., 2012. Chromosome numbers and polyploidy in Leontopodium (Asteraceae:Gnaphalieae) from the Qinghai-Tibet plateau of SW China. Caryologia, 65: 87-93. DOI:10.1080/00087114.2012.709779 |
Meng Y., Sun H., Yang Y.P., Nie Z.L., 2010. Polyploidy and new chromosome counts in Anaphalis (Asteraceae:Gnaphalieae) from the Qinghai-Tibet plateau of China. J. Syst. Evol., 48: 58-64. DOI:10.1111/jse.2010.48.issue-1 |
Meng Y., Yang Y.P., Deng T., Nie Z.L., 2016. New chromosome counts, polyploidy, and karyotype evolution in Aster L. (Asteraceae:Astereae) from the QinghaiTibet plateau. Caryologia, 69: 370-378. |
Meng Y., Yang Y.P., Sun H., Deng T., Nie Z.L., 2014. Chromosome numbers, karyotypes, and polyploidy evolution of Anaphalis species (Asteraceae:Gnaphalieae) from the Hengduan mountains, SW China. Caryologia, 67: 238-249. DOI:10.1080/0144235X.2014.974352 |
Myers N., Mittermeier R.A., Mittermeier C.G., Da Fonseca G.A., Kent J., 2000. Biodiversity hotspots for conservation priorities. Nature, 403: 853-858. DOI:10.1038/35002501 |
Nie Z.L., Wen J., Gu Z.J., Boufford D.E., Sun H., 2005. Polyploidy in the flora of the Hengduan Mountains hotspot, southwestern China. Ann. Mo. Bot. Gard., 92: 275-306. |
Olson D.M., Dinerstein E., 2002. The Global 200:priority ecoregions for global conservation. Ann. Mo. Bot. Gard., 89: 199-224. DOI:10.2307/3298564 |
Paszko B., 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst. Evol., 258: 39-48. DOI:10.1007/s00606-005-0389-2 |
Peruzzi L., Altınordu F., 2014. A proposal for a multivariate quantitative approach to infer karyological relationships among taxa. Comp. Cytogen., 8: 337. DOI:10.3897/CompCytogen.v8i4.8564 |
Peruzzi L., Eroğlu H.E., 2013. Karyotype asymmetry:again how to measure and what to measure?. Comp. Cytogen., 7(1): 1-9. DOI:10.3897/compcytogen.v7i1.4431 |
Peruzzi L., Leitch I.J., Caparelli K.F., 2009. Chromosome diversity and evolution in Liliaceae. Ann. Bot., 103: 459-475. DOI:10.1093/aob/mcn230 |
Razaq Z.A., Vahidy A.A., Ali S., 1994. Chromosome numbers in Compositae from Pakistan. Ann. Mo. Bot. Gard., 81: 800-808. DOI:10.2307/2399925 |
Russell A., Safer S., Weiss-Schneeweiss H., Temsch E., Stuppner H., Stuessy T.F., Samuel R., 2013. Chromosome counts and genome size of Leontopodium species(Asteraceae:Gnaphalieae) from south-western China. Bot. J. Linn. Soc., 171: 627-636. DOI:10.1111/boj.2013.171.issue-3 |
Semple, J. C., Watanabe, K., 2009. In: Funk, V. A., Susanna, A., Stuessy, T. F., Bayer, R. J. (Eds. ), A Review of Chromosome Numbers in Asteraceae with Hypotheses on Chromosomal Base Number Evolution. Systematics, Evolution, and Biography of Compositae. IAPT, Vienna, pp. 61-72.
|
Shi, Z., Chen, Y. L., Chen, Y. S., Lin, Y. R., Liu, S. W., Ge, X. J., Gao, T. G., Zhu, S. X., Liu, Y., Christopher, J. H., Yang, Q., Raab-Straube, E. V., Gilbert, M. G., Nordenstam, B., Kilian, N., Brouillet, L., Illarionova, I. D., Hind, D. J. N., Jeffrey, C., Bayer, R. J., Kirschner, J., Greuter, W., Anderberg, A. A., Semple, J. C., Stěpáek, J., Freire, S. E., Martins, L., Koyama, H., Kawahara, T., Vincent, L., Sukhorukov, A. P., Mavrodiev, E. V., Gottschlich, G., 2011. Astereae. In: Wu, Z. Y., Raven, P. H., Hong, D. Y. (Eds. ), Flora of China, pp. 545-652. Science Press (Beijing) & Missouri Botanical Garden Press (St. Louis).
|
Stebbins G.L., 1940. The significance of polyploidy in plant evolution. Am. Nat., 74: 54-66. DOI:10.1086/280872 |
Stebbins G.L., 1950. Variation and Evolution in Plants. New York: Columbia University Press.
|
Stebbins G.L., 1971. Chromosomal Evolution in Higher Plants. London: Addison-Wesley.
|
Sun H., 2002. Evolution of arctic-tertiary flora in Himalayan-Hengduan mountains. Acta Bot. Yunnan, 24: 671-688. |
Wang B., Yang H.L., Zheng T.S., Yen H.B., Xu X.J., 1999. Karyotype analysis of Carpesium cernuum. Zhong Yao Cai, 22: 163-165. |
Wang, W. T., 1994. Vascular Plants of the Hengduan Mountains, vol. 2. Science Press, Beijing.
|
Wang X., Liu B.B., Ma Y.Z., Xie P.H., He X.Y., Shang B.L., Wang Y.J., 2013. Chromosomal studies on the alpine genus Dolomiaea (Asteraceae:Cardueae) from the Qinghai-Tibet plateau and adjacent regions. Caryologia, 66: 186-193. DOI:10.1080/00087114.2013.823296 |
Wang Y.J., Liu J.Q., 2004. A preliminary investigation on the phylogeny of Saussurea(Asteraceae:Cardueae) based on chloroplast DNA trn LF sequences. Acta Phytotax. Sin., 42: 136-153. |
Wu Z.Y., 1988. Hengduan mountain flora and her significance. J. Jap. Bot., 63: 297-311. |
Xu B., Li Z.M., Sun H., 2014a. Plant diversity and floristic characters of the alpine subnival belt flora in the Hengduan Mountains, SW China. J. Syst. Evol., 52: 271-279. DOI:10.1111/jse.v52.3 |
Xu B., Li Z.M., Sun H., 2014b. Seed Plants of the Alpine Subnival Belt from the Hengduan Mountains, SW China. Beijing: Science Press.
|
Yuan Q., Yang Q.E., 2006. Polyploidy in Aconitum subgenus Lycoctonum (Ranunculaceae). Bot. J. Linn. Soc., 150: 343-353. DOI:10.1111/j.1095-8339.2006.00468.x |
Yuan Q., Yang Q.E., 2008. Low incidence of polyploids and high uniformity of karyotypes displayed by Delphinium (Ranunculaceae) in the Hengduan Mountains region of south-west China. Bot. J. Linn. Soc., 158: 172-188. DOI:10.1111/boj.2008.158.issue-1 |
Zhang D.C., Boufford D.E., Ree R.H., Sun H., 2009a. The 29°N latitudinal line:an important division in the Hengduan mountains a biodiversity hotspot in southwest China. Nord. J. Bot., 27: 405-412. DOI:10.1111/njb.2009.27.issue-5 |
Zhang D.C., Zhang Y.H., Boufford D.E., Sun H., 2009b. Elevational patterns of species richness and endemism for some important taxa in the Hengduan Mountains, southwestern China. Biodivers. Conserv., 18: 699-716. DOI:10.1007/s10531-008-9534-x |
Zhang J.W., Nie Z.L., Sun H., 2009c. Cytological study on the genus Syncalathium(Asteraceae-Lactuceae), an endemic taxon to alpine scree of the Sino-Himalayas. J. Syst. Evol., 47: 226-230. DOI:10.1111/jse.2009.47.issue-3 |
Zhang J.W., Nie Z.L., Wen J., Sun H., 2011. Molecular phylogeny and biogeography of three closely related genera, Soroseris, Stebbinsia, and Syncalathium(Asteraceae, Cichorieae), endemic to the Tibetan plateau, SW China. Taxon, 60: 15-26. |
Zhang J.W., Sun H., Nie Z.L., 2007. Karyological studies on the Sino-Himalayan endemic Soroseris and two related genera of tribe Lactuceae (Asteraceae). Bot.J. Linn. Soc., 154: 79-87. DOI:10.1111/j.1095-8339.2007.00644.x |



