b. Plant Germplasm and Genomics Center, The Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China;
c. University of the Chinese Academy of Sciences, Beijing 100049, China
Sugars provide carbon skeletons, energy, and act as signal molecules for plants during seed germination, seedling growth and development, flowering, fructification, and senescence (Rolland et al., 2006). Sucrose, an important disaccharide combination of the monosaccharides glucose and fructose, is the major intermediate product of photosynthesis in source tissues. Plants utilize a small part of the product of photosynthesis for source tissue development, and transfer the remaining carbohydrates to plant sink organs through the phloem in the form of sucrose (Aoki et al., 2003; Gong et al., 2015; Kuhn and Grof, 2010; Lemoine, 2000). Clearly, sucrose transport plays a pivotal role in the growth and development of plant organs.
Sucrose transporters (SUTs), or sucrose carriers (SUCs), are a super family belonging to the major facilitator superfamily. These proteins contain several hydrophobic regions that can be assigned to 12 membrane-spanning regions (Kuhn and Grof, 2010). Since Riesmeier isolated the first SUC gene from spinach using yeast complementation strategy in 1992 (Riesmeier et al., 1992), more sucrose transporters have been identified in a variety of plant species, such as Arabidopsis (Sauer et al., 2004), rice (Aoki et al., 2003), Theobroma cacao (Li et al., 2014), maize (Aoki et al., 1999), Brassica napus (Jian et al., 2016), and form a supergene family (Kuhn and Grof, 2010). These SUC genes can be divided into five clades (SUT1eSUT5) in a phylogenetic tree constructed by Kuhn and Grof (2010) using 62 plant sucrose transporter sequences and the genes. SUT1 clade was dicotyledon-specific, whereas clades SUT3 and SUT5 were monocotyledon-specific. SUT2 and SUT4 contained both dicotyledon and monocotyledon plant species (Kuhn and Grof, 2010).
Turnip (Brassica rapa var. rapa) is a cruciferous biennial plant belonging to B. rapa, together with B. rapa pekinensis and B. campestris L. Only the turnip is able to form a turgor taproot. The taproot, which is the principal and valuable storage organ of turnip, contains rich nutrient substances, such as vitamin C, vitamin K, calcium ions, and is fit for human consumption or for livestock feed. Recent studies have also found that the turnip possesses antihypoxic activity and can be used to prevent altitude stress (Chu et al., 2017; Ogawa et al., 2007; Xie et al., 2010). Therefore, understanding the mechanism of the taproot formation and improving the yield and quality of the turnip taproot are very important for the full use of the taproot.
In this paper, we focused on identifying SUC genes from turnip and analyzing their physicochemical properties and evolutionary status according to the deduced amino acid sequence. The expression patterns of BrrSUC genes in different tissues and at early stages of taproot formation were also analyzed. Our research provides gene resources, further insights on the mechanism of taproot formation, and potential clues for improving the yield and quality of the turnip taproot.
2. Materials and methods 2.1. Plant materialsSeeds of B. rapa var. rapa (KTRG-B17, Lhasa Tibet, China) were sown in plastic pots containing a mixture of perlite and organic soil mixture once a week at a fixed time for four weeks. The pots were placed in the field to ensure that turnips grew naturally. The hypocotyls from the five 7-, 14-, 21-, and 28-day-old turnip seedlings were collected, respectively. In addition, root, hypocotyl and leaf tissue, along with flowering organs, were harvested from five 21-day-old turnip seedlings. The samples were frozen in liquid nitrogen.
2.2. RNA extraction and cDNA preparationThe samples were ground in liquid nitrogen and the total RNA was extracted using the Eastep® Super Total RNA Extraction Kit according to the manufacturer's instructions (Promega, Madison, WI, USA). The quality and concentration of RNA were assessed by 0.8% agarose gel electrophoresis and NanoDrop1000 spectrophotometer. The firststrand cDNA was synthesized using 1-μg RNA and GoScript™ Reverse Transcriptase (Promega, Madison, WI, USA) in 20-μL reaction volumes according to the manufacturer's instructions.
2.3. Molecular cloning of SUC genes of turnipTwelve SUC gene sequences of B. rapa (BraSUCs) were searched from the Brassica database (http://brassicadb.org/brad/). Primers for the amplification of the full-length coding DNA sequence (CDS) of the turnip SUCs were designed by Oligo7 software based on the sequences of BraSUCs. The amplification primer sequences are listed in supplemental Table 1. Polymerase chain reaction (PCR) was performed in a 50-μL reaction mixture containing 2 μg cDNA and 1 μL Phanta Super-Fidelity DNA Polymerase (Vazyme Biotech Co., China, Nanjing) under the following PCR conditions: 3 min at 95 ℃ for pre-denaturation, 35 cycles of amplification (30 s at 95 ℃, 30 s at 56 ℃, and 90 s at 72 ℃), and a final extension at 72 ℃ for 7 min. A 1% agarose gel electrophoresis staining was used to confirm the size of the genes, and the EasyPure® Quick Gel Extraction kit was used for the purification and recovery of DNA from the agarose gel. The PCR products of the purification were cloned into the PMD18-T vectors (TaKaRa Biotechnology, Dalian, China) and sequenced by TsingKe (Tsingke Biological Technology Co., Beijing, China).
| Gene | CDS (bp) | MWKDa | pI | GRAVY | Instability index (%) |
| BrrSUCl.1 | 1544 | 55.09 | 9.34 | 0.497 | 34.63 |
| BrrSUC1.2 | 1527 | 54.20 | 9.14 | 0.547 | 37.05 |
| BrrSUC1.3 | 1542 | 54.66 | 9.27 | 0.556 | 36.72 |
| BrrSUC1.4 | 1538 | 54.91 | 9.38 | 0.472 | 60.26 |
| BrrSUC2.1 | 1542 | 54.54 | 9.20 | 0.477 | 29.86 |
| BrrSUC2.2 | 1527 | 54.02 | 9.19 | 0.468 | 28.99 |
| BrrSUC3.1 | 1775 | 63.23 | 5.82 | 0.376 | 38.84 |
| BrrSUC3.2 | 1719 | 61.09 | 6.15 | 0.432 | 34.66 |
| BrrSUC4.1 | 1506 | 53.68 | 9.27 | 0.475 | 38.66 |
| BrrSUC4.2 | 1524 | 54.37 | 9.40 | 0.524 | 35.25 |
| BrrSUC6.1 | 1434 | 50.91 | 8.84 | 0.622 | 33.31 |
| BrrSUC6.2 | 1480 | 52.79 | 9.21 | 0.589 | 31.71 |
BraSUCs were identified by performing a protein BLAST (BLASTP) analysis with the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) at a cut-off value of < E-20 using the Arabidopsis sucrose transporter amino acid sequences as the query sequences, respectively. After merging the sequence of genes, we performed a BLASTP analysis again using these BraSUCs as the query sequence, finally obtaining 12 BraSUC genes.
The cDNA of BrrSUCs were cloned and verified by sequencing, and were translated into amino acid sequences through DNAMAN software. The molecular mass calculation and physical and chemical properties of the predicted proteins were performed on the ExPASy-ProtParam online tool (http://expasy.org/tools). The conserved motifs of predicted proteins were analyzed by the MEME Suite (http://meme-suite.org/). The deduced amino acid sequences of the 12 BrrSUCs and 47 SUCs gene sequences from seven other plant species searched and obtained from GenBank were aligned using ClustalW. The phylogenetic tree was constructed using the neighbor-joining method (1000 bootstrap replicates) with the MEGA 7.0 software. FigTree1.4.0 software was used to adjust the phylogenetic tree.
2.5. Real-time quantitative PCRThe primers for real-time quantitative PCR (qRT-PCR) were designed on NCBI's Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized (Generay Biotech, Shanghai, China). The sequences of qRT-PCR primers are listed in supplemental Table 2. The cDNAs of four different tissues and four developmental phases were used as templates for qRT-PCR. A 20-μL qRT-PCR reaction volume containing 50 ng cDNA and FastStart Universal SYBR Green Master Mix (ROX) was prepared. Then, the qRT-PCR was performed on an Applied Biosystems 7500 Fast RealTime PCR System according to the manufacturer's instructions. Three independent biological replicates were performed for each sample and β-tubulin was chosen as a loading control. The qRT-PCR results were analyzed according to the 2-△△CT method (Schmittgen and Livak, 2008).
| Similarity | |||||||||||||
| BrrSUC1.1 | BrrSUC1.2 | BrrSUC1.3 | BrrSUC1.4 | BrrSUC2.1 | BrrSUC2.2 | BrrSUC3.1 | BrrSUC3.2 | BrrSUC4.1 | BrrSUC4.2 | BrrSUC6.1 | BrrSUC6.2 | ||
| MM(kDa) | 55.09 | 54.20 | 54.66 | 54.91 | 54.54 | 54.02 | 63.23 | 61.09 | 53.68 | 54.37 | 50.91 | 52.79 | |
| Identify | BrrSUCl.1 | 一 | 81.2 | 80.0 | 77.6 | 71.6 | 73.1 | 51.0 | 52.0 | 57.4 | 56.4 | 77.0 | 74.6 |
| BrrSUC1.2 | 78.0 | 一 | 95.3 | 89.4 | 86.3 | 87.3 | 56.6 | 60.3 | 62.9 | 59.9 | 85.0 | 84.0 | |
| BrrSUC1.3 | 74.8 | 90.7 | 一 | 91.1 | 88.4 | 87.6 | 55.7 | 57.4 | 63.1 | 60.3 | 85.0 | 83.7 | |
| BrrSUC1.4 | 71.0 | 83.8 | 83.7 | 一 | 83.7 | 83.1 | 55.4 | 56.8 | 65.0 | 63.1 | 84.0 | 82.7 | |
| BrrSUC2.1 | 62.4 | 76.4 | 76.9 | 74.4 | 一 | 97.2 | 58.6 | 60.0 | 62.7 | 60.5 | 82.3 | 81.3 | |
| BrrSUC2.2 | 63.1 | 77.1 | 76.9 | 74.8 | 94.3 | 一 | 56.9 | 58.9 | 62.9 | 61.4 | 82.9 | 80.8 | |
| BrrSUC3.1 | 35.0 | 40.8 | 38.8 | 39.6 | 40.4 | 39.8 | 一 | 92.9 | 54.3 | 55.3 | 56.4 | 58.0 | |
| BrrSUC3.2 | 35.2 | 41.2 | 39.0 | 40.1 | 40.7 | 40.6 | 90.1 | 一 | 56.2 | 55.8 | 58.4 | 57.7 | |
| BrrSUC4.1 | 43.5 | 49.4 | 48.7 | 49.8 | 46.9 | 47.9 | 40.9 | 42.2 | 一 | 93.8 | 64.8 | 63.6 | |
| BrrSUC4.2 | 43.2 | 47.9 | 47.5 | 49.7 | 46.2 | 48.1 | 41.0 | 41.3 | 91.0 | 一 | 63.1 | 64.6 | |
| BrrSUC6.1 | 68.0 | 77.8 | 75.8 | 74.1 | 72.1 | 71.9 | 39.5 | 40.1 | 50.1 | 48.8 | 一 | 90.9 | |
| BrrSUC6.2 | 65.9 | 76.5 | 74.9 | 72.5 | 72.3 | 71.2 | 39.7 | 38.9 | 48.9 | 49.6 | 85.0 | 一 | |
We identified 12 SUC genes from the Chinese cabbage Chiifu genome, and subjected them to BLASTP searches on the turnip genome. A total of 11 in silico candidate homologous SUC genes in turnip were identified, excluding BrrSUC1.4. To further verify the identity of these genes, we used cDNA from turnip flower, leaf, and root tissues to clone the full-length coding sequence for all 11 candidate genes. BrrSUC1.4 was not thought to be found in silico in turnip genome, Although BrrSUC1.4 was not found in silico from the turnip genome, we used primers based on the BrSUC1.4 sequence from Chinese cabbage to successfully clone the gene. The 12 cloned SUC genes were named as BrrSUC1.1, BrrSUC1.2, BrrSUC1.3, BrrSUC1.4, BrrSUC2.1, BrrSUC2.2, BrrSUC3.1, BrrSUC3.2, BrrSUC4.1, BrrSUC4.2, BrrSUC6.1 and BrrSUC6.2. The sequence length of BrrSUCs varies from 1434 bp to 1775 bp, and BrrSUC3.1/ BrrSUC3.2 are longer than other members. The protein sequences of the BrrSUC family genes were deduced by DNAMAN software, and the molecular mass, physical, and chemical properties of the deduced proteins were further analyzed by using the ExPASyProtParam tool as indicated in Table 1. The predicted molecular mass of BrrSUCs varied from 50.90 KDa (BrrSUC6.1) to 63 KDa (BrrSUC3.1). The pI values range from 5.82 (BrrSUC3.1) to 9.40 (BrrSUC4.2). The pI of BrrSUC3.1 and BrrSUC3.2 was < 7, whereas the others are > 8.5. The grand average of the hydropathy value of the BrrSUC proteins was positive, meaning that they are hydrophobic proteins.
3.2. Similarity and phylogenetic analysis of BrrSUC proteinsTo better understand the evolutionary relationships among turnip SUC proteins and other plant species SUC proteins, an unrooted phylogenetic analysis from Arabidopsis thaliana, Brassica oleracea, Sorghum bicolor, Beta vulgaris, Oryza sativa, Zea mays, and Triticum aestivum was created, as shown in Fig. 1. Previous studies indicated that the SUC proteins from monocotyledon and dicotyledon species can be divided into five clades. BrrSUC3.1 and BrrSUC3.2 belonged to the SUT2 clade and BrrSUC4.1 and BrrSUC4.2 belonged to SUT4 clade. The rest of the BrrSUC proteins belonged to the SUT1 clade. These turnip SUC proteins were clustered closely with the dicotyledon, and were significantly distant from the monocotyledon as indicated in Fig. 1.

|
| Fig. 1 The phylogenetic analysis of 53 SUC proteins from Brassica rapa var. rapa. The unrooted tree was constructed using the NJ tree with 1000 bootstraps based on the protein sequences. The SUC proteins were clustered into five clades explained in the text. Accession numbers of the SUCs shown are as follows: Arabidopsis thaliana: AtSUC1, At1g71880; AtSUC2, At1g22710; AtSUT2, At2g02860; AtSUT4, At1g09960; AtSUC9, At5g06170. Oryza sativa: OsSUT1, D87819; OsSUT2, HQ540307; OsSUT3, AB071809; OsSUT4, AB091673; OsSUT5, AB091674; Triticum aestivum: TaSUT1A, AAM13408; TaSUT1B, AAM13409; TaSUT1D, AAM13410; Zea mays: ZmSUT1, BAA83501; ZmSUT2, AAS91375; ZmSUT3, ACF86653; ZmSUT4, AAT51689; ZmSUT5, ACF85284; ZmSUT6, ACF85673; Sorghum bicolor: SbSUT1, Sb01g045720; SbSUT2, Sb04g038030; SbSUT3, Sb01g022430; SbSUT4, Sb08g023310; SbSUT5, Sb04g023860; SbSUT6, Sb07g028120. Beta vulgaris: BvSUT1.1, NP_001290021.1; BvSUT1.2, XP_010673573.1; BvSUT1.3, XP_010675270.1; BvSUT3, XP_010683228.1; BvSUT4.1, XP_010695439.1; BvSUT4.2, XP_010695440.1. Brassica oleracea: BoSUC1.1, XP_013595174.1; BoSUC1.2, XP_013588232.1; BoSUC1.3, XP_013612789.1; BoSUC1.4, XP_013592286.1; BoSUC2.1, XP_013598534.1; BoSUC2.2, XP_013586960.1; BoSUC3.1, XP_013599420.1; BoSUC3.2, XP_013621461.1; BoSUC4.1, XP_013601858.1; BoSUC4.2, XP_013637826.1; BoSUC6.1, XP_013623414.1; BoSUC6.2, XP_013609440.1. |
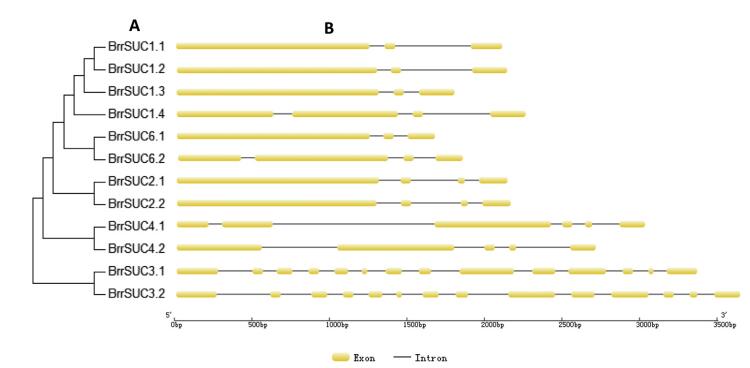
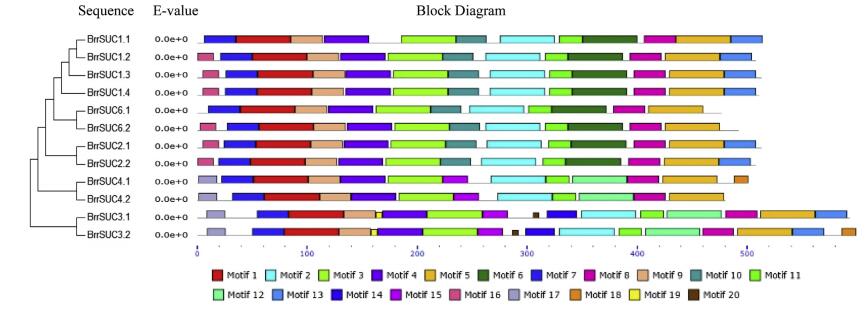
To estimate the homologies among the 12 BrrSUC proteins, the similarity/identity of the each amino acid sequence was performed using the EMBOSS-Needle program and is listed in Table 2. The results showed that the similarities in the amino acid sequence varied from 35.0% (BrrSUC1.1 and BrrSUC3.1) to 97.2% (BrrSUC2.1 and BrrSUC2.2), indicating that the BrrSUC family proteins have experienced significant differentiation over the evolutionary process. Positions of introns and exons within BrrSUC genes were determined and shown in Fig. 2. As the results show, closely related genes share similar gene structures. For example, SUT1 clade genes such as BrrSUC1 genes, BrrSUC6 genes and BrrSUC2 genes have 3-4 exons and 2-3 introns, and SUT4 clade genes (BrrSUC4.1 and BrrSUC4.2) have 5-6 exons and 4-5 introns. By contrast, SUT2 clade genes (BrrSUC3.1 and BrrSUC3.2) have more exons and introns (14 exons and 13 introns). Interestingly, the introns and exons within subclade genes also arranged similarly (Fig. 2). We further analyzed the putative protein motifs by MEME tool to compare the differences in the functional domains among these BrrSUC proteins. The results in Fig. 3 show that 20 putative protein motifs were identified for BrrSUC proteins. As mentioned above, the BrrSUC genes can be divided into three clades, as shown in the phylogenetic tree. Most of the closely clustered genes in each clade share common motif compositions, suggesting similar function among the BrrSUC proteins within the same clade. Generally, 12 or 13 motifs were observed in most BrrSUC proteins, whereas in BrrSUC3.1 and BrrSUC3.2, 16 or 17 motifs were found, respectively, suggesting that functional differentiations have occurred among the gene family. Though different in motif number, the arrangement of the motifs in these BrrSUC proteins was similar.
|
| Fig. 2 The exon-intron structure of the BrrSUC genes according to their phylogenetic relationships. (A). The phylogenetic tree was constructed using MAGA6.0 software based on the predicted amino acid sequences of BrrSUC. (B). The exon-intron structures were analyzed by the online tool Gene Structure Display Servers 2.0. |
|
| Fig. 3 The conserved motifs of BrrSUC proteins based on their phylogenetic relationship. The MEME Suite program was used to analyze the conserved motifs. Twenty conserved amino acid sequence blocks are displayed with different box colored. The 20 blocks are marked with numbers at the bottom. |
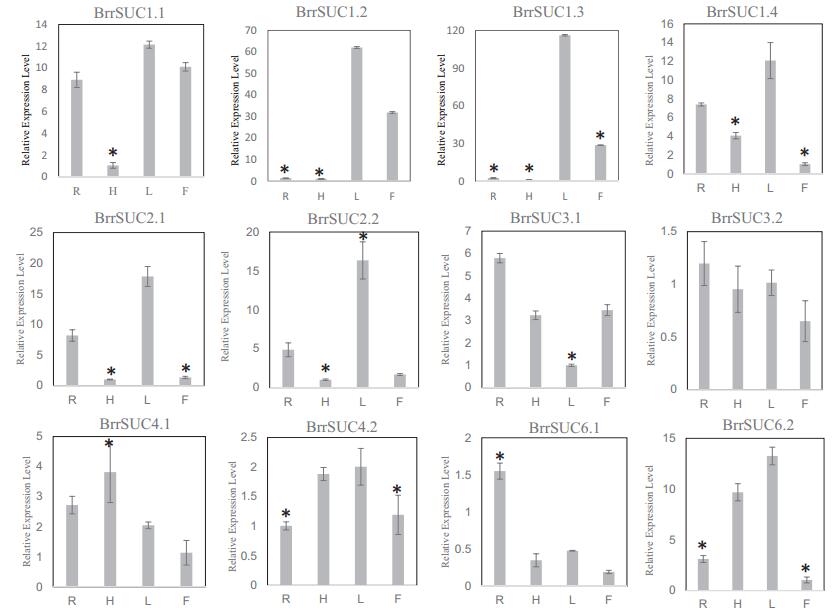
To explore the possible functions of the BrrSUC family, we monitored the expression of these SUC genes in different tissues. We collected leaves, hypocotyls, and roots from 21-day-old seedlings, together with the flowers in blooming stage for RNA isolation. After reverse transcription, we performed real-time PCR using these cDNA (Fig. 4). After flowering, the reproductive organ becomes the major sink tissue. For BrrSUC1 genes, BrrSUC1.2 and BrrSUC1.3 showed similar expression pattern, and were mainly expressed in leaves and flowers. BrrSUC1.1 exhibited lower expression in hypocotyl tissue in comparison to other tissues, whereas BrrSUC1.4 exhibited lower expression levels in flower tissue. BrrSUC2.1 and BrrSUC2.2 shared a similar expression pattern, and were mainly expressed in roots and leaves. BrrSUC3 and BrrSUC4 were both expressed in all tissues examined, whereas BrrSUC3.1 was expressed in all tissues except for leaves. BrrSUC6.1 was predominately expressed in roots, whereas BrrSUC6.2 was expressed mainly in the hypocotyl and leaves. The expression of the SUT1 clade, including BrrSUC1, BrrSUC2, and BrrSUC6, were highly tissue specific, whereas clade SUT2 (BrrSUC3.1 and BrrSUC3.2) and SUT4 (BrrSUC6.1 and BrrSUC6.2) were expressed in most tissues, suggesting that these two clades play important roles in plant development.
|
| Fig. 4 Relative expression analysis of twelve BrrSUC genes in turnip tissues. Relative transcript levels were analyzed by real-time RT-PCR. The constitutive β-tubulin gene was used as a control. R: roots; H: hypocotyls; L: leaves; F: flowers. The error bar represents SD for three independent assays. Asterisks indicate a significant difference (P < 0.01, Student's T-Test). |
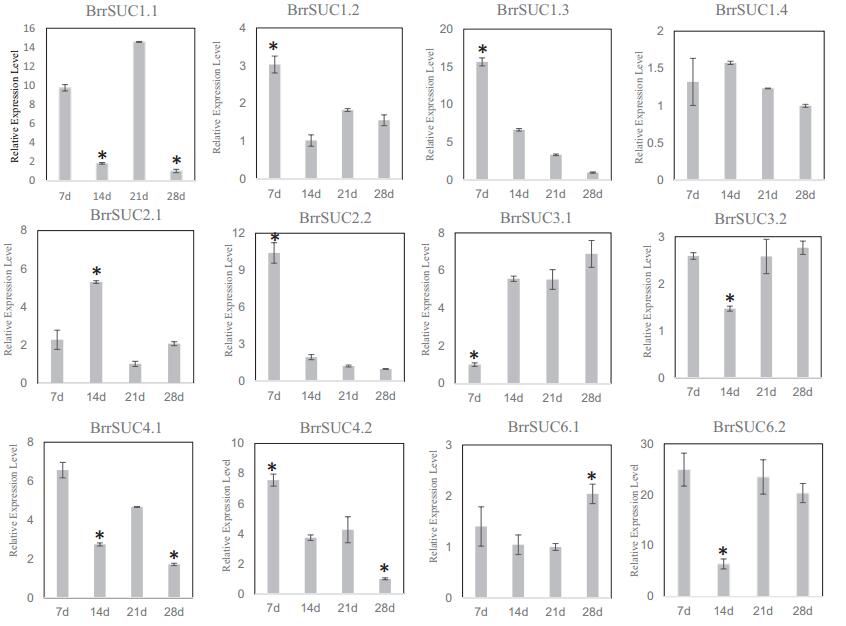
In contrast to other B. rapa, B. rapa var. rapa can form a taproot developed from hypocotyl and partial root organs. To determine whether the BrrSUC genes responded to this process, we collected hypocotyls from the four developmental stages of turnip (supplemental Fig. 1). The gene expression profiles of BrrSUC family in different stages were determined by qRT-PCR (Fig. 5). The results show that five genes (BrrSUC1.2, BrrSUC1.3, BrrSUC2.2, BrrSUC4.1, and BrrSUC4.2) were expressed at a higher level in 7-day-old seedlings (the first true leaves appeared), whereas the expression of these genes appeared to be downregulated afterwards. Conversely, the expression of BrrSUC3.1 was at a low level in 7-dayold seedlings, and increased as the turnip taproot formed (14-, 21-, and 28-day-old seedlings). The expression of the remaining BrrSUC genes shows no obvious changes (BrrSUC1.4, BrrSUC3.2, and BrrSUC6.1) or no significant correlation with the four stages of turnip taproot formation (BrrSUC1.1, BrrSUC2.1, and BrrSUC6.2).
|
| Fig. 5 Relative expression analysis of twelve BrrSUC genes during turnip taproot formation. Hypocotyls of turnip were harvested at 7, 14, 21, 28 days after seeds were sown. RTqPCR was performed to analyze relative transcript levels. The constitutive β-tubulin gene was used as a control. Error bars represent SD for three independent assays. Asterisks indicate a significant difference (P < 0.01, Student's T-Test). |
Sucrose transport is the main energy transport form in the plant sourceesink system, and plays important roles in plant growth and development. SUTs, also known as SUCs, are found in most plants, and function to distribute sucrose both intracellularly and at the whole plant level. SUCs have been extensively studied in Arabidopsis, rice (O. sativa L.), T. cacao, and other plant species (Kuhn and Grof, 2010). However, there have been relatively few studies of SUCs in Brassica species. In a recent study, the SUC gene family in oilseed rape (B. napus L.) was comprehensively discussed, and 22 SUC genes were identified from oilseed rape and nine SUC genes were identified from B. rapa (Jian et al., 2016). B. napus is an amphidiploid originating from the fusion of two diploid genomes, an A-genome progenitor (B. rapa) and a C-genome progenitor (B. oleracea) (Nagaharu, 1935). Obviously, more SUC genes remain to be identified in B. rapa. For more in-depth understanding of the genetic evolution and molecular function of the SUC gene family in B. rapa var. rapa, we identified 12 SUC genes in B. rapa pekinensis and confirmed their presence in the turnip by cloning and sequencing. Though the genome of B. rapa var. rapa was resequenced based on the B. rapa pekinensis genome, the turnip genome was not fully sequenced. Therefore, when the SUC genes of B. rapa pekinensis underwent BLASTP searches against the turnip genome, we only found 11 SUC genes. While we were able to isolate the SUC 1.4 gene from turnip, we have not been able to find its location on the turnip chromosome.
The SUC gene family can be divided into five clades in the plant species, and our results indicate that the BrrSUC genes are distributed in three of these five clades, which is similar to what previous studies have found (Kuhn and Grof, 2010). Interestingly, most of the SUC genes came in pairs, such as BrrSUC2.1 and BrrSUC2.2, BrrSUC3.1 and BrrSUC3.2, BrrSUC4.1 and BrrSUC4.2, and BrrSUC6.1 and BrrSUC6.2, whereas BrrSUC1 had four members. Moreover, these subclass member genes show more similar protein sequence and gene structures (or motifs, indicated by MEME). This phenomenon is caused by the ancestor of the Brassica species undergoing whole-genome triplication events followed by extensive diploidization (Cheng et al., 2016). However, the theoretical number of SUC genes was greater than the cloned SUC genes in this study, indicating the loss of many genes during Brassica evolution.
Several studies examined the expression of SUC family genes and revealed that these SUC genes show distinct expression patterns in a variety of plant species and play different roles in plant growth, development, and environment adaptation (Gottwald et al., 2000; Kuhn and Grof, 2010; Riesmeier et al., 1993, 1994; Weise et al., 2000). The expression pattern of the BrrSUC family genes also showed different expression patterns, which is consistent with previous studies. Single SUC genes often have more than one function. For example, SUC1 was reported to be mainly expressed in flowers and is important for pollen germination, and was also reported to be involved in long-distance sucrose transport and anthocyanin accumulation. Thus, the diverse expression patterns of BrrSUC genes assure the normal growth and adaptation of the turnip.
The hypocotyl is the most valuable tissue of the turnip, developing into and forming the edible taproot. The products of the photosynthesis in source tissue (mainly leaves) are distributed into the sink tissues, including the hypocotyl. The demand for carbohydrates is positively correlated with the sucrose transport capacity. We monitored the expression pattern of the 12 BrrSUC genes at the early stages of turnip taproot formation to check whether BrrSUC genes were responsive to this process, and found that only BrrSUC 3.1 was significantly upregulated during turnip taproot formation. Previous studies found that SUC3 was distinct from other family members in several features, such as open reading frame, the number of introns, and the codon usage bias. In our results, we also found that BrrSUC3 genes were distant from the other BrrSUC family members in phylogenetic tree, and the conserved motif of BrrSUC3 also significantly differed from other members. The functions of SUC3 genes are widely discussed in a variety of plant species for this reason. These studies indicate that SUC3 genes play an important role in a number of sink tissues, such as developing seeds in Arabidopsis, fruits in tomato (Bitterlich et al., 2014), and might function as the sucrose sensor to regulate the sucrose transport. The upregulated expression pattern of BrrSUC3.1 during turnip taproot formation provided us a clue that BrrSUC might be involved in taproot formation and function as the sucrose transporter or sucrose transport regulator in this process.
AcknowledgmentsWe thank Dr. Yunqiang Yang for assisting in the construction of the phylogenetic tree and data analysis. This work was supported by Major Program of National Natural Science Foundation of China (31590820, 31590823).
Appendix A. Supplementary dataSupplementary data related to this article can be found at http://dx.doi.org/10.1016/j.pld.2017.05.006.
Aoki N., Hirose T., Scofield G.N., Whitfeld P.R., Furbank R.T., 2003. The sucrose transporter gene family in rice. Plant Cell Physiol, 44: 223-232. DOI:10.1093/pcp/pcg030 |
Aoki N., Hirose T., Takahashi S., Ono K., Ishimaru K., Ohsugi R., 1999. Molecular cloning and expression analysis of a gene for a sucrose transporter maize (Zea mays L.). Plant Cell Physiol, 40: 1072-1078. DOI:10.1093/oxfordjournals.pcp.a029489 |
Bitterlich M., Krugel U., Boldt-Burisch K., Franken P., Kuhn C., 2014. The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J, 78: 877-889. DOI:10.1111/tpj.2014.78.issue-5 |
Cheng F., Sun R.F., Hou X.L., Zheng H.K., Zhang F.L., Zhang Y.Y., Liu B., Liang J.L., Zhuang M., Liu Y.X., Liu D.Y., Wang X.B., Li P.X., Liu Y.M., Lin K., Bucher J., Zhang N.W., Wang Y., Wang H., Deng J., Liao Y.C., Wei K.Y., Zhang X.M., Fu L.X., Hu Y.Y., Liu J.S., Cai C.C., Zhang S.J., Zhang S.F., Li F., Zhang H., Zhang J.F., Guo N., Liu Z.Y., Liu J., Sun C., Ma Y., Zhang H.J., Cui Y., Freeling M.R., Borm T., Bonnema G., Wu J., Wang X.W., 2016. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet, 48: 1218-1224. DOI:10.1038/ng.3634 |
Chu B., Chen C., Li J., Chen X., Li Y., Tang W., Jin L., Zhang Y., 2017. Effects of Tibetan turnip (Brassica rapa L.) on promoting hypoxia-tolerance in healthy humans. J. Ethnopharmacol, 195: 246-254. |
Gong X., Liu M.L., Zhang L.J., Ruan Y.Y., Ding R., Ji Y.Q., Zhang N., Zhang S.B., Farmer J., Wang C., 2015. Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiol. Plant, 153: 119-136. DOI:10.1111/ppl.2015.153.issue-1 |
Gottwald J.R., Krysan P.J., Young J.C., Evert R.F., Sussman M.R., 2000. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. U. S. A, 97: 13979-13984. DOI:10.1073/pnas.250473797 |
Jian, H. J. , Lu, K. , Yang, B. , Wang, T. Y. , Zhang, L. , Zhang, A. X. , Wang, J. , Liu, L. Z. , Qu, C. M. , Li, J. N. , 2016. Genome-wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L. ). Front. Plant Sci. 7.
|
Kuhn C., Grof C.P.L., 2010. Sucrose transporters of higher plants. Curr. Opin. Plant Biol, 13: 287-298. DOI:10.1016/j.pbi.2010.02.001 |
Lemoine R., 2000. Sucrose transporters in plants: update on function and structure. Biochim. Biophy. Acta, 1465: 246-262. DOI:10.1016/S0005-2736(00)00142-5 |
Li F.P., Wu B.D., Qin X.W., Yan L., Hao C.Y., Tan L.H., Lai J.X., 2014. Molecular cloning and expression analysis of the sucrose transporter gene family from Theobroma cacao L. Gene, 546: 336-341. DOI:10.1016/j.gene.2014.05.056 |
Nagaharu U., 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot, 7: 389-452. |
Ogawa Y., Omata Y., Nishio K., Saito Y., Yoshida Y., Niki E., 2007. Assessment of antioxidative activity of extract from fermented grain food mixture using chemical and cellular systems. Biofactors, 31: 237-248. DOI:10.1002/biof.v31:3/4 |
Riesmeier J.W., Hirner B., Frommer W.B., 1993. Potato sucrose transporter expression in minor veins indicates a role in phloem loading. Plant Cell, 5: 1591-1598. DOI:10.1105/tpc.5.11.1591 |
Riesmeier J.W., Willmitzer L., Frommer W.B., 1992. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. Embo J, 11: 4705-4713. |
Riesmeier J.W., Willmitzer L., Frommer W.B., 1994. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. Embo J, 13: 1-7. |
Rolland F., Baena-Gonzalez E., Sheen J., 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol, 57: 675-709. DOI:10.1146/annurev.arplant.57.032905.105441 |
Sauer N., Ludwig A., Knoblauch A., Rothe P., Gahrtz M., Klebl F., 2004. AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J, 40: 120-130. DOI:10.1111/tpj.2004.40.issue-1 |
Schmittgen T.D., Livak K.J., 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc, 3: 1101-1108. DOI:10.1038/nprot.2008.73 |
Weise A., Barker L., Kuhn C., Lalonde S., Buschmann H., Frommer W.B., Ward J.M., 2000. A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell, 12: 1345-1355. DOI:10.1105/tpc.12.8.1345 |
Xie Y., Jiang S., Su D., Pi N., Ma C., Gao P., 2010. Composition analysis and antihypoxia activity of polysaccharide from Brassica rapa L. Int. J. Biol. Macromol, 47: 528-533. DOI:10.1016/j.ijbiomac.2010.07.008 |



