Cinnamomum chago is an extremely rare species from the genus Cinnamomum of the family Lauraceae. This species is not well known and has not yet been recorded in Flora of China (Wu, 1991; Li et al., 2008). C. chago is a morphologically distinct, large evergreen tree that grows approximately 25 m tall, with a trunk diameter of 40 cm; leaves opposite or nearly opposite; leaf blade green or yellow-green, ovate-elliptic, 5-13 cm × 2.5-4.5 cm, leathery; venation pinnate with stout midrid, 7e9 pairs of lateral veins, axillary fossa absent or inconspicuous; petiole, 1.8 cm long; panicle terminal or sometimes axillary at the upper part of twig, 5e10 cm long; pedicels 5 mm long, glabrous; flowers yellow-green or redish at the upper part of the perianth lobes; perianth lobes elliptic, ca. 6 mm long; Fertile stamens 9, ca. 5 mm long; filaments pubescent; fruit subglobose or ovoid, green-yellow at maturity, 3 cm long and 2.5 cm in diameter, much bigger in this genus; Perianth cup is flat or discoid, 1.2e1.4 cm in diameter, leathery. Inside the flesh mesocarp, the hard leathery black-brown endocarp is pointed oval, with two hemispheric and white flesh cotyledons inside its embryo (Sun and Zhao, 1991). C. chago is considerably different from other Cinnamomun species because of the following: the perianth and the stamen are glabrous except the filament, the inner side of the perianth lobes is sparsely pubescent, the leaves are opposite but with pinnate venation, the fruit is bigger that grows approximately 3 cm long and 2.5 cm in diameter, and its typical discoid fruit cup. In the recently reported Cinnamomun phylogeny, including C. chago (we provided the material but the name was miswritten as Cinnamomum chaogo), supports the consideration of C. chago as a distinct species, and situates its phylogenetic status between the two sections of the Asian Cinnamomum plants (Sect. Camphora (Trew) Meissn. and Sect. Cinnamomum) (Huang et al., 2016).
Our field investigation found that C. chago is distributed along the Lancang River upstream of the tributary mountains in Yunlong County, Yunnan Province. It exhibits an extremely limited distribution with a fragmented pattern. Furthermore, no seedlings were found near the adult trees during our field survey. Although the species is narrowly distributed and has a low survival rate, its biological characteristics and conservation biology have been scarcely been reported, despite the fact that practical conservation strategies for endangered and rare plants are based on the understanding of biological characteristics and survival status of these plants (Sun et al., 2006; Shen et al., 2009). The present study thus investigated the habitat, biological characteristics, and genetic diversity of C. chago. Furthermore, corresponding conservation and utilization strategies have been proposed based on our results.
2. Materials and methods 2.1. Field investigation of the populationsTo investigate the distribution and the habitat of C. chago, we reviewed several studies and herbarium records and then performed a comprehensive field investigation during the flowering and the fruiting seasons of C. chago in 2014 and 2015. During our field survey, a global positioning device (GPS) was used to record the precise location of the detected species. The altitude, latitude, and longitude of each site were also recorded.
2.2. Biological characteristicsWe collected flowers and fruits during the flowering and the fruiting season, respectively, and then we immediately placed them in FAA to preserve their original characteristics. Using light microscopy, we observed the anatomy of C. chago. Scanning electron microscopy (SEM) was employed to observe pollen morphology. Thirty samples were selected randomly for observation of biological characteristics. These data were consolidated with observations from previous studies.
2.3. Analysis of genetic diversityOur field investigation revealed that the five remaining C. chago populations are distributed in Yunlong and Yangbi County in Yunnan Province. In 2014 and 2015, 54 C. chago samples were collected from these populations (Table 1). The distance between the collected individual samples was at least 15 m. Fresh young leaves were removed from the shoots, and then dried in silica gel and stored at -20 ℃ prior to DNA extraction. Detailed informations regarding the locations and the population codes of the samples are shown in Table 1.
| Population | Number | Longitude | Latitude | Altitude (m) |
| NMP | 10 | 99°16'35.03" | 25°33'46.79" | 2357 |
| DSB | 8 | 99°10'24.24" | 25°45'49.7" | 2317 |
| SBX | 12 | 99°56'28.35" | 25°34'13.27" | 2249 |
| XC | 12 | 99°56'33.40" | 25°34'8.23" | 2296 |
| LG | 12 | 99°55'09.2" | 25°33'08.9' | 2310 |
| NMP: NanMuPing; DSB: DaShiBa; SBX: ShunBiXiang; XC: XingCun; LG: LaGuo. | ||||
DNA was extracted from dried leaves using a modified CTAB method (Doyle and Doyle, 1988). Purified DNA was detected by 1.0% agarose gel electrophoresis and stored at -20 ℃ until use.
2.5. ISSR PCR amplificationDNA samples were randomly selected from mixed DNA of each population. The samples were screened using 100 primers obtained from the University of British Columbia Biotechnology Lab (UBCBL). Each randomly selected sample that has a mixed DNA from each population was used to screen 100 primers obtained from the University of British Columbia Biotechnology Lab (UBCBL). Finally, the 14 primers that generated clear bands with high polymorphism were selected for PCR amplification using a final volume of 20 ml. The reaction mixture consisted of 40 ng of template DNA, 1.5 ml of dNTP, 2 ml of 10x buffer, 1 ml of primer, and 0.3 ml of Taq DNA polymerase. The reaction underwent an initial denaturation for 7 min at 94 ℃, followed by 45 cycles of PCR, which includes 30 s of denaturation at 94 ℃, 45 s of annealing at appropriate annealing temperature, 90 s of extension at 72 ℃, and a final extension step of 7 min at 72 ℃. Subsequently, 5 ml of amplified products were visualized on a 1.5% agarose gels after electrophoresis in 0.5 × TBE at voltage 100 V for 80 min. The gels were photographed using a UV gel imaging system.
2.6. Data analysisThe individuals were scored for the presence (1) or absence (0) of amplified bands. The percentage of the polymorphic loci (PPB), effective number of alleles (Ne), Nei's genetic diversity (H), Shannon's information index (I), estimate of gene flow (Nm), total gene diversity (Ht), variability within populations (Hs), and coefficient of genetic differentiation (Gst) were calculated using POPGENE version 1.32 (Yeh et al., 1999) and GenAlEx version 6.501 (Peakall and Smouse, 2012) with manual corrections.
Analysis of molecular variance (AMOVA) was conducted to calculate the extent of genetic variation between and within the two populations by using GenALEx version 6.501 (Peakall and Smouse, 2012).
We conducted Bayesian analysis of the population structures using STRUCTURE version 2.2 (Pritchard et al., 2000). A total of 20 independent runs were performed for each set with K ranging from 1 to 20, a burn-in of 1 × 105 iterations, and 1 × 105 subsequent Markov Chain Monte Carlo steps. The combination of the admixture and the correlated allele frequency models was also analyzed. The second-order rate of change in the log probability of the data with respect to the number of clusters (ΔK) was also used to estimate the number of genetic clusters (Evanno et al., 2005). The bestfit number of groupings was evaluated using ΔK through STRUCTURE HARVESTER version 0.6.8 (Earl and von Holdt, 2012).
3. Results 3.1. Distribution and habitat of C. chagoInvestigating populations of C. chago during the flowering and fruiting seasons of 2014 and 2015, we found that this species is mainly distributed along the Lancang River upstream of the tributary mountains in Yunlong County, Yunnan Province. The species occurs in five isolated, fragmented populations: ShunBiXiang (SBX), XingCun (XC), LaGuo (LG), DaShBa (DSB), NanMuPing (NMP) individuals were sampled, respectively. SBX, XC, and LG populations of C. chago are distributed near local villages in YangBi County and are exposed to human activities. In contrast, the DSB and NMP populations are distributed in secondary evergreen broad-leaved forest in YunLong County at altitudes between 2100 m and 2400 m. All populations sampled have been subjected to high-frequency anthropogenic interference, particularly during the fruiting seasons when fruits are harvested by local villagers. Thus, the seeds were unable to grow into new seedlings.
3.2. Biological characteristicsPrevious research found that the leaves of C. chago are opposite or nearly opposite. The veins are pinnate and the midrib is burly and evident while the lateral veins are thin and have no glandular fossa. The flowers are bisexual, and greenewhite or yellowish. The perianth is glabrous or puberulent outside and densely pubescent inside. It has 12 stamens in total, with 4 regularly arranged whorls. There are fertile stamens 9 (of 1st, 2nd, and 3rd whorl) and regressive stamens 3 (of 4th whorl) (Sun and Zhao, 1991). Our field work confirmed these observations. We also noted that the pollen surface has small punch and triangular spike grain with cushion bumps at the base. The fruit is drupe, purple-black (color), globular, and large with a diameter of 2.5 cm when fully mature (Fig. 1). The seeds contain various nutrients, including sugar, protein, crude fat, and amino acids, and have been regarded as an ideal nut (unpublished data). The flowering time of C. chago varies slightly between different populations and microhabitats, whereas the period of flowering and fruiting is generally from April to October. Its special morphological feature combination (opposite leaves, pinnate leaf veins, no glandular fossa, drupe large, and pollen surface with triangular spike grain, with cushion bumps at the base, and small punch) indicated that C. chago is a key phylogenetic taxon between the two sections of Asian Cinnamomum plants (Sect. Camphora (Trew) Meissn. and Sect. Cinnamomum).

|
| Fig. 1 Biological characteristic and field investigation of Cinnamomum chago populations. a. habitat; b. Plant morphological; c. flowers; d. pollen; e. fruit; f. seed; g. field work. |
A total of 109 bands were generated by the 14 selected primers that produced 6-13 bands each, producing an average of 7.8 bands. All bands were polymorphic and completely accounted (Table 2). We observed a moderately high level of genetic diversity at the population and species level (population level: Ne=1.629, H=0.348, I=0.504, and PPB=83.3%; species level: Ne=1.864, H=0.460, I=0.652, and PPB=100%), as shown in Table 3. The genetic diversities within species (Ht) and within populations (Hs) were 0.4453 and 0.3485, respectively (Table 4). The genetic differentiation between the populations (Gst) was 0.2174. Based on the Gst value, the level of gene flow (Nm) was estimated at 1.7999 (Nm > 1). These results indicated high gene flow and low differentiation between the extant populations.
| Primer code | Sequence (5' to 3') | Scored bands | No. of polymorphic bands | Percentage of polymorphic bands |
| 811 | GAGAGAGAGAGAGAGAC | 6 | 6 | 100% |
| 815 | CTCTCTCTCTCTCTCTG | 7 | 7 | 100% |
| 834 | AGAGAGAGAGAGAGAGYT | 6 | 6 | 100% |
| 836 | AGAGAGAGAGAGAGAGYA | 6 | 6 | 100% |
| 840 | GAGAGAGAGAGAGAGAYT | 6 | 6 | 100% |
| 841 | GAGAGAGAGAGAGAGAYC | 7 | 7 | 100% |
| 843 | GAGAGAGAGAGAGAGAYG | 11 | 11 | 100% |
| 853 | TCTCTCTCTCTCTCTCRT | 7 | 7 | 100% |
| 854 | TCTCTCTCTCTCTCTCRG | 8 | 8 | 100% |
| 855 | ACACACACACACACACYT | 6 | 6 | 100% |
| 857 | ACACACACACACACACYG | 7 | 7 | 100% |
| 873 | GACAGACAGACAGACA | 13 | 13 | 100% |
| 880 | GGAGAGGAGAGGAGA | 11 | 11 | 100% |
| 881 | GGGTGGGGTGGGGTG | 8 | 8 | 100% |
| Mean | 7.8 | 7.8 | 100% | |
| Species level | 109 | 109 | 100% |
| Population | PPB (%) | Na | Ne | H | I |
| SBX | 75.23 | 1.752 | 1.583 | 0.320 | 0.460 |
| XC | 78.90 | 1.789 | 1.626 | 0.344 | 0.493 |
| LG | 74.31 | 1.743 | 1.529 | 0.300 | 0.438 |
| NMP | 92.66 | 1.927 | 1.711 | 0.389 | 0.562 |
| DSB | 95.41 | 1.954 | 1.693 | 0.389 | 0.567 |
| Mean | 83.30 | 1.833 | 1.629 | 0.348 | 0.504 |
| Species level | 100 | 2.000 | 1.864 | 0.460 | 0.652 |
| Notes: Na, observed number of alleles; Ne, effective number of alleles Kimura and Crow (1964); H, Nei's (1973) gene diversity; I, Shannon's Information index; P, the percentage of polymorphic loci. | |||||
| Ht | Hs | Gst | Nm | |
| Species level | 0.4453 | 0.3485 | 0.2174 | 1.7999 |
| Standard deviation | 0.0034 | 0.0089 | ||
| Notes: Ht, total variability; Hs, variability within populations; Gst, coefficient of genetic differentiation; Nm, estimate of gene flow. | ||||
Analysis of molecular variance results revealed that 17% of the genetic variation was partitioned between the populations and 83% occurred within the populations based on inter-simple sequence repeat (ISSR) markers (Table 5). These results indicated low genetic variation levels between the five populations.
| Source of variation | df | Sum of squares | Variation components | Percentage of variation (%) |
| Among populations | 4 | 228.120 | 3.625 | 17% |
| Within populations | 49 | 886.417 | 18.090 | 83% |
| Total | 53 | 1114.537 | 21.716 | - |
STUCTURE analysis based on the ΔK method revealed that DK was 56.18 for K=5 and ΔK was < 56.18 for all of the values of K (ranging from 1 to 20, except 5) as shown in Fig. 2a and b. Therefore, the optimal ΔK for K=5 provided strong evidence for the presence of five independent populations, which is consistent with the five natural populations sampled here (Fig. 2c).
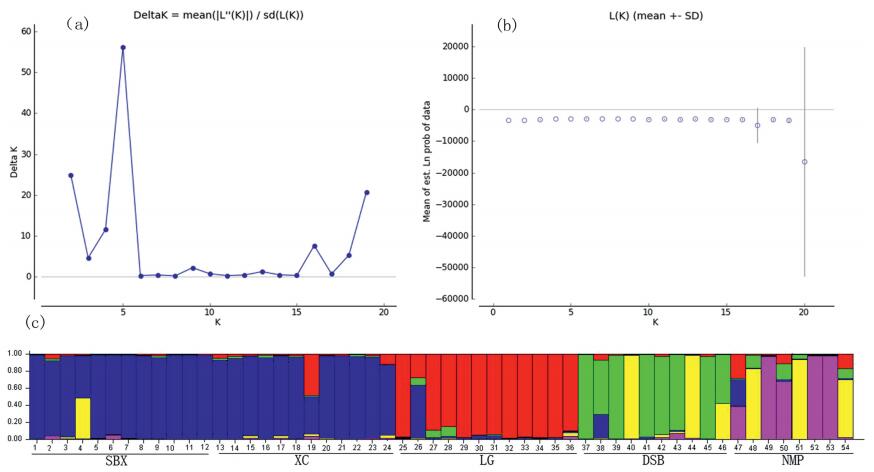
|
| Fig. 2 Results of Bayesian model-based clustering STRUCTURE analysis of 54 individuals of C. chago. (a) The probability of the data Lnp (D) (±SD) analysis the number of K clusters, and increase of LnP (D) given K, calculated as (LnP (D) k-LnP (D) k-1). (b) Delta K values from the mean log-likelihood probabilities from STRUCRURE runs where inferred clusters (K) ranged from 1 to 20. (c) Estimated genetic clustering (K ¼ 5) obtained with the STRUCTURE program for 54 individuals. Individuals are separated according to the population and black vertical line in the bar chart is population identifier. |
Our field survey revealed that distribution of C. chago is restricted to the Lacang River upstream of the tributary mountains in Yunlong County, Yunnan. These remaining populations not only have a limited geographical reach but are fragmented and highly isolated. It is well known that habitat is essential for the long-term persistence and survival of endemic and rare species (Kalliovirta et al., 2006; Shen et al., 2009). Importantly, our field surveys revealed that all the populations of C. chago occur in unprotected areas outside of nature reserves and are frequently affected by human interference. The survival status of C. chago is similar to other critically endangered plants such as Euryodendron excelsum (Shen et al., 2009) and Manglietiastrum sinicum (Tian et al., 2002; Sun et al., 2015). Our study also found that an additional threat to the survival and natural proliferation of the species is unrestrained harvesting of C. chago fruits for their nuts by local people. Thus, we suggest that this species be designated as a Plant Species with Extremely Small Populations (PSESP) in China and be given special attention and protection.
4.2. Genetic diversity of C. chagoThe genetic diversity of a species in small populations is lower than that in large populations due to genetic drift and inbreeding (Willi et al., 2006; Li et al., 2012). As a result, rare and endangered species with narrow geographical distributions harbor lower genetic diversity to a greater extent than similar species with broad geographical distributions (Hamrick and Godt, 1990). In the present study, genetic diversity within the C. chago species was detected using ISSR markers. However, our study showed that C. chago showed a high level of genetic diversity (Ne=1.629, H=0.348, I=0.504, PPB=83.3%) at the species level.
In general, the level of genetic diversity is influenced by several factors, including reproductive mode, biological traits and breeding system. Outcrossing species generally exhibit considerably higher levels of genetic diversity than selfing species (Hamrick and Godt, 1989; Nybom, 2004). The reproductive biology and breeding system of C. chago were not examined in this study, although, based on the high level of genetic diversity, we hypothesize that it is an outcrossing species. This hypothesis is also consistent with Rohwer's finding (1993) that plants from the family Lauraceae tend to be outcrossing.
High genetic variation enables a species to adapt to various environments (Zhao et al., 2012). High genetic diversity in the five remaining populations of C. chago indicate that the species is not endangered because of genetic factors (e.g. genetic diversity decline, genetic drift, and inbreeding). The main threat to this plant species may include habitat specialization, anthropogenic interference and its limited distribution. Regardless, the factors that lead to the “endangered” status of this species must be further elucidated.
4.3. Genetic structure of C. chagoGenetic structure is affected by several factors, including breeding system, genetic drift, population size, seed dispersal, gene flow, evolutionary history, and natural selection (Hamrick and Godt, 1990). Our analyses of the genetic structure of C. chago showed that the 54 individuals formed 5 populations (Fig. 2c). Molecular variance analysis showed that genetic variation mostly occurred within populations (83%). The coefficients of genetic differentiation (Gst) and the gene flow (Nm) between the five extant populations were 0.2174 and 1.7999, respectively. These results indicate that the frequency of gene flow between the groups is sufficient to prevent differentiation between populations caused by genetic drift. This might be explained by long-distance dispersal of pollen and/or seeds which are know to result in low genetic differentiation and high gene flow between the populations of the same species (Yao et al., 2007; Zhao et al., 2012). At present, there has been little research on C. chago and the mechanisms of seed and pollen dispersal have not been elucidated for this species. Thus, the factors that facilitate frequent and continual gene flow between the five populations of C. chago are still unknown. In order to address these issues, future research on C. chago reproductive biology is needed.
4.4. Conservation implicationsEndemic plant species with limited distribution face both internal and external threats to their survival (Zu et al., 1999). Our study indicates that while the distribution of C. chago populations are small, isolated and fragmented, this species still has a relatively high level of genetic diversity. According to Williamson and Werth (1999), the main threats to species with limited distribution but high genetic diversity are external, such as habitat fragmentation, geographical isolation, and human interference. We suggest intensified protection of the remaining populations of C. chago in situ and recommend prioritizing conservation of the DSB and NMP populations, which have the highest levels of genetic diversity. In addition, we suggest that local people be encouraged to grow more C. chago and to harvest more nuts. Seed collection for germplasm storage and artificial seedlings for ex situ conservation is also recommend. Population recovery and reintroductions of C. chago must also be conducted to facilitate recovery of wild populations.
5. AcknowledgmentsThis study was financially supported by grant 31560224 and 31360074 from the National Natural Science Foundation of China and grant 2015J002 from the Graduate Science of foundation projects of Yunnan Educational Committee.
Doyle J.J., Doyle J.L., 1988. Isolation of plant DNA from fresh tissue. Focus, 12: 13-15. |
Earl D.A., vonHoldt B.M., 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Reso, 4: 359-361. DOI:10.1007/s12686-011-9548-7 |
Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol, 14: 2611-2620. DOI:10.1111/mec.2005.14.issue-8 |
Hamrick, J.L., Godt, M.J.W., 1989. Allozyme diversity in plant species. In: Brown, H.D., Clegg, M.T., Kahler, A.L. (Eds.), Plant Population Genetics, Breeding and Genetic Resources. Sinauer Associates, Inc, Sunderland, MA, pp. 43-46.
|
Hamrick, J.L., Godt, M.J., 1990. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer, Sunderland, MA, pp. 43-63.
|
Huang J.F., Li L., van der Werff H., et al, 2016. Origins and evolution of cinnamon and camphor: a phylogenetic and historical biogeographical analysis of the Cinnamomum group (Lauraceae). Mol. Phylogenet. Evol, 96: 33-44. DOI:10.1016/j.ympev.2015.12.007 |
Kalliovirta M., Ryttari T., Heikkinen R.K., 2006. Population structure of a threatened plant, Pulsatilla patens, in boreal forests: modeling relationships to overgrowth and site closure. Biodivers. Conserv, 15: 3095-3108. DOI:10.1007/s10531-005-5403-z |
Kimura M., Crow J.F., 1964. The number of alleles that can be maintained in a finite population. Genetics, 49: 725-738. |
Li, H.W., Li, J., Huang, P.H., Wei, F.N., Cui, H.B., van der Werff, H., 2008. Lauraceae. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds.), Flora of China, vol. 7. Science Press and Missouri Botanical Garden Press, Beijing, China, St. Louis, Missouri, USA.
|
Li Y.Y., Guan S.M., Yang S.Z., et al, 2012. Genetic decline and inbreeding depression in an extremely rare tree. Conserv. Genet, 13: 343-347. DOI:10.1007/s10592-011-0286-x |
Nei M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA, 70: 3321-3323. DOI:10.1073/pnas.70.12.3321 |
Nybom H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol, 13: 1143-1155. DOI:10.1111/mec.2004.13.issue-5 |
Peakall R., Smouse P.E., 2012. GenAlEx 6., 5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics, 28: 2537-2539. DOI:10.1093/bioinformatics/bts460 |
Pritchard J.K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. |
Rohwer J.G., 1993. Lauraceae. In: Kubitzki K., Rohwer J.G., Brittrich, V. (Eds.), The Families and Genera of Flowering Plants. Berlin: Springer-Verlag: 426-437.
|
Shen S.K., Wang Y.H., Wang B.Y., et al, 2009. Distribution, stand characteristics and habitat of a critically endangered plant Euryodendron excelsum H T Chang (Theaceae): implications for conservation. Plant Spec. Biol, 24: 133-138. DOI:10.1111/psb.2009.24.issue-2 |
Sun B.X., Zhao H.L., 1991. A new species of cinnamomum from Yunnan. Journal of Yunnan University, 13: 93-94. |
Sun W., Zhou Y., Han C., et al, 2006. Status and conservation of Trigonobalanus doichangensis (Fagaceae). Biodivers. Conserv, 15: 1303-1318. |
Sun, W.B., Ma, Y., Chen, G., et al., 2015. Rescuing Magnolia sinica (Magnoliaceae), a critically endangered species endemic to Yunnan, China. Oryx 1-4.
|
Tian K., Zhang G., Cheng X., et al, 2002. The habitat fragility of Manglietiastrum sinicum. Acta Bot. Yunnanica, 25: 551-556. |
Williamson P.S., Werth C.R., 1999. Levels and patterns of genetic variation in the endangered species Abronia macrocarpa (Nyctaginaceae). Am. J. Bot, 86: 293-301. DOI:10.2307/2656946 |
Willi Y., Van Buskirk J., Hoffmann A.A., 2006. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst, 37: 433-458. DOI:10.1146/annurev.ecolsys.37.091305.110145 |
Wu Z.Y., 1991. Flora of Yunnan. Beijing: Science Press.
|
Yao X.H., Ye Q.G., Kang M., et al, 2007. Microsatellite analysis reveals interpopulation differentiation and gene flow in endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China. New Phytol, 176: 472-480. DOI:10.1111/j.1469-8137.2007.02175.x |
Yeh, F.C., Yang, R.C., Boyle, T., 1999. POPGENE VERSION 1.31: Microsoft Windowbased Free Software for Population Genetic Analysis. ftp://ftp.microsoft.com/Softlib/HPGL.EXE.
|
Zhao X.F., Ma Y.P., Sun W.B., et al, 2012. High genetic diversity and low differentiation of Michelia coriacea (Magnoliaceae), a critically endangered endemic in southeast Yunnan, China. Int. J. Mol. Sci, 13: 4396-4411. DOI:10.3390/ijms13044396 |
Zu Y.G., Zhang W.H., Yan X.F., 1999. Conservation Biology of the Endangered Plant Adenophora lobophylla Hong. Beijing: Science Press.
|



