b. College of Crop Science, Fujian Agriculture & Forestry University, Fuzhou, 350002, China
The dwarf phenotype in plants provides an opportunity for studying regulatory mechanisms of plant growth and development. This character is also favored in breeding. Dwarf mutants have been discovered in many species, and the regulatory mechanisms underlying this trait have been partially revealed (Daviere and Achard, 2013; Waldie et al., 2014; Zhang et al., 2014). Various factors can result in dwarf phenotypes associated with the gibberellin (Daviere and Achard, 2013), brassinosteroid (Zhang et al., 2014) and/or strigolactone pathways (Waldie et al., 2014).
In rice, at least 20 dwarf genes have been cloned, and most of them are involved in the synthesis and signal transduction of gibberellin, brassinosteroid and/or strigolactone (http://www.ricedata.cn/gene/gene_sd.htm). Rice dwarf mutant Daikoku, which carries the d1 gene, was first isolated as a spontaneous mutant (Ashikari et al., 1999; Fujisawa et al., 1999). The d1 mutant shows abnormal gross morphologies, such as dwarfism, shortened internodes, small-rounded seeds and erect leaves (Ashikari et al., 1999; Fujisawa et al., 1999). The D1 gene encodes a subunit of a G-protein, the GTP-hydrolysing Gα, the mutation of which alters responses to both gibberellin and brassinosteroid (Oki et al., 2009c; Ueguchi-Tanaka et al., 2000). G-proteins are membrane-associated heterotrimers consisting of Gα, Gβ and Gγ subunits. The Gβγ dimer binds tightly to GDP-bound Gα, enhancing the coupling of Gα to the G-protein-coupled receptor. Upon receptor stimulation by a signal molecule, the state of the receptor changes and Gα dissociates from the receptor and Gβγ, while GTP is exchanged for the bound GDP, which leads to Gα activation. Then, Gα goes on to activate other molecules in the cell (Urano and Jones, 2014).
Transposable elements are DNA fragments that are able to move around the genome, and have been found in virtually all eukaryotic species investigated so far. They are divided into two classes based on their means of transposition and replication. Class Ⅰ retrotransposons are transposed using a "copy and paste" mechanism through RNA intermediates. Class Ⅱ DNA transposons are directly transposed from one site to another using the "cut and paste" mechanism without any RNA intermediates (Grandbastien, 2015; Kumar and Bennetzen, 1999).
Long terminal repeat (LTR) retrotransposons are class Ⅰ transposable elements and are the most common type. They are ubiquitous in the plant kingdom, and are the main constituents of large plant genomes (Kumar and Bennetzen, 1999; Ma et al., 2004). For example, LTR-retrotransposons comprise more than 50% of the maize genome (Kumar and Bennetzen, 1999) and more than 22% of rice genome (Ma et al., 2004). About 41 families, including 13, 000 elements, have been revealed in rice (Vitte et al., 2007). However, only Tos elements have been studied extensively, and Tos17 was applied to make rice insertion mutant database (Grandbastien, 2015; Hirochika et al., 1996).
The complete copies of LTR retrotransposons consist of two LTRs and an internal region. LTR sequences contain signals for transcription, initiation and termination, while the internal region encodes the proteins that are necessary for the retro-transposition cycle. LTR retrotransposons can be subdivided into copia-type and gypsy-type elements (Grandbastien, 2015).
Many mutants have been caused by retrotransposon insertion. However, most of the mutants were caused by Tos17 insertion, and only a few mutants inserted by other retrotransposons have been identified (Table S1).
We fortuitously found a dwarf mutant in a paddy field. Here we report that the gene is located near the D1 gene on chromosome 5, and the dwarf mutant is the result of a copia-type LTRretrotransposon insertion of the D1 gene. This copia-type LTRretrotransposon belongs to the Osr4 group, and the characteristics of Osr4 members were analyzed in this study.
2. Materials and methods 2.1. Plant materials and genetic mappingThe dwarf mutant was found in the Min Nong 1 rice paddy field. To identify the dwarf gene, two F2 populations from crosses between the mutant and Nipponbare or LPBG08 plants were produced for genetic mapping. DNA was extracted from fresh leaves from each of the F2 plants using the SDS method (Zheng et al., 1995) with a few modifications. SSRs were amplified for use as molecular markers. BSA (Michelmore et al., 1991) was used to find the markers that linked to the target gene.
2.2. Identification of the mutant gene and the insertTotal RNA was extracted from rice leaves and flowers following Bilgin's method (Bilgin et al., 2009). Total RNA (1 mg) was reversetranscribed using M-MLV reverse transcriptase in a final volume of 20 ml to obtain cDNA. RT-PCR was used to detect gene expression and to find the deletion of the mutant gene.
A set of PCR was done with the primers on D1 gene. Tail-PCR was performed to determine the insert using Wang's method (Wang et al., 2011). All primers are shown in Table 2.
| Structure | No. of elements |
| Intact elements with TSDs | 33 |
| Intact elements without TSDs | 7 |
| One LTR partially deleted | 8 |
| Both LTR partially deleted | 17 |
| Name | Primer sequence | Purpose |
| P1 | TTACCTTGTTCCGTTGCTTT | Determine insert position |
| P2 | CTGCAACTTCCTGATTGTGC | Determine insert position |
| P3 | GCACAATCAGGAAGTTGCAG | Determine insert position |
| P4 | CATGGACAGCACACCTTTGT | Determine insert position |
| P5 | AGCAGTTTTGCACCCTATGT | Determine insert position |
| P6 | CCATTTGTCCGTACTCTTGC | Determine insert position |
| D1-R2-2 | CCAGCCTTTCAAACGAACCA | Tail-PCR |
| D1-R2 | AGCAGTTTTGCACCCTATGT | Tail-PCR |
| D1-EXON6-R1 | CCTTCGTTATGTAGACTGCGT | Tail-PCR |
| CT-R2 | ACACAGTACCGACGTCTTGT | Osr4 expression |
| T-F2 | GCCGAGGAGATCACTAAGCA | Osr4 expression |
| CT-R3 | TCCTTCTACCTGGGGATCGA | Osr4 expression |
| T-F1 | AAGCCATGTCCTCCCAAAGA | Osr4 expression |
| D1-cDNA-F1 | ACGTGCTTCCTGGAAAGAGA | D1 cDNA |
| D1-cDNA-R2 | AGACCTGAACAGCCCACAGT | D1 cDNA |
| RT-F | TGCTGCCATTAGCGAATATG | D1 gene expression |
| RT-R | GGCTTGCTGCTCTGGAAGTA | D1 gene expression |
We used Chr4-1 as a query to search the rice genome sequence (IRGSO-1.0) with Blastn (http://rapdb.dna.affrc.go.jp/tools/blast) to identify the Osr4 LTR-retrotransposons in Nipponbare. The Highscoring Segment Pairs in the Blast result, whose sequence identities were more than 80% and lengths more than 350bp, were selected. The homologous retrotransposon sequences, LTR mutations, CDS mutations and directed repeats were manually analyzed and revealed. The alignment and phylogenetic tree were constructed with MEGA6 (http://megasoftware.net/). To find the Osr4 LTR-retrotransposons in other species, we used Chr4-1 as query to search a nr/nt database, and the High-scoring Segment Pairs whose sequence identities are more than 80% and lengths are more than 4000bp were manually analyzed, and their CDSs were used to construct the phylogenetic tree.
3. Results 3.1. Mapping the dwarf geneA spontaneous dwarf mutant which shows broad, dark green leaves, compact panicles, and short, round grains was found in Min Nong 1 paddy field (Fig. 1). Each internode of the mutant is shorter than in the wildtype, and the second internode is extremely short (Fig. 1C and D). However, the plant heights of the wildtype and the mutant are not obviously different at the tillering stage (Fig. 1E).
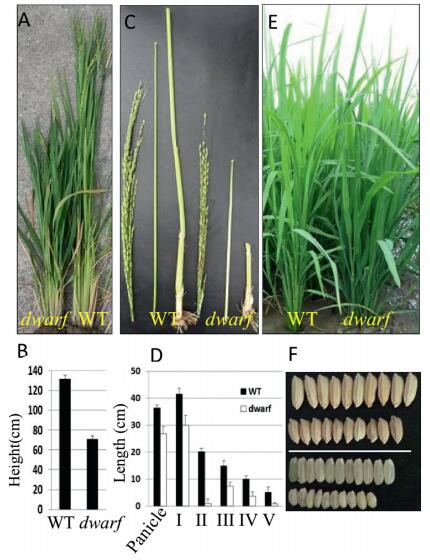
|
| Fig. 1 Phenotypes of the wild-type and dwarf mutant. (A) and (B) The plant heights of the dwarf and wildtype at heading stage. (C) and (D) The lengths of the panicles and internodes of the wildtype and dwarf mutant. (E) The morphological phenotypes of the dwarf and wildtype at tillering stage. (F) Seeds of wildtype (Upper) and dwarf (Lower). |
The segregation ratio of dwarf plants to wildtype plants fits to 1:3 in the single Mendelian factor model, which indicates that one recessive gene controls the dwarf phenotype. We made crosses between Nipponbare and the mutant, and between the mutant and LPBG08. We tested the segregation ratios, and both crosses segregated according to the Mendelian factor model.
We developed two DNA pools: wildtype pool and dwarf pool. For the dwarf pool, nine dwarf plants were selected from the F2 individuals of the cross between Nipponbare and the dwarf mutant; their DNA was extracted and pooled. For the wildtype pool, the DNAs from nine F2 wildtype individuals were mixed. We selected a total of 364 SSR markers that were evenly distributed on the 12 chromosomes, and then detected their polymorphisms among the two DNA pools and the parents. The SSR markers RM289, RM509, RM430 and RM163 on chromosome 5 exhibited polymorphisms between the two DNA pools, and these results indicated that the gene controlling the dwarf trait was most likely linked to the four markers. Further analysis showed that the dwarf gene is located between RM289 and RM430 and co-segregates with RM509 (Fig. 2).
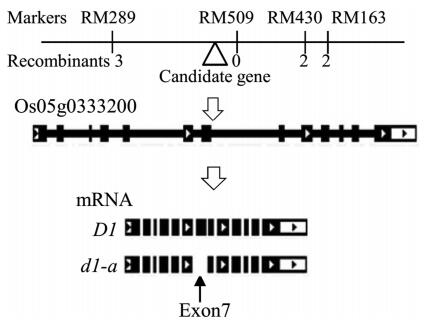
|
| Fig. 2 Mapping and cloning of the dwarf gene, and the structure of D1 and d1-a genes. |
The phenotype of our mutant is very similar to the dwarf 1 mutant (Ashikari et al., 1999; Fujisawa et al., 1999, 2001; Miura et al., 2009; Oki et al., 2009b; Ueguchi-Tanaka et al., 2000), which is located on chromosome 5 near SSR marker RM509. We speculated that our dwarf mutant was controlled by an allele of the D1 gene. To confirm this, we detected the D1 cDNA with PCR, and found that the PCR product from the dwarf mutant is smaller than that from the wildtype (Fig. 3A). We sequenced the PCR products, and discovered a 102-bp deletion in our dwarf mutant gene, covering all of exon 7 (Fig. 2), but didn't reveal an expression difference between the wildtype and the mutant (Fig. 3B). We termed this mutant gene d1-a.
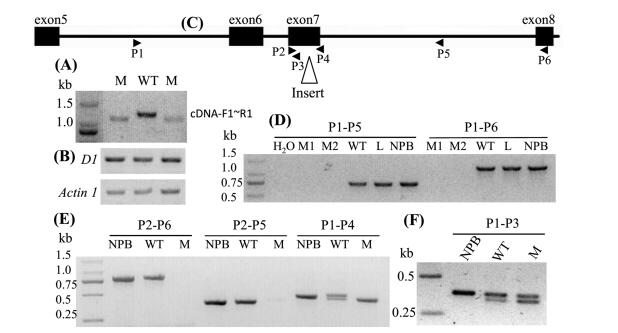
|
| Fig. 3 (A) The different sizes of D1 cDNA between wildtype and the mutant. (B) The D1 gene expression level in wildtype and mutant. (C) The relative positions of the primers and position of the insert. P, primer. (D), (E) and (F) The results of the PCR products with different primer pairs. WT, wildtype. M, mutant. NPB, Nipponbare. L, LPBG08. |
To analyze the exact length of the deletion in the genome, we amplified genomic DNA that included exon 7, but we did not obtain PCR products in the mutant (Fig. 3D). We hypothesized that there was an insert fragment in the D1 gene of the mutant. To validate this hypothesis, we performed several PCRs, and found that the insert should be in exon 7 (Fig. 3C-E). However, to our surprise, we obtained double PCR product bands in the wildtype, and one smaller band in the mutant, and one bigger band in Nipponbare with primer P1 and P4 (Fig. 3E). The D1 gene was previously thought to be a single copy gene in the rice genome (Ishikawa et al., 1995). However, our results did not support this assumption and implied that there may be other D1 variants in rice plants.
In order to identify the variants, we performed PCR with primer P1 and P3, which were on the same side of the insert (Fig. 3C). The results exhibited two bands in the wildtype and the mutant, but one bigger band in Nipponbare (Fig. 3F). It is difficult to explain these results. Fortunately, using Blastn, we identified a truncated D1 gene which was also on chromosome 5 and about 394.7 kb distant from the D1 gene in Nipponbare (Fig. 4A). This truncated gene, which we termed D1-like, contained seven exons of the D1 gene (Fig. 4B). We checked Min Nong 1 and RP Bio-226, and found that both of them contained the D1-like gene. However, the D1-like gene was longer in Nipponbare (japonica) than in Min Nong 1 and RP Bio-226 (indica). In addition, the P1eP3 and P1eP4 regions of the D1-like gene were almost identical to the same regions in the D1 gene in Nipponbare, RP Bio-226 and the wildtype (Fig. 4C). The differences led to the results shown in Fig. 3E and F. The P1eP4 region included an insert in d1-a, so only the shorter band could be obtained (Fig. 3E). In addition, the D1-like gene contained many point mutations compared with the D1 gene.

|
| Fig. 4 A comparison between D1 and D1-like gene. (A) The relative position of D1 and D1-like gene in Nipponbare. (B) A comparison of D1 and D1-like gene structure. (C) The deletion of D1-like gene in Wildtype and RP Bio-226(Bio). |
The D1-like gene was a section of Os05t0341300 gene (http://rapdb.dna.affrc.go.jp/viewer/gene_detail/irgsp1?name=Os05t0341300), and exons 1, 2, 3, 4, 5, 6 and 7 of D1-like were also exons of Os05t0341300. The gene prediction of Os05t0341300 was supported by the transcript AK241716, but no start codon was indicted in the prediction. We used the DNA sequence of AK241716 as query to perform a Blastx search, and only D1 (Gα) proteins in different species were hit. The homologous regions covered the partial region of AK241716, and contained a stop codon. These results suggest that Os05t0341300 may be a pseudogene. Moreover, we also found at least two remains of LTRs in intron 8 of Os05t0341300 (Fig. 4B).
3.3. An active LTR-retrotransposon inserted rice D1 geneWe used a tail PCR to reveal the insert sequence. The results showed that the sequence of the tail-PCR product was about the same as the partial sequence of an LTR-retrotransposon. We designed primers according to the LTR-retrotransposon sequence (ref|NC_008397.2|: 31604515 to 31610215), and performed PCR with one primer on the LTR-retrotransposon and one primer on D1 gene, and then sequenced the PCR products. The results showed that the sequences of both sides of the insert were nearly identical to that of the sides of an LTR-retrotransposon (ref|NC_008397.2|: 31604515 to 31610215). The insert position is shown in Fig. 5A. However, it is difficult to determine whether the insert contains a complete LTR-retrotransposon. Further analysis showed that the LTR-retrotransposon (ref|NC_008397.2|: 31604515 to 31610215) belongs to the Osr4 group (McCarthy et al., 2002; Vitte et al., 2007). We checked the expression of the LTR-retrotransposon, and found that the expression in leaves was much higher than in flower tissues (Fig. 5C). To further investigate the expression of the LTRretrotransposon in other tissues, we used the sequence of the LTR-retrotransposon as the query to search PLEXdb (http://www.plexdb.org/modules/tools/plexdb_blast.php), and obtained a probe set (ID-Os.45957.3.S1_x_at), which matches the LTR-region. The expression profile of Os.45957.3.S1_x_at is shown in Fig. S1, which partially reflects the expression of the LTR-retrotransposon. The LTR-retrotransposon is expressed in all tissues, with higher expression levels in some seeds and leaves. These results suggested that at least one of the Osr4 members should be active.
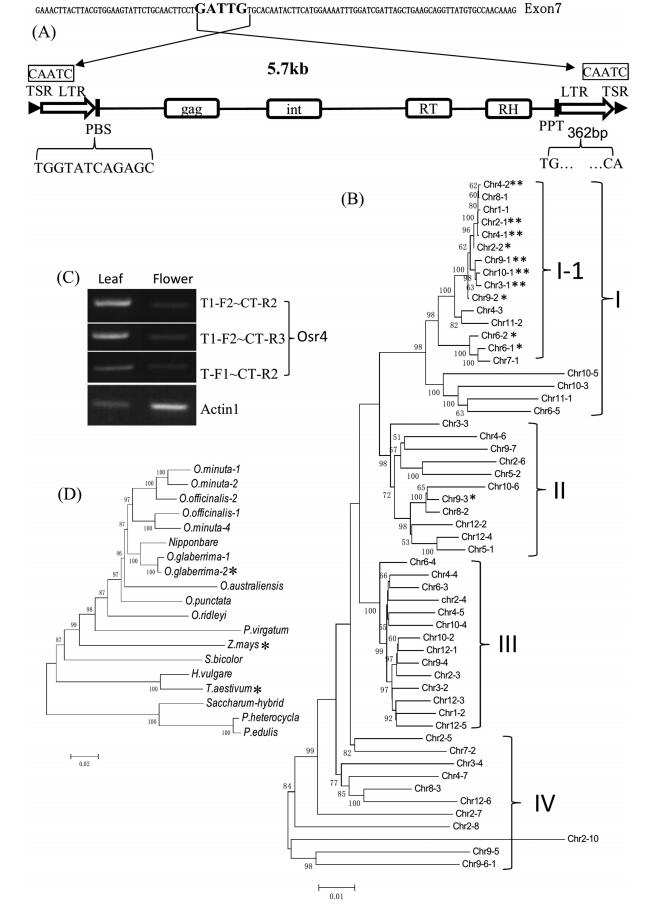
|
| Fig. 5 (A) The exact position of the insert and a typical Osr4 LTR retrotransposon. LTR (Long terminal repeat) with TG at the 5' extremity and CA at the 3' extremity; TSR (target site repeat) is a 5bp short direct repeat string flanking the 5' and 3' extremities of an element; PBS, a sequence complemented to the 3' tail of some tRNA; PPT (polypurine tract) is a short rich purine segment. Protein domains: The gag gene encodes proteins involved in maturation, packaging of retrotransposon RNA and proteins into a form suitable for integration into the genome. Int, integrase; RT, reverse transcriptase; RH, RNase H. (B) The Neighbor-joining tree of 65 LTR-retrotransposon homologs with their DNA sequences. The scale bar indicates the simple matching distance. *Indicate the retrotransposon with a complete CDS. **Indicate the possible functional retrotransposons. (C) The expression of the retrotransposon was detected with RT-PCR. Three different pairs of primers were used for retrotransposon. (D) The Neighbor-joining tree of LTR-retrotransposons in different species with their coding region sequences. *Indicates retrotransposons with a complete CDS. |
We found 65 members of the Osr4 LTR-retrotransposons in the Nipponbare genome, and revealed 11 retrotransposons with a complete CDSs (Coding DNA sequence) in addition to 54 with mutant CDSs (Table 1 and S2). We also found some point mutations in the 11 retrotransposons, but did not determine whether the mutant sites affect their function. Comparing the CDSs of retrotransposons, we revealed many more changes in the LTR region (data not shown) thought to regulate expression (Grandbastien, 2015). Short directed repeats were detected at the insert sites in 35 LTR-retrotransposons, while they were absent in another 30 LTR-retrotransposons (Table S2).
We classified the 65 homologs into 4 groups according to a phylogenetic tree. Fifteen members in group Ⅰ-1 showed high similarity with each other, including 10 LTR-retrotransposons with a complete CDS within the group (Table S2 and Fig. 5B). The insert of d1 gene in the mutant also exhibited high similarity with Chr4-1 in group Ⅰ-1. These results implied that the members of group Ⅰ-1, which showed higher activity, may have undergone transpositions frequently and recently.
The Osr4 members were distributed over all chromosomes (Fig. S1), and most of them were located in introns (33) and intergenic regions (29), while only 3 members were found in exons (Table S2). Some Osr4 members were located in promoters (Table S2), and might affect downstream gene expression.
The cis-regulation of LTR-retrotransposon gene expression mainly located in the LTR region, and some regulatory motifs in LTR play a key role in the expression (Dhadi et al., 2015; Grandbastien, 2015; Takeda et al., 1999). We checked the left LTR region, and revealed two new types of putative regulatory motifs: Box-A and Box-B (Fig. 6A). There were two Box-A motifs and three Box-B motifs in the LTR region. Only eight out of 65 Osr4 members contained all five motifs. Six of these are predicted to be functional retrotransposons containing the five motifs, a TATA box, PBS (a ~18bp sequence complemented to the 3' tail of some tRNA, which is very important because tRNA binding process is the first step of initiating reverse transcription.), a complete CDS and polypurine tract (Fig. 6B). In addition, some functional retrotransposons lacked the short directed repeats at the insert sites (Table S2 and Fig. 6B).
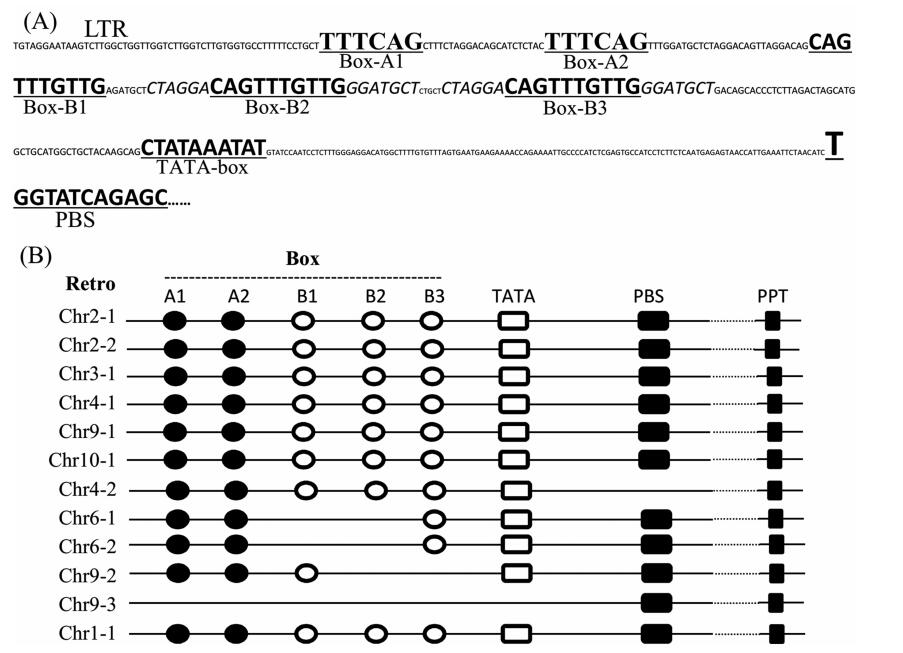
|
| Fig. 6 The possible regulatory motifs in left LTR. (A) The possible regulatory motifs of Chr4-1 left LTR. (B) The motifs in left LTRs of some Osr4 typical members. |
We searched the NCBI nr/nt database with the Chr4-1 DNA sequence, and only discovered the Osr4 members in monocots. However, most of them contained frameshift mutations or stop codons in the CDS, while some of them lost short directed repeats. We found only one LTR-retrotransposon in some species and more in other species. It is not certain whether there were active LTRretrotransposons in the species. The phylogenic tree of the LTRretrotransposons in different species is shown in Fig. 5D.
4. DiscussionIn rice plants, d1 mutants were originally identified in 1999 (Ashikari et al., 1999; Fujisawa et al., 1999), and have been used in subsequent G-protein studies (Oki et al., 2009a, 2009c; Suharsono et al., 2002; Ueguchi-Tanaka et al., 2000). Thereafter, many other d1 mutants which contained base substitutions, insertions or deletions in the d1 gene have been identified and show very similar phenotypes (Ashikari et al., 1999; Fujisawa et al., 1999; Oki et al., 2009b). This implies there may be several mutation hotspots in the D1 gene region. The d1-a gene lost its exon 7 of mRNA, and exon 7 encodes 34 amino acid residues (161e194), which includes one GoLoco binding site. GoLoco binding sites of Gα protein are responsible for binding with regulatory proteins possessing GoLoco motifs, which maintain G-protein subunit dissociation in the absence of Gα activation (Kimple et al., 2002). This implied the Gα protein in the d1-a mutant lost its activity completely or partially. Our mutant showed a similar phenotype as other d1 mutants, and the mutant gene was located at the same position as the D1 gene. These results suggested that the dwarf mutant phenotype in our study was caused by a mutation in the d1-a gene.
Previous studies showed that the d1-4 mutant exhibited a mild phenotype, characterized by bigger seeds and higher plants in contrast to other mutants (Oki et al., 2009b). Most d1 mutants contain deletions (Ashikari et al., 1999; Chen et al., 2011; Fujisawa et al., 1999; Oki et al., 2009b), and three d1 mutants contained stop codons (Oki et al., 2009b). We found a novel d1 mutant, which contained an Ors4 copia retrotransposon in exon 7. This insert resulted in loss of exon 7 in d1 mRNA of the d1-a mutant. Compared with some severe d1 mutants (d1-1, -2, -3, -5 etc.) (Oki et al., 2009b), the d1-a mutant is predicted to be a mild mutant, and we found no obvious difference in plant height between d1-a mutant and wildtype plants at the tillering stage (Fig. 1E). Our results revealed a D1-like gene followed by two LTR-retrotransposon remnants (Fig. 4B), which implied that the D1-like gene may have been truncated by recombination between retrotransposons after D1 gene duplication. The D1-like gene may exist in all cultivated rice.
Osr4 LTR-retrotransposon was firstly identified in 2002 (McCarthy et al., 2002). The length of the LTR retrotransposon reference copy is 5665bp, and its LTR length is 350bp (McCarthy et al., 2002; Vitte et al., 2007). However, up to this date, the Osr4 copies have not been characterized. We carefully analyzed Osr4 retrotransposons in Nipponbare, and revealed 65 homologs. We predicted six possible active retrotransposons, which contained all motifs in LTR, a complete CDS, PBS and PPT (Table S2 and Fig. 6B). The members of group I-1 could transpose recently, and included all possible active retrotransposons. One of Osr4 retrotransposons inserted into D1 gene in the d1-a mutant, and resulted in loss of gene function. The Osr4 retrotransposon is expressed in all tissues. These findings suggested that at least one Osr4 retrotransposon was activated. Osr4 retrotransposon expression in some seeds and leaves was higher than in other tissues (Fig. S2), but the transposition that occurred in somatic cells may have produced plant mosaics, and only the transposition which occurred in sexual cells could be inherited. Osr4 retrotransposon maintained a certain level of expression in pollens and ovaries (Fig. S2), and the heritability of the novel transposon should not be too low. The high similarities among the members in group Ⅰ-1, which included all predicted functional retrotransposons, support these findings.
Author's contributionsXS conceived the study and designed the experiments. JC and HZ identified the gene and the insert. JC, HZ, ZS, XZ and XS analyzed the data. KL and YG completed the field experiments. XS drafted the manuscript.
AcknowledgementsThis work was supported by National and Fujian Provincial Natural Science Foundation (No. 31371557 and K55NI921A), and Fujian Provincial Department of Education Project (K58MLV03A) and a startup fund from Fujian Agriculture and Forestry University (http://www.fafu.edu.cn/) to X.S.
Appendix A. Supplementary dataSupplementary data related to this article can be found at http://dx.doi.org/10.1016/j.pld.2017.01.003.
Ashikari M., Wu J., Yano M., Sasaki T., Yoshimura A., 1999. Rice gibberellininsensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTPbinding protein. Proc. Natl. Acad. Sci. U.S.A, 96: 10284-10289. DOI:10.1073/pnas.96.18.10284 |
Bilgin D.D., DeLucia E.H., Clough S.J., 2009. A robust plant RNA isolation method suitable for Affymetrix GeneChip analysis and quantitative real-time RT-PCR. Nat. Protoc, 4: 333-340. DOI:10.1038/nprot.2008.249 |
Chen H.-X., Zhou C.-B., Xing Y.-Z., 2011. A new rice dwarf1 mutant caused by a frame-shift mutation. Hered. (Beijing), 33: 397-403. DOI:10.3724/SP.J.1005.2011.00397 |
Daviere J.M., Achard P., 2013. Gibberellin signaling in plants. Development, 140: 1447-1151. |
Dhadi S.R., Xu Z., Shaik R., Driscoll K., Ramakrishna W., 2015. Differential regulation of genes by retrotransposons in rice promoters. Plant Mol. Biol, 87: 603-613. DOI:10.1007/s11103-015-0300-7 |
Fujisawa Y., Kato H., Iwasaki Y., 2001. Structure and function of heterotrimeric G proteins in plants. Plant Cell Physiol, 42: 789-794. DOI:10.1093/pcp/pce111 |
Fujisawa Y., Kato T., Ohki S., Ishikawa A., Kitano H., Sasaki T., Asahi T., Iwasaki Y., 1999. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. U.S.A, 96: 7575-7580. DOI:10.1073/pnas.96.13.7575 |
Grandbastien M.A., 2015. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta, 1849: 403-416. DOI:10.1016/j.bbagrm.2014.07.017 |
Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M., 1996. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U.S.A, 93: 7783-7788. DOI:10.1073/pnas.93.15.7783 |
Ishikawa A., Tsubouchi H., Iwasaki Y., Asahi T., 1995. Molecular cloning and characterization of a cDNA for the alpha subunit of a G protein from rice. Plant Cell Physiol, 36: 353-359. DOI:10.1093/oxfordjournals.pcp.a078767 |
Kimple R.J., Kimple M.E., Betts L., Sondek J., Siderovski D.P., 2002. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature, 416: 878-881. DOI:10.1038/416878a |
Kumar A., Bennetzen J.L., 1999. Plant retrotransposons. Annu. Rev. Genet, 33: 479-532. DOI:10.1146/annurev.genet.33.1.479 |
Ma J., Devos K.M., Bennetzen J.L., 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res, 14: 860-869. DOI:10.1101/gr.1466204 |
McCarthy E.M., Liu J., Lizhi G., McDonald J.F., 2002. Long terminal repeat retrotransposons of Oryza sativa. Genome Biol, 3(0053): 1-11. |
Michelmore R.W., Paran I., Kesseli R.V., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A, 88: 9828-9832. DOI:10.1073/pnas.88.21.9828 |
Miura K., Agetsuma M., Kitano H., Yoshimura A., Matsuoka M., Jacobsen S.E., Ashikari M., 2009. A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc. Natl. Acad. Sci. U.S.A, 106: 11218-11223. DOI:10.1073/pnas.0901942106 |
Oki K., Kitagawa K., Fujisawa Y., Kato H., Iwasaki Y., 2009a. Function of alpha subunit of heterotrimeric G protein in brassinosteroid response of rice plants. Plant Signal Behav, 4: 126-128. DOI:10.4161/psb.4.2.7627 |
Oki K., Inaba N., Kitano H., Takahashi S., Fujisawa Y., Kato H., Iwasaki Y., 2009b. Study of novel d1 alleles, defective mutants of the alpha subunit of heterotrimeric G-protein in rice. Genes Genet. Syst, 84: 35-42. DOI:10.1266/ggs.84.35 |
Oki K., Inaba N., Kitagawa K., Fujioka S., Kitano H., Fujisawa Y., Kato H., Iwasaki Y., 2009c. Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol, 50: 161-172. DOI:10.1093/pcp/pcn182 |
Suharsono U., Fujisawa Y., Kawasaki T., Iwasaki Y., Satoh H., Shimamoto K., 2002. The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. U.S.A, 99: 13307-13312. DOI:10.1073/pnas.192244099 |
Takeda S., Sugimoto K., Otsuki H., Hirochika H., 1999. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J, 18: 383-393. DOI:10.1046/j.1365-313X.1999.00460.x |
Ueguchi-Tanaka M., Fujisawa Y., Kobayashi M., Ashikari M., Iwasaki Y., Kitano H., Matsuoka M., 2000. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. U.S.A, 97: 11638-11643. DOI:10.1073/pnas.97.21.11638 |
Urano D., Jones A.M., 2014. Heterotrimeric G protein-coupled signaling in plants. Annu. Rev. Plant Biol, 65: 365-384. DOI:10.1146/annurev-arplant-050213-040133 |
Vitte C., Panaud O., Quesneville H., 2007. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics, 8: 218. DOI:10.1186/1471-2164-8-218 |
Waldie T., McCulloch H., Leyser O., 2014. Strigolactones and the control of plant development: lessons from shoot branching. Plant J, 79: 607-622. DOI:10.1111/tpj.2014.79.issue-4 |
Wang Z., Ye S., Li J., Zheng B., Bao M., Ning G., 2011. Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol, 11: 109. DOI:10.1186/1472-6750-11-109 |
Zhang C., Bai M.Y., Chong K., 2014. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep, 33: 683-696. DOI:10.1007/s00299-014-1578-7 |
Zheng K., Subudhi P.K., Domingo J., Maopantay G., Huang N., 1995. Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet. Newslett, 12: 48. |



