b. Biological Chemistry and Crop Protection, Rothamsted Research, Harpenden, UK
Endophytic fungi are microorganisms that establish an endosymbiotic relationship with their host plants. These plants benefit ecologically because their growth and tolerance to environmental stresses are increased by the presence of those symbionts. Endophytic fungi can also be important and novel sources of natural bioactive compounds with potential applications in agricultural, medicinal, and food industries (Strobel, 2003). Plant-associated fungi might also produce the same bioactive compounds as their host plants. This has been demonstrated by gibberellins generated in Fusarium fujikuroi and taxol obtained from endophytic fungi associated with Taxus brevifolia (Strobel, 2003).
Endophytic fungi can be isolated from surface-sterilized plant parts or internal tissues (Vesterlund et al., 2011). Among the large number of endophytes already isolated, some fungi produce bioactive substances that are involved in the relationship between endophyte and host. These fungi are ubiquitous in algae (Hawksworth, 1987), lichens (Li et al., 2007), and mosses (Schulz et al., 1993), as well as in conifer species (Carroll and Carroll, 1978) and angiosperms (Gond et al., 2007). However, only 6-7% of such fungi have been identified within this microbial world. Although numerous studies have focused on endophytic fungi in temperate plants, the fungal endophytes of tropical plants must be explored if we are to identify the remaining 93% (Murali et al., 2007).
Both culture-dependent and-independent approaches are often taken to examine the structure of plant-associated endophytic communities and isolate endophytic fungi. However, culturedependent methods are somewhat limited when studying biodiversity because only a small fraction of the fungal populations can be recovered when using various culture media. Alternatively, culture-independent techniques are based on the analysis of DNA extracted directly from plant samples, thereby overcoming such limitations. Despite these advances in identifying endophytes, however, fast-growing fungi are still isolated preferentially while unculturable and/or slow-growing fungi can escape detection (Duong et al., 2006). Nevertheless, culture-independent methods are currently the most effective means for screening fungal diversity from natural samples.
Medicinal plants are of great interest to many researchers because their compounds account for a large proportion of the current pharmacopoeia. Natural products are the main source of active ingredients in medicines. Over 80% of all drug substances, including most anti-cancer and anti-infective agents, are either natural products or inspired by a natural compound (McChesney et al., 2007). Paris polyphylla (Liliaceae) is highly valued because its rhizomes are widely used in traditional Chinese medicine for treating various inflammations and injuries. As an important component of Chinese patent medicines, such as "Biyan Qingdu Keli", these plants help treat chronic rhinitis and nasopharyngeal cancer (Zhao et al., 2010). In India, this species is found in Manipur, Uttarakhand, Himachal Pradesh, Lushai, and the Aka Hills (Tiwari et al., 2010). The main active ingredients of P. polyphylla are steroidal saponins, of which at least 30 have now been isolated from rhizomes through phytochemical methods (Zhao et al., 2010). Although the demand for these rhizomes has been increasing in China, natural populations are declining significantly due to slow plant growth and over-harvesting. This species is at high risk of extinction (Zhang et al., 2011). Therefore, to meet commercial needs, it is urgent that researchers develop an effective way to increase rhizome yields or saponin concentrations through artificial cultivation. If new procedures can be devised for producing the endophytic fungi associated with this species, the dependence on wild resources can be decreased.
Saponin concentrations in P. polyphylla increase as plants age (Conglong et al., 2011). The diversity of endophytic fungi in this species has been investigated using cultural methods (Li et al., 2008). However, that study did not indicate the age of the samples nor consider any link between endophyte diversity and plant maturation. Therefore, the objective of our experiments was to elucidate the changes in diversity of endophyte fungi while P. polyphylla rhizomes are developing and to determine whether this diversity is correlated with saponin levels.
2. Materials and methods 2.1. Plant materialsHealthy rhizomes were collected in August 2014 from four-, six-, and eight-year-old plants of P. polyphylla that were grown in the germplasm resource garden of Yunnan Agriculture University, Kunming, China. The exact age is guaranteed because of their growth from seeding. Triplicate sampling was done with three plants per age class. All samples were placed in a cooler and immediately transported to the laboratory where they were refrigerated at 4 ℃. The rhizomes were processed within 48 h of being collected, and taxonomic identifications of all plant materials were performed by Prof. Zhongjun He (Yunnan Agriculture University). Voucher specimens were deposited in the Faculty of Agronomy and Biotechnology, Yunnan Agriculture University.
2.2. Fungal isolation and identificationEndophytic fungi were isolated based on the procedures described by Li et al. (2007). After the samples were cleaned and cut into segments (5 mm × 5 mm × 5 mm), they were surfacedisinfected by washing in 75% ethanol for 1 min, then rinsed twice in sterile distilled water, and placed in a 0.05 mg L-1 sodium hypochlorite solution for 3 min before being rinsed several times in sterile distilled water. The surface-sterilized samples were placed on plates of potato dextrose agar medium (PDA; HiMedia) containing a final concentration of 50 mg L-1 of the antibiotic streptomycin sulphate. After incubation in the dark at 28 ℃ for approximately one month, the segments were examined periodically. When the colonies developed, they were transferred to new Petri dishes filled with PDA. The fungi were isolated and subcultured to obtain pure cultures, which were then allowed to grow for 14 d before being subjected to morphological examination. The fungal specimens were deposited at Yunnan Agricultural University.
2.3. DNA extraction, PCR amplifications, and phylogenetic analysisThe endopytic fungi genomic DNA was extracted from the mycelia using a DNeasy Plant Mini Kit (Qiagen, USA) according to the manufacturer's protocol. The DNA samples were diluted in 50 mL of TE buffer containing RNase (10 mg mL-1), incubated at 37 ℃ for 2 h, and examined on a 1% agarose gel containing ethidium bromide. Total DNA was stored at-20 ℃. The ITS region in the ribosomal DNA was amplified using forward primer ITSF (5'-CTTGGTCATTTAGAGGAAGTAA-3') and reverse primer ITSR (5'-TCCTCCGCTTATTGATATG-3') (Julian and Alga, 2006). Each PCR reaction was prepared in a mixture (50 mL total volume) containing 10 mM for each dNTP, 1 × PCR buffer with MgCl2, 0.6 U of Taq DNA polymerase, 0.8 mM for each primer, and 20 ng of total DNA template. All PCR amplifications were performed in a Thermal Cycler (Bio-Rad, USA) under parameters that included an initial denaturation at 95 ℃ for 5 min; then 35 cycles of denaturation at 95 ℃ for 35 s, annealing at 55 ℃ for 55 s, and extension at 72 ℃ for 2 min; followed by a final extension at 72 ℃ for 10 min. Products were analyzed on a 1% agarose gel and visualized under a UV transilluminator. The PCR fragments of the expected size (about 500 bp) were excised from the gel with a clean, sharp scalpel and sent to the Beijing Genomics Institute (BGI) for sequencing. Chromatograms were visually checked and edited using BioEdit software. Homology searches of sequences were conducted with BLAST algorithm software at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). Multiple sequence alignments of the obtained sequences and reference sequences from GenBank were performed using CLUSTALW.
2.4. Plant metagenomic DNA extraction and retrieval of ITS sequencesA Qiagen Stool Kit (QIAGEN Inc.) was used to extract total metagenomic DNA from the P. polyphylla samples, following the manufacturer's instructions. We used PCR to amplify the endophytic fungi ITS with the extracted metagenomic DNA (combined to form one per age class). The universal primers included ITS1f (5'-TGGTCATTTAGAGGAAGTAAAA-3') and ITS1r (5'-GCTGCGTTCTTCATCGATGC-3') for the fungal ITS fragments (Julian and Alga, 2006). The reaction program was as follows: 94 ℃ for 5 min; then 35 cycles of 94 ℃ for 30 s, 55 ℃ for 45 s, and 72 ℃ for 90 s; then holding at 4 ℃ in the 50-μL PCR volume. Products were examined on a 1% agarose gel. The PCR amplicon was purified with a Qiagen Gel and PCR Purification Kit and sequenced by BGI according to the Illumina paired-end protocol to generate millions of reads.
2.5. Sequence analysisFor analyzing fungal diversity, generated ITS sequences that were less than 140 bp long after quality-trimming were excluded. The Fungal ITS Extractor 1.1 (Nilsson et al., 2009) was applied to obtain the variable ITS subregion of fungal ITS sequences and to exclude any portions of neighboring ribosomal genes. Raw sequences without pyrotags and forward primer sequences, as well as non-fungal ITS sequences, were also excluded. Any ITS chimeric sequences were removed with a BLAST-based open source software package. The resultant sequences for individual samples were subjected to the pyrosequencing analysis pipeline at the UNITE database, and diversity within the fungal community was examined based on species available from the NCBI website.
3. Results and discussionWe used culture-dependent and-independent techniques with the Hiseq2000 platform to examine possible connections between the high diversity of endophytic microorganisms and any agerelated variations in steroidal saponin concentrations in P. polyphylla. The culture-based approach involved 270 segments (90 per age class) and revealed 147 isolates belonging to 18 well-identified taxa of endophytic fungi. Testing of our three age classes indicated significant variations in colonization frequency. The largest number of endophytes was recovered from eight-year-old rhizomes (49%), followed by four-year-old (30%) and six-year-old (21%) rhizomes (Fig. 1). The richness and number of endophyte isolates were highest in the older rhizomes. This was possibly due to longer colonization times and more favorable habitats for fungal establishment.

|
| Fig. 1 Recovery of endophytic fungal isolates from Paris polyphylla rhizomes at different ages. |
Numerous biotic and abiotic factors influence endophytic infections and colonization, including the chemistry and structure of the host tissue and the environmental conditions to which a plant is exposed (Sanchez-Azofeifa et al., 2012). Therefore, higher concentrations of anti-fungal and anti-herbivore substances may have reduced the opportunity for colonization in our younger P. polyphylla rhizomes. Similar observations have been reported for the leaves of Bauhinia brevipes (Hilarino et al., 2011). Among the taxa identified here, Fusarium oxysporum (46.55%), Leptodontidium sp. (8.66%), Trichoderma viride (6.81%), Penicillium chrysogenum (5.55%), Paraphaeosphaeria sporulosa (4.54%), and Plectosphaerella cucumerina (4.08%) were the most frequently isolated species (Table 1). As also concluded by Li et al. (2008), F. oxysporum is one of the most diverse phyto-pathogenic fungi and is widely dispersed throughout the soil, where it degrades lignin and complex carbohydrate molecules associated with soil debris. This fungus has also been isolated from healthy rhizomes of Dioscorea zingiberensis (Zhang et al., 2010), Ginkgo biloba (Yuna et al., 2012), and Cinnamomum kanehirae (Quan-Xin et al., 2011). Bazin et al. (1990) reported that many soil bacteria and fungi are able to colonize epidermal and outer cortical cells of healthy roots inter-and intracellularly. That may be the reason why the great richness of F. oxysporum is in according with the soil.
| Sr. No. | Fungus | Relative frequency (%) | ||
| 4 years | 6 years | 8 years | ||
| 1 | Alternaria sp. | 4.55 | ||
| 2 | Cylindrocarpon sp. | 9.09 | ||
| 3 | Chaetomium globosum | 3.70 | ||
| 4 | Chaetomium sp. | 6.82 | ||
| 5 | Fusarium oxysporum | 48.00 | 66.67 | 25.00 |
| 6 | Fusarium redolens | 9.26 | ||
| 7 | Fusariumsp. | 8.00 | ||
| 8 | Leptodontidiumsp. | 26.00 | ||
| 9 | Plectosphaerella cucumerina | 4.00 | 3.70 | 4.55 |
| 10 | Paraphaeosphaeria sporulosa | 13.64 | ||
| 11 | Pyrenochaeta sp. | 4.55 | ||
| 12 | Penicillium chrysogenum | 16.67 | ||
| 13 | Penicillium swiecickii | 6.82 | ||
| 14 | Trichoderma viride | 20.45 | ||
| 15 | Trichoderma ovalisporum | 4.00 | 2.27 | |
| 16 | Trichoderma gamsii | 6.00 | ||
| 17 | Truncatella angustata | 2.27 | ||
| 18 | Trichocladium opacum | 4.00 | ||
In contrast to the fungal populations reported by Li et al. (2008), we noted that P. polyphylla was associated with other taxa (in addition to F. oxysporum). This discrepancy might be explained by differences in growth environments and experimental materials between the two studies, because endophyte diversity is easily affected by environment. For example, the fungal colony in Truncatella angustata was the least common, with a colonization frequency of only 0.75%. This species has also been isolated from healthy vines of grape (Vitis vinifera), and is sometimes regarded as a pathogen in that plant (Diaz et al., 2011).
Some fungal taxa associated with our P. polyphylla samples were specific to age class. These included Fusarium sp., Leptodontidium sp., Trichoderma gamsii, and Trichocladium opacum, which were found only with four-year-old rhizomes. Likewise, Chaetomium globosum, F. redolens, and P. chrysogenum occurred only with six-year-old rhizomes, while Alternaria sp., Cylindrocarpon sp., Chaetomium sp., P. sporulosa, Pyrenochaeta sp., Penicillium swiecickii, T. viride, and T. angustata were specific to eight-year-old rhizomes. Several factors may affect plant-associated microbial communities, such as anthropic factors (Rasche et al., 2006), plant physiology (Islam et al., 2010), environmental conditions (Yousaf et al., 2010), and pathogen infections (Bulgari et al., 2011). Moreover, a shift in the composition of those communities can be driven by genetic and physiological diversity (Manter et al., 2010). Thus, alterations in the developmental physiology of the rhizomes may change the compositional diversity of endophytic fungi in plants at different stages of maturity (c.f., Conglong et al., 2011). Our eight-year-old rhizomes had special endophytic fungi, e.g., Alternaria sp., Cylindrocarpon sp., Chaetomium sp., P. sporulosa, Pyrenochaetasp., P. swiecickii, T. viride, and T. angustata. The most abundant fungus, T. viride, is free-living in the soil and root ecosystems, and can produce a wide range of antibiotic substances (Woo et al., 2006). Although we found no direct evidence that these fungi were responsible for elevated saponin production, our results suggest that fungal diversity is accompanied by higher concentrations of saponin.
Overall, 51.51% of the isolates identified here belonged to the Sordariomycetes, 9.09% to the Dothideomycetes, 6.06% to the Eurotiomycetes, and 3.03% to the Ascomycetes (Fig. 2). However, we did not recover any Ascomycetes species from six-and eight-yearold rhizomes, nor did we find any Dothideomycetes in four-and six-year-old plants, or any Eurotiomycetes in four-year-old plants. By comparison, members of Sordariomycetes were present in all three age classes. These high frequencies demonstrated how easily fungi can colonize roots and shoots and efficiently communicate with the host plant via chemical signals, such as sugar, hormone et al. (Hilarino et al., 2011). Some strains may be associated with the stimulation of plant growth or biomass production, as well as protection against pathogens, the supply of nutrients and hormones to host plants, suppression of diseases caused by pathogens, or alleviation of abiotic stress (Hermosa et al., 2012). Among the Ascomycetes, we found only Leptodontidium sp. in four-year-old rhizomes (Table 1). Porras-Alfaro and Bayman (2011) have suggested that L. orchidicola belongs to an artificial group, i.e., the "dark septate endophytes". This fungus can promote plant growth and protect its host from various stresses by secreting secondary metabolites. Eurotiomycetes are common fungal endophytes. However, we found Dothideomycetes only in the eight-year-old rhizomes. Some members of this group can produce a variety of metabolites (Samson and Frisvad, 2004). For example, Paraphaeosphaeria quadriseptata in Dothideomycetes has potential anticancer activity (Turbyville et al., 2006). Thus, it is possible that these fungi have an important role in secondary metabolism in the rhizomes, but their functions, especially those of the Dothideomycetes, will require further study.
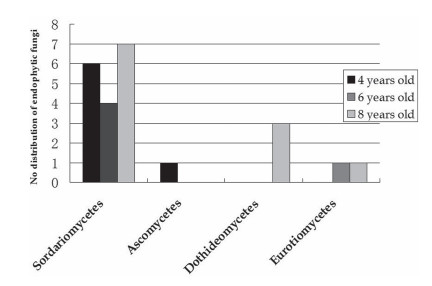
|
| Fig. 2 Distribution of endophytic fungi sampled from rhizomes of Paris polyphylla at different ages. |
To improve our understanding of the interactions between plants and fungal communities in the rhizomes of P. polyphylla, we used ITS pyrosequencing analysis of the polymerase chain reaction (PCR)-amplified internal transcribed spacer (ITS) gene. This approach is designed to provide an overview of fungal community profiles that encompass a range of fundamental processes associated with ecosystem functioning, such as organic matter decomposition, nutrient cycling, and plant growth stimulation (U'Ren et al., 2014). Here, Good's coverage values (calculated at a 97% similarity cut-off) indicated that the number of pyrosequencing reads obtained from our samples was sufficient to capture the microorganism diversity inP. polyphylla. Various genera, including Alternaria, Chaetomium, Fusarium, Plectosphaerella, Pyrenochaeta, and Penicillium were detected by both isolation and ITS pyrosequencing methods. The latter means revealed a more complex microbiota than that obtained through fungal cultures. However, ITS pyrosequencing analysis of total fungal DNA did not present any bands corresponding to several representative species, including isolates for Cylindrocarpon sp., Leptodontidium sp., P. sporulosa, T. viride, T. ovalisporum, T. gamsii, T. angustata, and T. opacum (Table 2). This was the case even though our colony cultures had indicated that those species were predominant members of the microbiota. Biases in total DNA extraction from these strains was likely the reason that those were not recovered from the rhizome samples. Muyzer et al. (1993) have also shown that some species representing very small communities are not visible in profiles of microbial communities. It is possible that the enrichment of fungi in the growth medium may result in the proliferation of only a few original samples at the expense of reduced competition and stimulated metabolism. In contrast, we were unable to recover Neonectria sp. with our cultivation techniques even though it often occurs in plants and exhibits strong antimicrobial activity (Bigelis et al., 2006). We hypothesize that some species that were detected by sequencing but not isolated by plating may play an important role in secondary metabolism in the rhizomes. Therefore, meta-transcriptomic analysis may allow us to identify key genes that are expressed during various processes of plant growth and development.
| Sr. No. | Fungus | Relative Frequency (%) | ||
| 4 years | 6 years | 8 years | ||
| 1 | Unknown | 79.248 | 76.636 | 67.578 |
| 2 | Acremonium | 0.008 | 0 | 0.118 |
| 3 | Alternaria | 0.006 | 0 | 0.010 |
| 4 | Archaeorhizomyces | 0.030 | 0 | 0 |
| 5 | Aspergillus | 0.118 | 0 | 0.174 |
| 6 | Bionectria | 0 | 0.008 | 0.064 |
| 7 | Candida | 0.032 | 0 | 0 |
| 8 | Chaetomium | 0.004 | 0.002 | 0.024 |
| 9 | Cladosporium | 0.018 | 0.008 | 0.032 |
| 10 | Dipodascus | 0.036 | 0 | 0 |
| 11 | Doratomyces | 0 | 0.002 | 0 |
| 12 | Fusarium | 18.070 | 14.944 | 11.120 |
| 13 | Galactomyces | 0.038 | 0 | 0 |
| 14 | Gibberella | 0 | 0.356 | 0.336 |
| 15 | Gliomastix | 0 | 0 | 0.046 |
| 16 | Hirsutella | 0 | 0.004 | 0.024 |
| 17 | Ilyonectria | 0 | 0 | 0.004 |
| 18 | Kodamaea | 0 | 0 | 0.028 |
| 19 | Lophiostoma | 0.002 | 0 | 0.024 |
| 20 | Microdochium | 0 | 0.038 | 0 |
| 21 | Myrothecium | 0.016 | 0.016 | 0 |
| 22 | Nectria | 0 | 0 | 0.018 |
| 23 | Neonectria | 0.046 | 0.048 | 0.988 |
| 24 | Penicillium | 0.088 | 0.010 | 0.332 |
| 25 | Pichia | 0.006 | 0 | 0.010 |
| 26 | Plectosphaerella | 02.164 | 07.866 | 18.548 |
| 27 | Pyrenochaeta | 0 | 0.052 | 0.422 |
| 28 | Rhodotorula | 0 | 0.002 | 0.056 |
| 29 | Schizophyllum | 0.002 | 0.008 | 0.028 |
| 30 | Spencermartinsia | 0.006 | 0 | 0 |
| 31 | Stachybotrys | 0.062 | 0 | 0 |
| 32 | Thyrostroma | 0 | 0 | 0.016 |
Older plants of P. polyphylla have higher levels of saponin (Conglong et al., 2011). Increased concentrations in our eight-year-old rhizomes might have been associated with enhanced microorganism abundance and activity, even though we could not absolutely demonstrate that here. Endophytes can promote plant health and growth, elicit plant metabolism, or directly produce biotechnologically relevant compounds (Brader et al., 2014). Furthermore, Zhao et al. (2010) have described the potential of developing metabolites from the endophyte P. guilliermondii as antimicrobial agents. Their report indirectly supports our hypothesis. In this context, the characterization of uncultivable microorganism communities is a fundamental step in strengthening our understanding of the important role that endophytes have in the metabolism and accumulation of secondary metabolites. Thus, our findings offer a preliminary but very promising prospect for improving the cultivation of P. polyphylla through the use of endophytes.
AcknowledgementsThis study was supported by grants from the National Natural Science Foundation of China (81473310, 31260075, 31560085).
Bazin, M. J. , Markham, P. , Scott, E. M. , Lynch, J. M. , 1990. Population dynamics and rhizosphere interactions. In: Lynch, J. M. (Ed. ), The Rhizosphere. Wiley, Chichester, UK, pp. 99-127.
|
Bigelis R., He H.Y., Yang H.Y., Chang L.P., Greenstein M., 2006. Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J. Ind. Microbiol. Biotechnol, 33: 815-826. |
Brader G., Compant S., Mitter B., Trognitz F., Sessitsch A., 2014. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol, 27: 30-37. DOI:10.1016/j.copbio.2013.09.012 |
Bulgari D., Casati P., Crepaldi P., Daffonchio D., Quaglino F., Brusetti L., AttilioBianco P., 2011. Restructuring of endophytic bacterial communities in grapevine yellows-diseased and recovered Vitis vinifera L. plants. Appl. Environ. Microbiol, 77: 5018-5022. DOI:10.1128/AEM.00051-11 |
Carroll G.C., Carroll F.E., 1978. Studies on the incidence of coniferous needle endophytes in the Pacific Northwest. Can. J. Bot, 56: 3034-3043. DOI:10.1139/b78-367 |
Conglong X., Jie Z., Shuangshuang L., 2011. HPLC fingerprints of Paris polyphylla Smith var. yunnanensis of different growing years. Chin. J. Mod. Appl. Pharm, 28: 515-519. |
Diaz G.A., Prehn D., Besoain X., Chavez E.R., 2011. Neofusicoccum parvum associated with grapevine trunk diseases in Chile. Plant Dis, 95: 1032-1033. |
Duong L.M., Jeewon R., Lumyong S., Hyde K.D., 2006. DGGE coupled with ribosomal DNA gene phylogenies reveal uncharacterized fungal phylotypes. Fungal Divers, 23: 121-138. |
Gond S.K., Verma V.C., Kumar A., Kumar V., Kharwar R.N., 2007. Study of endophytic fungal community from different parts of Aegle marmelos Correae (Rutaceae) from Varanasi (India). World J. Microbiol. Biotechnol, 23: 1371-1375. DOI:10.1007/s11274-007-9375-x |
Hawksworth D.L., 1987. Observations on three algicolus microfungi. Notes R. Bot. Gard. Edinb, 44: 549-560. |
Hermosa R., Viterbo A., Chet I., Monte E., 2012. Plant-beneficial effects of Trichoderma and of its genes. Microbiology, 158: 17-25. DOI:10.1099/mic.0.052274-0 |
Hilarino M.P.A., Silveira F.A.O., Oki Y., Rodrigues L., Santos J.C., Junior A.C., Fernandes G.W., Rosa C.A., 2011. Distribution of the endophytic fungi community in leaves of Bauhinia brevipes (Fabaceae). Acta Bot. Bras, 25: 815-821. |
Islam S.M.A., Math R.K., Kim J.M., Yun M.G., Cho J.J., Kim E.J., Lee Y.H., Yun H.D., 2010. Effect of plant age on endophytic bacterial diversity of balloon flower (Platycodon grandiflorum) root and their antimicrobial activities. Curr. Microbiol, 61: 346-356. DOI:10.1007/s00284-010-9618-1 |
Julian I.M., Alga Z., 2006. Sequences, the environment and fungi. Mycologist, 20: 62-74. |
Li J., Zhao J.L., Xu L.J., 2008. Endophytic fungi from rhizomes of Paris polyphylla var. yunnanensis. World J. Microbiol. Biotechnol, 24: 733-737. |
Li W.C., Zhou J., Guo S.Y., Guo L.D., 2007. Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Divers, 25: 69-80. |
Manter D.K., Delgado J.A., Holm D.G., Stong R.A., 2010. Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb. Ecol, 60: 157-166. DOI:10.1007/s00248-010-9658-x |
McChesney J.D., Venkataraman S.K., Henri J.T., 2007. Plant natural products: back to the future or into extinction?. Phytochemistry, 68: 2015-2022. DOI:10.1016/j.phytochem.2007.04.032 |
Murali T.S., Suryanarayanan T.S., Venkatesan G., 2007. Fungal endophyte communities in two tropical forests of southern India: diversity and host affiliation. Mycol. Prog, 6: 191-199. DOI:10.1007/s11557-007-0540-2 |
Muyzer G., De Waal E.C., Uitterlinden A.G., 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol, 59: 695-700. |
Nilsson R.H., Bok G., Ryberg M., Kristiansson E., Hallenberg N., 2009. A software pipeline for processing and identification of fungal ITS sequences. Source Code Biol. Med, 4: 1751-1763. |
Porras-Alfaro A., Bayman P., 2011. Hidden fungi, emergent properties: endophytes and microbiomes. Annu. Rev. Phytopathol, 49: 291-315. DOI:10.1146/annurev-phyto-080508-081831 |
Quan-Xin W., Sai-Fei L., Feng Z., 2011. Chemical constituents from endophytic fungus Fusarium oxysporum. Fitoterapia, 82: 777-781. DOI:10.1016/j.fitote.2011.04.002 |
Rasche F., Hodl V., Poll C., Kandeler E., Gerzabek M.H., van Elsas J.D., Sessitsch A., 2006. Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol. Ecol, 56: 219-235. |
Samson, R. A. , Frisvad, J. C. , 2004. Penicillium subgenus Penicillium: New Taxonomic Schemes, Mycotoxins and Other Extrolites. In: Studies in Mycology, No. 49. Centraalbureau voor Schimmelcultures, Utrecht, p. 260.
|
Sanchez-Azofeifa A., Oki Y., Fernandes G.W., 2012. Relationships between endophyte diversity and leaf optical properties. Trees, 26: 291-299. DOI:10.1007/s00468-011-0591-5 |
Schulz B., Wanke U., Draeger S., Aust H.J., 1993. Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycol. Res, 97: 1447-1450. |
Strobel G.A., 2003. Endophytes as sources of bioactive products. Microb. Infect, 5: 535-544. |
Tiwari J.K., Ballabha R., Tiwari P., 2010. Ethnopaediatrics in Garhwal Himalaya, Uttarakhand, India (psychomedicine and medicine). N. Y. Sci. J, 3: 123-126. |
Turbyville T.J., Wijeratne E.M., Liu M.X., Burns A.M., Seliqa C.J., Luevano L.A., Faeth S.H., Whitesell L., Gunatilaka A.A., 2006. Search for Hsp90 inhibitors with potential anticancer activity: isolation and SAR studies of radicicol and monocillin I from two plant-associated fungi of the Sonoran desert. J. Nat. Prod, 69: 178-184. DOI:10.1021/np058095b |
U'Ren J.M., Riddle J.M., Monacell J.T., Carbone I., Miadlikowska J., Arnold A.E., 2014. Tissue storage and primer selection influence pyrosequencing-based inferences of diversity and community composition of endolichenic and endophytic fungi. Mol. Ecol. Res, 14: 1032-1048. |
Vesterlund S.R., Helander M., Faeth S.H., 2011. Environmental conditions and host plant origin override endophyte effects on invertebrate communities' structure and guilds. Fungal Divers, 47: 109-118. DOI:10.1007/s13225-011-0089-x |
Woo S.L., Scala F., Ruocco M., Lorito M., 2006. The molecular biology of the interactions between Trichoderma spp, phytopathogenic fungi and plants. Phytopathology, 96: 181-185. DOI:10.1094/PHYTO-96-0181 |
Yousaf S., Andria V., Reichenauer T.G., Smalla K., Sessitsch A., 2010. Phylogenetic and functional diversity of alkane degrading bacteria associated with Italian ryegrass (Lolium multiflorum) and Birdsfoot trefoil (Lotus corniculatus) in a petroleum oil-contaminated environment. J. Hazard Mater, 184: 523-532. DOI:10.1016/j.jhazmat.2010.08.067 |
Yuna C., Dawei Y., Xiufeng B., Sun B., Zhao Y., Zhang Y., 2012. Ginkgolide B produced endophytic fungus (Fusarium oxysporum) isolated from Ginkgo biloba. Fitoterapia, 83: 913-920. DOI:10.1016/j.fitote.2012.04.009 |
Zhang M., Li Y.W., Li Z.Y., Huang X.L., Zhu D., Liu Q.S., 2011. Progress on studies of endangered ethno-medicine of Rhizoma Paris. J. Minzu Univ. China, 20: 65-69. |
Zhang R., Li P., Zhao J., Shan T., Yin C., Zhou L., 2010. Endophytic fungi from Dioscorea zingiberensis and their effects on the growth and diosgenin production of the host plant cultures. Nat. Prod. Res. Dev, 22: 11-15. |
Zhao J.L., Mou Y., Shan T.J., 2010. Antimicrobial metabolites from the endophytic fungus Pichia guilliermondii isolated from Paris polyphylla var. yunnanensis. Molecules, 15: 7961-7970. |



