b. Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Menglun, Mengla, Yunnan, 666303, China;
c. CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, No. 132, Lanhei Road, Kunming, Yunnan, 650201, China;
d. University of Chinese Academy of Sciences, Beijing, 100049, China;
e. State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Yunnan, 650091, China
Plants undergo developmental and physiological changes throughout their life history, ending with senescence and death (Lim et al., 2005). Leaf senescence is an important part of plant development and increases reproductive success. During senescence, plants relocate mobilizable nutrients from older leaves to other developing organs, including seeds, stems and roots (Lim et al., 2005). Under optimal conditions, the onset of leaf senescence is normally initiated in an age-dependent manner; however, it can also be triggered by environmental changes that are integrated into the developmental aging program (Buchanan-Wollaston et al., 2005; Lim et al., 2007). Plant senescence represents one of the adaptive mechanisms that plants possess that may help to increase survival in a given ecological niche.
Leaf senescence is a highly coordinated and sophisticated cellular process during which leaf cells undergo active degenerative alterations. Furthermore, this biochemical process is closely associated with the increased expression of numerous senescence-associated genes (SAGs), including SAG12 and SAG29, but reduced expression of photosynthetic genes, such as CAB1 and RBCS1A (Gan and Amasino, 1995; Hortensteiner, 2006; Park et al., 1998; Qi et al., 2015; Weaver et al., 1998). Studies have also demonstrated that a distinct set of phytohormones play critical roles during leaf senescence, with jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), salicylic acid (SA), brassinosteroids accelerating senescence, while auxin and cytokinins delay leaf aging (Jibran et al., 2013). Interestingly, gibberellin (GA) was recently revealed to be a crucial growth regulator that positively modulates leaf senescence in Arabidopsis (Chen et al., 2014, 2017).
Temporal profiling has revealed that leaf senescence involves the coordinated expression of thousands of genes. Notably, numerous WRKY genes are strongly expressed in senescing leaves, suggesting that WRKY genes play a role in leaf senescence (Guo et al., 2004). Recent studies have provided evidence that WRKY proteins function as critical components in senescence-associated regulatory pathways. For example, WRKY57 was shown to function as a common component of JA- and auxin-mediated signaling pathways to modulate JA-induced leaf senescence (Jiang et al., 2014). In addition, WRKY75, together with SA and reactive oxygen species (ROS), has been shown to form a tripartite amplification loop to promote leaf senescence (Guo et al., 2017). Furthermore, our recent study demonstrated that WRKY45 directly binds several SAG promoters to activate their expression, thereby activating GA-mediated leaf senescence (Chen et al., 2017).
Here, we show that WRKY75 transcript and protein levels increased during leaf senescence. We found that age-triggered leaf senescence was delayed in WRKY75 mutants, but accelerated in plants overexpressing WRKY75. Further investigation indicated that WRKY75 participates in GA-mediated leaf senescence by directly regulating the expression of several downstream SAGs. We also show that the ability of WRKY75 to activate transcription can be repressed by both GAI and RGL1, and that GAI gain-of-function or RGL1 overexpression can partially rescue the early-senescence phenotype caused by WRKY75 overexpression. Our findings thus demonstrate that WRKY75 may participate in GA-mediated leaf senescence in Arabidopsis.
2. Materials and methods 2.1. Materials and plant growth conditionsAll mutant and transgenic plants were in the Col-0 background. The WRKY mutants (wrky75-1 and wrky75-25), WRKY75 and RGL1 transgenic over-expressing plants, and Myc-WRKY75/RGL1 plants have been described in our previous studies (Chen et al., 2017; Zhang et al., 2018). Arabidopsis plants were grown at 22 ℃ under a 16-h-light/8-h-dark cycle. All chemicals were purchased from Takara Biotechnology (Dalian, China).
2.2. qRT-PCR analysisQuantitative RT-PCR was conducted as described in Chen et al. (2013). The gene-specific primers for qRT-PCR were WRKY75-F (5′-ATATGGCCAAAAGGCCGTCA-3′) and WRKY75-R (5′-TGCTCGAAGTTTTCGGTGGA-3′), SAG12-F (5′-ATCCAAAAGCAACTTCTATTACAGG-3′) and SAG12-R (5′-CCACTGCCTTCATCAGTGC-3′), SAG29-F (5′-GCCACCAGGGAGAAAAGG-3′) and.
SAG29-R (5′-CCACGAAATGTGTTACCATTAGAA-3′), ACTIN2-F (5′-TGTGCCAATCTACGAGGGTTT-3′) and ACTIN2-R (5′-TTTCCCGCTCTGCTGTTGT-3′).
2.3. Measurements of chlorophyll content and ion leakageChlorophyll was extracted from detached leaves with 80% acetone. Chlorophyll content was determined at 663 and 645 nm according to Lichtenthaler (1987).
To measure ion leakage, we incubated detached leaves in deionized water for at least 2 h (less than 10 h) and then determined conductivities (C1) of the solutions. The samples were then boiled in the same deionized water for 15 min. After cooling, the conductivities (C2) of the solutions were measured again (Li et al., 2013). The degree of ion leakage was calculated as the ratio of C1:C2.
2.4. ChIP assaysWRKY75:YFP-WRKY75:3′-WRKY75, Myc-WRKY75/RGL1 and Col-0 leaves were harvested for ChIP experiments as described in Saleh et al. (2008). The GFP or Myc antibody was used to immunoprecipitate the protein–DNA complex, and the precipitated DNA was purified using a PCR purification kit for real-time qPCR analysis. The ChIP experiments were performed three times. Chromatin precipitated without antibody was used as the negative control, while isolated chromatin before precipitation was used as the input control. ChIP results are presented as a percentage of input DNA.
2.5. Transient expression assaysTransient expression assays were performed in Nicotiana benthamiana leaves as described by Chen et al. (2017). Briefly, NLS was fused with a GFP reporter gene behind the native promoter of SAG12 or SAG29.The CaMV 35S promoter was used to drive full-length coding sequences of RGL1, GAI, GUS, and WRKY75. These constructs were then introduced into Agrobacterium tumefaciens (strain EHA105). Expression of GFP was determined 48 h after infiltration. Experiments were repeated three times with similar results.
2.6. Determination of YFPSenescing leaves of WRKY75:YFP-WRKY75:3′-WRKY75 plants were detached and then YFP was observed under a confocal laser scanning microscope (Olympus).
2.7. Treatment of GAWe treated seedlings with GA3 as described in Chen et al. (2017). Myc-WRKY75/RGL1 seedlings were harvested after treatment with 50 μM GA3 or mock for 8 h.
2.8. Accession numbersThe following genes were detected in this work: WRKY75 (At5G13080), WRKY45 (At3G0 1970), RGL1 (At1G66350), GAI (At1G14920), SAG12 (At5G45890), SAG29 (At5g13070), and ACTIN2 (At3G18780).
3. Results 3.1. Expression of WRKY75 in senescing leavesWRKY75 was previously found to play important roles in phosphate starvation, defense responses, flowering initiation, and root hair initiation (Devaiah et al., 2007; Encinas-Villarejo et al., 2009; Zhang et al., 2018; Rishmawi et al., 2014). An Arabidopsis microarray database indicates that WRKY75 is highly expressed in senescent leaves (Winter et al., 2007; Fig. S1). Our data further confirm these results (Fig. 1), and imply that WRKY75 may function as a SAG that is up-regulated during senescence. WRKY75 expression increased greatly in leaves during early senescence (ES), and showed even stronger expression at late senescence (LS) (Fig. 1A and Fig. S2A). This same pattern of WRKY75 expression was observed across the senescence gradient from the tip to the base of leaves (Fig. 1B and Fig. S2B).
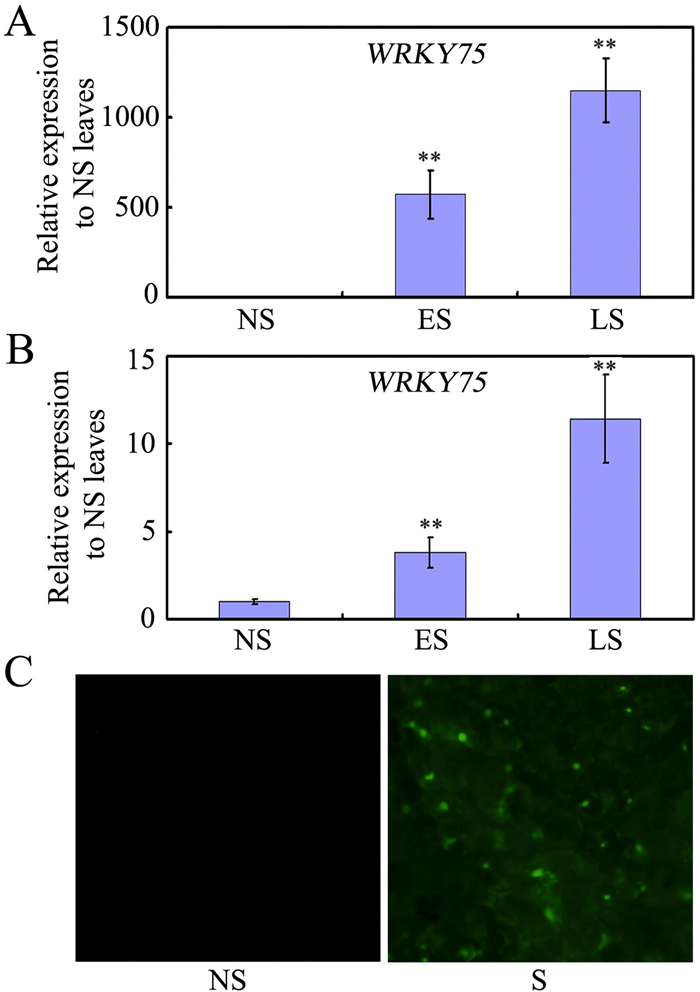
|
| Fig. 1 Expression of WRKY75 in senescing leaves. (A) qRT-PCR analysis of WRKY75 transcript levels in wild-type leaves at different developmental stages. Transcript levels of WRKY75 in non-senescent (NS) leaves were arbitrarily set to 1. Values are means ± SD of three independent biological replicates. **P < 0.01, Student's t-test compared with NS leaves. (B) qRT-PCR analysis of WRKY75 transcript levels in different parts of a senescing wild-type leaf. Transcript levels of WRKY75 in NS leaves were arbitrarily set to 1. Values are means ± SD of three independent biological replicates. **P < 0.01, Student's t-test compared with NS parts of a senescing wild-type leaf. (C) YFP detection of WRKY75 inwrky75-25 mutant background that harbors the WRKY75:YFP-WRKY75:3′-WRKY75. YFP signals were observed in senescing leaves of the WRKY75:YFP-WRKY75:3′-WRKY75 transgenic plants. These experiments were performed three times with similar results. |
To further understand WRKY75 expression patterns, we measured WRKY75 protein accumulation during leaf senescence by visualizing yellow fluorescent protein (YFP) in leaves of WRKY75pro: YFP-WRKY75:3′WRKY75 plants. No YFP signal was observed in non-senescing leaves, whereas strong YFP signals were observed in senescing leaves (Fig. 1C). These findings show that both transcript and protein levels of WRKY75 increase during leaf senescence, suggesting that WRKY75 plays a role in leaf senescence.
3.2. Altered age-triggered leaf senescence resulting from knock-down or ectopic expression of WRKY75High expression levels of WRKY75 in senescing leaves prompted us to examine the role of WRKY75 in senescence using WRKY75 mutants (wrky75-1 and wrky75-25) (Zhang et al., 2018). Analysis of the rosettes of 7-week-old plants revealed that leaf senescence was delayed in WRKY75 mutants compared to in WT plants (Fig. 2A).This finding is consistent with that of two recent reports (Guo et al., 2017; Zhang et al., 2017). Chlorophyll content in these plants indicated that chlorophyll was degraded more slowly in WRKY75 mutants than in WT plants (Fig. 2B). Correspondingly, both ion leakage and expression of representative senescence-induced SAGs (e.g., SAG12 and SAG29) were lower in WRKY75 mutants than in WT plants (Fig. 2C–E). These findings indicate that disruption of WRKY75 delays the senescence process of plants.

|
| Fig. 2 WRKY75 mutation delays age-triggered leaf senescence. (A) The senescence phenotypes of 7-week-old Col-0 andwrky75 mutant plants. (B) Relative chlorophyll content of the Col-0 and wrky75 mutant plants. (C) Membrane ion leakage of the Col-0 and wrky75 mutant plants. (D) and (E) qRT-PCR analysis of transcript levels of senescence marker genes in age-triggered senescing leaves of Col-0 and wrky75 mutant plants. For B-E, values are means ± SD of three independent biological replicates. **P < 0.01, Student's t-test compared with Col-0. These experiments were performed three times with similar results. |
To further determine the role of WRKY75 in leaf senescence, we again used 35S: WRKY75 transgenic Arabidopsis plants (Zhang et al., 2018). Compared with WT plants, 35S: WRKY75 transgenic plants showed early onset senescence (Fig. S3A). Consistent with this finding, transgenic plants had lower chlorophyll contents, but higher ion leakage and stronger SAG expression (Fig. S3B–E). Together, these observations suggest that over-expression of WRKY75 promotes leaf senescence.
3.3. In vivo interactions between WRKY75 and its target promotersPrevious studies have revealed that WRKY proteins function by directly binding to a cis-acting DNA element, namely W-box (T/CTGACC/T), present in promoters of their target genes (Eulgem et al., 2000; Ulker and Somssich, 2004). We provide evidence that WRKY75 functions as an activator in age-triggered leaf senescence by promoting the expression of senescence-associated genes. We found several putative W-box elements in promoters of bothSAG12 and SAG29 (Fig. 3A), indicating that WRKY75 may directly regulate their expression, thus promoting leaf senescence. To determine whether WRKY75 can directly regulate SAG12 or SAG29 expression, chromatin immunoprecipitation (ChIP) experiments were performed using WRKY75:YFP-WRKY75:3′-WRKY75 plants (Rishmawi et al., 2014). These experiments showed that WRKY75 can interact with the SAG12 and SAG29 promoters (pSAG12-3, pSAG29-2, and pSAG29-3) via the W-box sequence (Fig. 3B), suggesting that WRKY75 directly regulates their transcription.
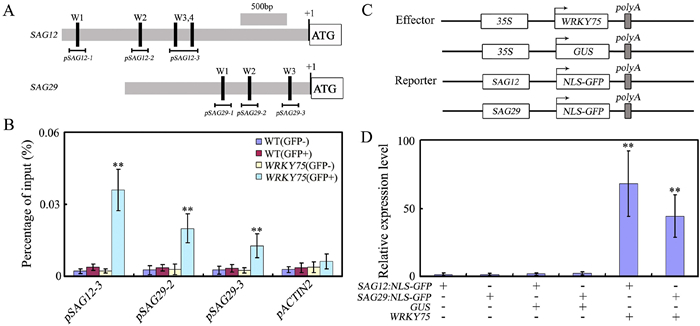
|
| Fig. 3 WRKY75 directly regulates the expression of SAG12 and SAG29. (A) The promoter structure of the SAG12 and SAG29 genes and fragment used in the ChIP assay. (B) WRKY75 directly binds to the promoters of SAG12 and SAG29. ChIP assays were performed with chromatin prepared from WRKY75:YFP-WRKY75:3′-WRKY75 transgenic plants, using an anti-GFP antibody (IP). ChIP results are presented as a percentage of input DNA. Values are mean ± SD of three independent biological replicates. Asterisks indicate Student's t-test significant differences as compared to controls, **P < 0.01. (C) Schematic of theSAG12:NLS-GFP and SAG29:NLS-GFP reporters and WRKY75 and GUS effectors. (D) Transient expression assays showed that WRKY75 activates the expression of SAG12 and SAG29 determined by qRT-PCR analysis. Values are means ± SD of three independent biological replicates. **P < 0.01, Student's t-test compared with controls. These experiments were performed three times with similar results. |
To further determine the regulatory function of WRKY75, we performed transient expression assays in tobacco (N. benthamiana) leaves. We fused the promoters of SAG12 and SAG29-both direct targets of WRKY75-to a reporter construct, the nuclear localization signal (NLS)-GFP gene (SAG12:NLS-GFP and SAG29:NLS-GFP) (Fig. 3C). The effector plasmids had a GUS or WRKY75 gene under the control of the CaMV 35S promoter (35S: GUS and 35S: WRKY75) (Fig. 3C). Compared to controls, co-expression of WRKY75 in the effector plasmids greatly increased GFP reporter expression (Fig. 3D), suggesting that WRKY75 functions as an activator during age-triggered leaf senescence.
3.4. RGL1 and GAI inhibit WRKY75-mediated transcriptional activationWe previously reported that WRKY75 can physically interact with DELLA proteins (Zhang et al., 2018), prompting us to hypothesize that WRKY75 may participate in senescence through the GA pathway. We also speculated that physical interactions between WRKY75 and DELLA proteins inhibit the ability of WRKY75 to activate transcription of target genes. To test these hypotheses, we again performed transient expression assays in tobacco (N. benthamiana) leaves with 35S: WRKY75, 35S: RGL1, 35S: GAI and 35S: GUS as effectors and SAG12:NLS-GFP as a reporter (Fig. 4A). We found that WRKY75 expression greatly increases the expression of GFP driven by the SAG12 promoter. However, co-expression of RGL1 or GAI with WRKY75 markedly reduces GFP expression in comparison with expression of WRKY75 or GUS alone (Fig. 4B). These results demonstrate that both GAI and RGL1 can repress WRKY75-mediated transcriptional activation.

|
| Fig. 4 GAI and RGL1 repress WRKY75 activation ability. (A) Schematic of the SAG12:NLS-GFP reporter and WRKY75, RGL1, GAI and GUS effectors. (B) Transient expression assays showed that RGL1 and GAI repress transcriptional activation of WRKY75 determined by qRT-PCR analysis. Values are mean ± SD of three independent biological replicates. Asterisks indicate Student's t-test significant differences as compared to controls, **P < 0.01. (C) RGL1 interferes the binding of WRKY75 to its target genes (shown in Fig. 4A). ChIP assays were performed with chromatin prepared fromMyc-WRKY75/RGL1 plants, using an anti-Myc antibody (IP). ChIP results are presented as a percentage of input DNA. Values are mean ± SD of three independent biological replicates. Asterisks indicate Student's t-test significant differences as compared to controls, **P < 0.01. These experiments were performed three times with similar results. |
To determine whether the ability of WRKY75 to bind to target genes is affected by interactions with DELLA proteins, we performed ChIP assays using Myc-WRKY75/RGL1 plants. When we treated plants with GA, the binding efficiency of WRKY75 to the SAG12 promoter increased greatly, implying that RGL1 may reduce the binding ability of WRKY75 to its target genes in vivo.
3.5. Repression of age-triggered leaf senescence by GAI gain-of-function or RGL1 overexpressionBecause several DELLAs physically associate with WRKY75 and modulate its transcriptional ability, we then wondered whether GAI gain-of-function or RGL1 overexpression could rescue the earlier senescence phenotype caused by WRKY75 overexpression. Then both Myc-WRKY75/gai-1 and Myc-WRKY75/RGL1 plants were used (Zhang et al., 2018). Senescence occurred earlier in Myc-WRKY75/gai-1 plants than in gai-1 plants (Fig. 5A). Furthermore, Myc-WRKY75/gai-1 plants had lower chlorophyll content and higher levels of SAG expression than gai-1 plants (Fig. 5B–E). Myc-WRKY75/RGL1 plants showed similar phenotypes (Fig. S4). These findings indicate that GAI gain-of-function or RGL1 overexpression may at least partially delay the early senescence phenotype caused by WRKY75 overexpression.
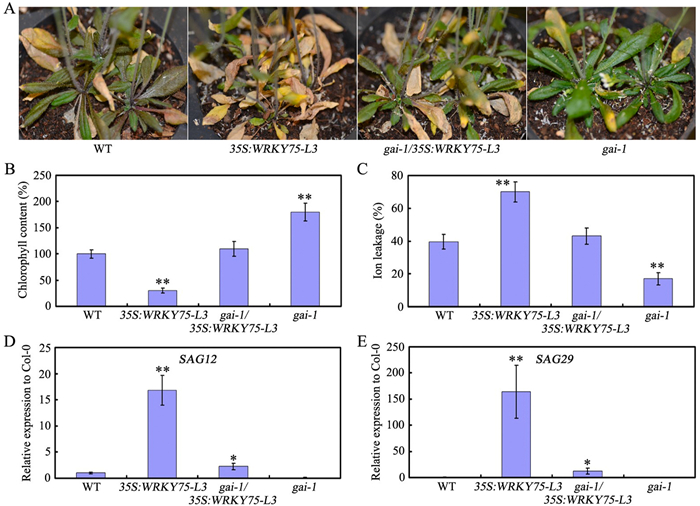
|
| Fig. 5 GAI gain-of-function partially rescues the early-senescence phenotype of WRKY75 overexpressing plants. (A) The senescence phenotypes of the indicated genotypes. (B) Relative chlorophyll content of the indicated genotypes. (C) Membrane ion leakage of the indicated genotypes. (D) and (E) qRT-PCR analysis of transcript levels of senescence marker genes in the indicated genotypes. For B-D, values are means ± SD of three independent biological replicates. *P < 0.05, **P < 0.01, Student's t-test compared with Col-0. These experiments were performed three times with similar results. |
To further determine the role of WRKY75 in GA-mediated leaf senescence, we compared the GA response in wrky75-1, wrky75-25, WT, 35S: WRKY75-L3, and 35S: WRKY75-L6 plants. We previously demonstrated that GA induces slight increases in WRKY75 gene expression (Zhang et al., 2018). Here, we confirm that WRKY75 expression is promoted in della but reduced in ga1 or gid1a/gid1b/gid1c plants (Fig. 6A). Furthermore, when we treated plants with GA, chlorophyll content and SAG12 expression indicated that GA-mediated leaf senescence was delayed in wrky75 mutants but accelerated in WRKY75 overexpressing plants. Combined, these results suggest that WRKY75 participates in the modulation of leaf senescence through the GA pathway.

|
| Fig. 6 Effects of GA on wrky75 and WRKY75-overexpressing plants. (A) qRT-PCR analysis of WRKY75 transcript levels in ga1, gid1agid1bgid1c, and della mutant plants. (B) GA response of the indicated genotypes. (C) Relative chlorophyll content of the indicated genotypes as shown in (B). (D) Relative expression of SAG12 in the indicated genotypes as treated in (B). For A, C and D, values are means ± SD of three independent biological replicates. **P < 0.01, Student's t-test compared with Col-0 or Ler. These experiments were performed three times with similar results. |
Our previous study revealed that WRKY45 functions as a novel activator of the senescence transcriptional network (Chen et al., 2017). Thus, we wondered whether WRKY75 would affect the expression of WRKY45 during leaf senescence. As shown in Fig. 7A, WRKY45 expression was inhibited in the wrky75 mutants but was activated in WRKY75 over-expressing plants (Fig. 7A). To test whether WRKY45 is a direct target of WRKY75, ChIP assays were preformed using WRKY75:YFP-WRKY75:3′-WRKY75 plants. WRKY75 can directly bind to the WRKY45 promoter through the W-box cis-element (pWRKT45-2) (Fig. 7B and C). These results demonstrate that WRKY75 directly regulates WRKY45 expression during leaf senescence, and also imply that there may exist extensive cross-regulation among WRKY members during leaf senescence, which contributes to the facilitation of senescence-associated transcriptional reprogramming.
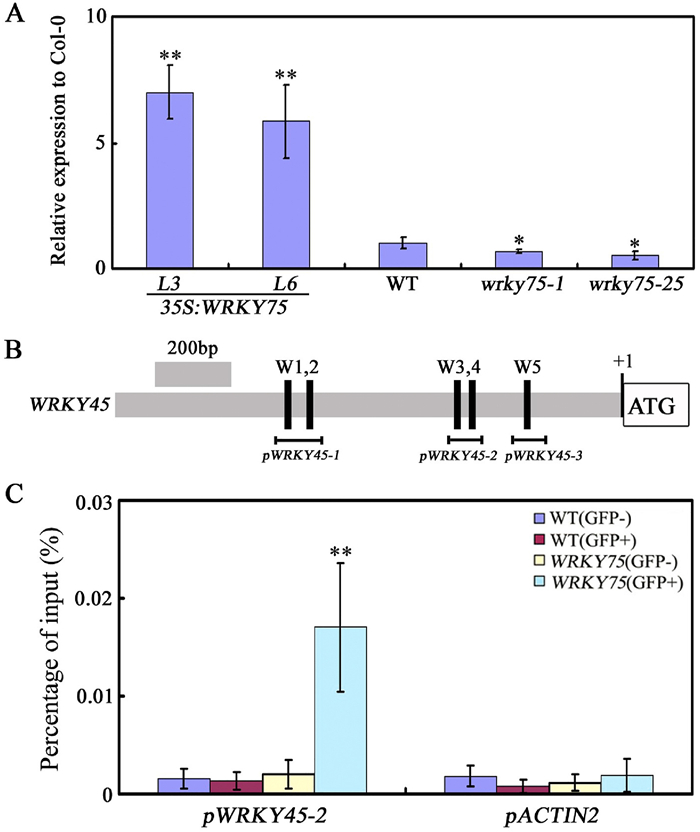
|
| Fig. 7 WRKY75 positively regulates WRKY45 expression during leaf senescence. (A) qRT-PCR analysis of transcript levels of WRKY45 in the indicated genotypes. Values are means ± SD of three independent biological replicates. *P < 0.05, **P < 0.01, Student's t-test compared with Col-0. (B) The promoter structure of WRKY45 gene and fragment used in the ChIP assay. (C) WRKY75 directly binds to the promoter of WRKY45. ChIP assays were performed with chromatin prepared from WRKY75:YFP-WRKY75:3′-WRKY75 transgenic plants, using an anti-GFP antibody (IP). ChIP results are presented as a percentage of input DNA. Values are mean ± SD of three independent biological replicates. Asterisks indicate Student's t-test significant differences as compared to controls, **P < 0.01. These experiments were performed three times with similar results. |
Plant senescence is tightly controlled by the temporal coordinated expression of numerous SAGs. Over the past decades, our understanding of plant senescence has improved immensely. Microarray expression profiling has identified WRKY factors as the second largest transcription factor group in the senescent transcriptome (Guo et al., 2004), suggesting that these genes play central roles in modulating transcriptional changes during senescence. However, the biological function of WRKY factors during leaf senescence remains largely unknown. Recently, several Arabidopsis WRKY proteins have been shown to play important roles during leaf senescence, including WRKY6, WRKY22, WRKY45, WRKY53, WRKY54, WRKY57, and WRKY70 (Robatzek and Somssich 2002; Zhou et al., 2011; Chen et al., 2017; Miao and Zentgraf, 2007; Jiang et al., 2014; Ulker et al., 2007). Here, we provide further evidence that WRKY75 may function as a new component that positively regulates GA-mediated leaf senescence.
We found that WRKY75 is strongly expressed in senescing leaves at both mRNA and protein levels, when compared with young leaves (Fig. 1), implying that WRKY75 functions as a SAG to modulate the senescence. Phenotypic analysis using wrky75 T-DNA mutants and WRKY75-overexpressing plants demonstrated thatWRKY75 acts as a positive regulator during age-triggered leaf senescence (Fig. 2 and Fig. S3). Further investigation revealed that WRKY75 participates in senescence through the direct activation of several SAGs, including SAG12 and SAG29 (Fig. 3). A recent study also found that WRKY75 directly promotes SA INDUCTION-DEFICIENT 2 (SID2) expression, but suppresses CATALASE 2 (CAT2) transcription during leaf senescence (Guo et al., 2017). Therefore, WRKY75 appears to function as both an activator and repressor to finely modulate leaf senescence. Similarly, our previous study demonstrated that WRKY8 controls plant defense responses to viral infection by positively regulating ABI4 while negatively regulating ACS6 (Chen et al., 2013). Taken together, these findings indicate that WRKY factors act as both positive and negative regulators that fine-tune signaling and transcriptional networks involved in mediating plant growth and stress responses.
GA, as an essential hormone, modulates diverse aspects of plant development. Recent studies have demonstrated that GA signaling may have a positive effect on leaf senescence (Chen et al., 2014, 2017·bib_Chen_et_al_2014·bib_Chen_et_al_2017); however, the specific mechanisms by which GA affects this progress have yet to be determined. Numerous studies have suggested that WRKY proteins function as key regulators during leaf senescence. We previously found that WRKY45 regulates age-triggered leaf senescence through a physiological interaction with the DELLA protein RGL1. Other senescence-associated WRKY transcription factors may also be involved in GA-mediated age-dependent leaf senescence. Here, we provide evidence that WRKY75 may positively regulate leaf senescence through the GA pathway.
DELLAs function as crucial transcriptional repressors of the GA pathway, and have been shown to modulate GA-mediated response through interactions with downstream transcription factors. We previously found that GA affects floral initiation via interactions between DELLA and CO or WRKY proteins, including WRKY12, WRKY13 and WRKY75 (Li et al., 2016; Wang et al., 2016; Zhang et al., 2018). We also found that GA modulates aged-triggered leaf senescence through a physiological interaction with WRKY45 (Chen et al., 2017). The expression of WRKY75 is markedly elevated at both mRNA and protein levels in senescing leaves, and the physiological interaction between WRKY75 and DELLA proteins may interfere with WRKY75-mediated transcriptional activation (Fig. 4). Furthermore, RGL1 can disrupt the association of WRKY75 and its target genes in vivo (Fig. 4), and GAI gain-of-function and RGL1 overexpression can partially delay the precocious senescence phenotype caused by WRKY75 overexpression (Figs. 5 and S4). Together, these data suggest that WRKY75 may function downstream of DELLAs to modulate leaf senescence.
In Arabidopsis, leaf senescence is tightly associated with flowering, and a delay in flowering always delays senescence. Previous studies have shown that DELLA proteins retard both flowering and senescence in Arabidopsis, and that GA signaling positively regulates flowering and senescence by degrading DELLA proteins (Wilson et al., 1992; Achard et al., 2007; Chen et al., 2017). The elimination of DELLA inhibition is thought to accelerate the onset of the reproductive stage, subsequently promoting leaf senescence. Our findings support these results. Specifically, we found that the life cycle of della mutants was shortened, whereas the lifespans of the GA biosynthesis mutant ga1 and the gid1a/gid1b/gid1c triple mutants were prolonged. Combined with our previous findings (Zhang et al., 2018), we have demonstrated that flowering and leaf senescence are delayed in WRKY75 knockout plants and accelerated in plants overexpressing WRKY75. We also found that WRKY75 can regulate flowering and leaf senescence through the GA pathway, indicating that WRKY75 may act as a common component of the GA-mediated regulatory network for both flowering and senescence. Identification of regulatory factors like WRKY75 improves our understanding of plant flowering and senescence processes.
5. ConclusionsIn this study, we provide evidence that WRKY75 acts as a positive regulator in age-triggered leaf senescence. Our findings suggest that WRKY75 may modulate the onset and progression of leaf senescence within a senescence-associated transcriptional network by integrating both age and GA signaling. These findings increase our understanding of senescence-associated signaling and transcriptional reprogramming controlled by WRKY proteins.
Author contributionsLGC conceived the project and designed the experiments. HYZ, LPZ, SGW and YLC performed the experiments. LGC wrote the article. All authors interpreted and discussed the data.
Declaration of Competing InterestThe authors declare no conflict of interest.
AcknowledgmentsWe thank Dong-Tao Ren and Martin Hülskamp for kindly providing the wrky75-1 and wrky75-25 mutant seeds, respectively. This work was funded by the National Key R & D Plan (2016YFD0101006), Natural Science Foundation of China (31671275), and Yunnan Fundamental Research Projects (2019FA010).
Appendix A. Supplementary dataSupplementary data to this article can be found online at _aaaaaa_paichu__.
Achard P., Baghour M., Chapple A., et al, 2007. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA, 104: 6484-6489. DOI:10.1073/pnas.0610717104 |
Buchanan-Wollaston V., Page T., Harrison E., et al, 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J., 42: 567-585. DOI:10.1111/j.1365-313X.2005.02399.x |
Chen L.G., Xiang S.Y., Chen Y.L., et al, 2017. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant., 10: 1174-1189. DOI:10.1016/j.molp.2017.07.008 |
Chen L.G., Zhang L.P., Li D.B., et al, 2013. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 110, E1963-E1971.. Proc. Natl. Acad. Sci. USA, 110: E1963-E1971. DOI:10.1073/pnas.1221347110 |
Chen M.X., Maodzeka A., Zhou L.H., et al, 2014. Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci., 219-220: 26-34. DOI:10.1016/j.plantsci.2013.11.016 |
Devaiah B.N., Karthikeyan A.S., Raghothama K.G., 2007. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol., 143: 1789-1801. DOI:10.1104/pp.106.093971 |
Encinas-Villarejo S., Maldonado A.M., Amil-Ruiz F., et al, 2009. Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) Fa WRKY1 and Arabidopsis at WRKY75 proteins in resistance. J. Exp. Bot., 60: 3043-3065. DOI:10.1093/jxb/erp152 |
Eulgem T., Rushton P.J., Robatzek S., et al, 2000. The WRKY superfamily of plant transcription factors. Trends Plant Sci., 5: 199-206. DOI:10.1016/S1360-1385(00)01600-9 |
Gan S., Amasino R.M., 1995. Inhibition of leaf senescence by autoregulated production of cytokinin. Science, 270: 1986-1988. DOI:10.1126/science.270.5244.1986 |
Guo P.G., Li Z.H., Huang P.X., et al, 2017. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29 : 2854-2870.. Plant Cell, 29: 2854-2870. DOI:10.1105/tpc.17.00438 |
Guo Y., Cai Z., Gan S., 2004. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ., 27: 521-549. DOI:10.1111/j.1365-3040.2003.01158.x |
Hortensteiner S., 2006. Chlorophyll degradation during senescence. Annu. Rev.Plant Biol., 57: 55-77. DOI:10.1146/annurev.arplant.57.032905.105212 |
Jibran R., A Hunter D., P Dijkwel P., 2013. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol., 82: 547-561. DOI:10.1007/s11103-013-0043-2 |
Jiang Y.J., Liang G., Yang S.Z., et al, 2014. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell, 26: 230-245. DOI:10.1105/tpc.113.117838 |
Li Z., Peng J., Wen X., et al, 2013. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell., 25: 3311-3328. DOI:10.1105/tpc.113.113340 |
Lim P.O., Kim H.J., Nam H.G., 2005. Leaf senescence. Annu. Rev. Plant Biol., 58: 115-136. |
Li W., Wang H.P., Yu D.Q., 2016. The Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant, 9: 1492-1503. DOI:10.1016/j.molp.2016.08.003 |
Lichtenthaler H.K., 1987. Chlorophylls and carotenoids pigments of photosynthetic biomembranes. Method. Enzymol., 148: 350-382. |
Miao Y., Zentgraf U., 2007. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell, 19: 819-830. DOI:10.1105/tpc.106.042705 |
Park J.H., Oh S.A., Kim Y.H., et al, 1998. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol. Biol., 37: 445-454. DOI:10.1023/A:1005958300951 |
Qi T.C., Wang J.J., Huang H., et al, 2015. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup Ⅲe and Ⅲd factors in Arabidopsis. Plant Cell, 27: 1634-1649. DOI:10.1105/tpc.15.00110 |
Rishmawi L., Pesch M., Juengst C., et al, 2014. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol., 165: 186-195. DOI:10.1104/pp.113.233775 |
Robatzek S., Somssich I.E., 2002. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev., 16: 1139-1149. DOI:10.1101/gad.222702 |
Saleh A., Alvarez-Venegas R., Avramova A., 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc., 3: 1018-1025. DOI:10.1038/nprot.2008.66 |
Ulker B., Shahid Mukhtar M., Somssich I.E., 2007. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta, 226: 125-137. DOI:10.1007/s00425-006-0474-y |
Ulker B., Somssich I.E., 2004. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol., 7: 491-498. DOI:10.1016/j.pbi.2004.07.012 |
Wang H., Pan J.J., Li Y., et al, 2016. The DELLA-CONSTANS transcription factor cascade integrates gibberelic acid and photoperiod signaling to regulate flowering. Plant Physiol., 172: 479-488. DOI:10.1104/pp.16.00891 |
Weaver L.M., Gan S., Quirino B., et al, 1998. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol., 37: 455-469. DOI:10.1023/A:1005934428906 |
Wilson R.N., Heckman J.W., Somerville C.R., 1992. Gibberellin is required for floweringin Arabidopsis thaliana under short days. Plant Physiol., 100: 403-408. DOI:10.1104/pp.100.1.403 |
Winter D., Vinegar B., Nahal H., et al, 2007. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PloS One, 2: e718. DOI:10.1371/journal.pone.0000718 |
Zhang L.P., Chen L.G., Yu D.Q., 2018. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol., 176: 790-803. DOI:10.1104/pp.17.00657 |
Zhang S.C., Li C., Wang R., et al, 2017. The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol., 173: 2294-2307. DOI:10.1104/pp.16.00008 |
Zhou X., Jiang Y.J., Yu D.Q., 2011. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells, 31: 303-313. DOI:10.1007/s10059-011-0047-1 |



