b. College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, Guangdong, China;
c. College of Life Sciences, Anhui Normal University, Wuhu, 241000, Anhui, China;
d. College of Landscape Architecture, Fujian Agriculture and Forestry University, Fuzhou, 350002, Fujian, China
Caryophyllaceae is the largest family in Caryophyllales with ca. 100 genera and 3000 species (Hernández-Ledesma et al., 2015). Traditionally, the family has been divided into three subfamilies according to the morphological characters of the stipules and sepals as well as the position of stamens in flowers (Lu et al., 2001). However, recent molecular phylogenetic analyses of these three subfamilies indicated that although the monophyly of the family was strongly supported, none of the three subfamilies were monophyletic (Harbaugh et al., 2010; Greenberg and Donoghue, 2011; Sadeghian et al., 2015). Based on molecular evidence, 11 tribes are now circumscribed and widely accepted in Caryophyllaceae, although relationships among them as of yet have not been well resolved (Harbaugh et al., 2010; Greenberg and Donoghue, 2011). In the last decade, a series of molecular phylogenetic studies have also made great progress in generic delimitation within Caryophyllaceae: some genera have been recircumscribed, such as Arenaria L., Minuartia L., Pseudostellaria Pax and Stellaria L. (Dillenberger and Kadereit, 2014; Pusalkar and Singh, 2015; Sadeghian et al., 2015; Zhang et al., 2017; Sharples and Tripp, 2019); some new genera have been described, such asMinuartiella Dillenb. & Kadereit, Mcneillia Dillenb. & Kadereit, Shivparvatia Pusalkar & D. K. Singh, Hartmaniella M. L. Zhang & Rabeler, Nubelaria M. T. Sharples & E. Tripp and Rabelera (L.) M. T. Sharples & E. Tripp (Dillenberger and Kadereit, 2014; Pusalkar and Singh, 2015; Zhang et al., 2017; Sharples and Tripp, 2019); and some genera have been reinstated, such as Cherleria L., Eremogone Fenzl, Facchinia Rchb., Odontostemma Benth. ex G. Don and Sabulina Rchb (Dillenberger and Kadereit, 2014; Sadeghian et al., 2015). These studies dramatically improved our understanding of the evolutionary relationships among Caryophyllaceae members.
The monotypic genus Pseudocerastium C.Y. Wu, X.H. Guo & X.P. Zhang was described in 1998 based on a collection (X.H. Guo 951054, ANUB & KUN) from Anhui province, China (Zhang and Guo, 1998), and it is one of the two Caryopyllaceae genera endemic to China (the other is Psammosilene W.C. Wu & C.Y. Wu) (Lu et al., 2001; Yu et al., 2016). Psammosilene was well studied in recent phylogenetic analyses, and its phylogenetic position within Caryophyllaceae has been well clarified with its generic status well supported (Greenberg and Donoghue, 2011; Li et al., 2019). However, the genus Pseudocerastium has never been sampled in phylogenetic studies. Morphological analysis suggested thatPseudocerastium is closely related to Cerastium L., Myosoton Moench and Stellaria, which are included in the tribe Alsineae as traditionally circumscribed (Zhang and Guo, 1998). However, the Alsineae has been shown to be polyphyletic, and, under the current classification of Caryophyllaceae, has subsequently been subdivided into at least four tribes (viz. Alsineae, Sclerantheae, Sagineae, Sperguleae) (Harbaugh et al., 2010; Greenberg and Donoghue, 2011). Thus, the phylogenetic position of Pseudocerastium within Caryophyllaceae is still unclear, and a complete phylogenetic framework of the family is necessary to resolve this issue.
In the present study, we explored the phylogenetic position and taxonomic status of the genus Pseudocerastium within Caryophyllaceae. For this purpose, phylogenetic analyses were conducted that sampled Pseudocerastium stellarioides X.H. Guo et X.P. Zhang and representatives of Caryophyllaceae reported in previous molecular phylogenetic studies (Greenberg and Donoghue, 2011; Zhang et al., 2017). To better understand the relationships among Pseudocerastium and its relatives, we used our new phylogenetic framework to analyze morphological character evolution.
2. Material and methods 2.1. Taxon sampling and DNA sequencesTo avoid misidentification of Pseudocerastium stellarioides, a small dried leaf from one isotype (X. H. Guo 951054, ANUB-13023625) deposited in the ANUB herbarium was sampled. Total DNA was extracted and then sequenced by genome skimming following the protocol of Zeng et al. (2018). Plastid and nuclear ribosomal internal transcribed spacer (ITS) sequence reads were assembled using the software GetOrganelle (Jin et al., 2020), with the reference plastid genome of Colobanthus quitensis (Kunth) Bartl (GenBank accession number: NC_028080) and ITS sequence of Stellaria media (L.) Vill. (MK044722), respectively. All of the obtained genes in the plastid genome were annotated in the software PGA (Qu et al., 2019).
The phylogenetic analyses of this study were done using two data sets. First, a plastid phylogenomic analysis of Caryophyllaceae was conducted to reveal the approximate position at the tribe level of Pseudocerastium within the family. Twenty species representing 12 genera and six tribes of Caryophyllaceae were sampled, and seven species from Achatocarpaceae, Amaranthaceae and Gisekiaceae were selected as outgroups based on previous phylogenetic relationships reported in Yao et al. (2019). Detailed information of all species sampled are provided in Table 1. Based on the results from the first data set, we used five DNA regions (matK, rbcL, rps16 intron, trnL-F and ITS) to conduct a phylogenetic study of the tribe Alsineae with enlarged taxon sampling (57 species belonging to 13 genera). Arenaria serpyllifolia L. from the tribe Arenarieae was selected as outgroup based on the phylogenetic framework reported in Greenberg and Donoghue (2011). Sequences of the four plastid regions of P. stellarioides were extracted from the assembled whole plastid genome. Detailed information of all species sampled and sequences used are available in Table 2.
| Taxa | GAN | Taxa | GAN |
| Agrostemma githago L. | NC_023357 | Psammosilene tunicoides W.C. Wu & C.Y. Wu | NC_045947 |
| Cerastium arvense L. | MH627219 | Pseudocerastium stellarioides X.H. Guo et X.P. Zhang | MT507771 |
| Colobanthus apetalus (Labill.) Druce | NC_036424 | Pseudostellaria heterophylla (Miq.) Pax | MK801111 |
| Colobanthus quitensis (Kunth) Bartl. | NC_028080 | Pseudostellaria okamotoi Ohwi | NC039974 |
| Dianthus caryophyllus L. | NC_039650 | Pseudostellaria longipedicellata S. Lee, K. Heo & S.C. Kim | NC_039454 |
| Dianthus gratianopolitanus Vill. | LN877395 | Pseudostellaria palibiniana (Takeda) Ohwi | NC041166 |
| Dianthus longicalyx Miq. | KM668208 | Silene aprica Turcz. | MK397897 |
| Gymnocarpos przewalskii Bunge ex Maxim. | NC036812 | Silene capitata Kom. | NC_035226 |
| Gypsophila vaccaria (L.) Sm. | NC_040936 | Silene psammitis Link ex Spreng. | MN365990 |
| Lychnis wilfordii (Regel) Maxim. | NC035225 | Spergula arvensis L. | NC041240 |
| Outgroups | |||
| Achatocarpus pubescens C.H. Wright | NC_040947 | Gisekia pharnaceoides L. | NC_041296 |
| Amaranthus hypochondriacus L. | NC_030770 | Phaulothamnus spinescens A. Gray | MH286322 |
| Alternanthera philoxeroides (Mart.) Griseb. | MK450441 | Ptilotus polystachyus (Gaudich.) F. Muell. | NC_046575 |
| Celosia argentea L. | NC_041294 | ||
| Taxa | GenBank accession numbers | ||||
| matK | rbcL | rps16 intron | trnL-F | nrITS | |
| Cerastium arvense L. | AY936295 | JX848446 | MH243535 | FJ404976 | MH219805 |
| Cerastium beeringianum Cham. & Schltdl. | KC474448 | KC482422 | – | AY521318, AY521365 | MG236459 |
| Cerastium brachypetalum Pers. | – | KF997372 | – | – | – |
| Cerastium davuricum Fisch. ex Spreng. | KX158358 | KX158395 | KX158432 | – | KX158321 |
| Cerastium dichotomum L. subsp. inflatum (Link) Cullen | KX158359 | KX158396 | KX158433 | – | KX158322 |
| Cerastium dinaricum Beck & Szyszył. | – | – | – | KJ716526 | KJ716515 |
| Cerastium fontanum Baumg. | KX821263 | KF602216 | FJ404899 | FJ404977 | GU444015 |
| Cerastium furcatum Cham. & Schltdl. | MH116578 | MH116103 | – | – | MH117479 |
| Cerastium glomeratum Thuill. | JN895359 | HM849882 | – | KY697436 | AY857977 |
| Cerastium latifolium L. | – | KF602212 | – | AY521301, AY521348 | – |
| Cerastium nigrescens (H.C. Watson) Edmondston ex H.C. Watson | – | KF997275 | – | AY521315, AY521362 | KX165939 |
| Cerastium pusillum Ser. | JN589226 | – | – | JN589683 | JN589112 |
| Cerastium regelii Ostenf. | KC474450 | KC482424 | – | AY521317, AY521364 | MG236500 |
| Cerastium subtriflorum Dalla Torre & Sarnth. | – | – | – | KJ716527 | MH537035 |
| Cerastium szechuense F.N. Williams | – | – | – | JN589674 | JN589116 |
| Cerastium tomentosum L. | JN589244 | KF997321 | MH243538 | AY521310, AY521357 | JN589031 |
| Dichodon cerastoides (L.) Rchb. | – | MG249356 | MH243542 | AY521340, AY521388 | MH219812 |
| Dichodon dubium (Bastard) Ikonn. | – | – | MH243544 | AY521341, AY521389 | MH219815 |
| Hartmaniella oxyphylla (B.L. Rob.) M.L. Zhang | KX158348 | KX158385 | KX158422 | – | KX158311 |
| Hartmaniella sierra (Rabeler & R.L. Hartm.) M.L. Zhang | KX158351 | KX158388 | KX158425 | – | KX158314 |
| Holosteum marginatum C.A. Mey. | JN589261 | – | – | JN589732 | JN589093 |
| Holosteum umbellatum L. | MK520188 | MK525977 | FJ404909 | JN589655 | JN589051 |
| Lepyrodiclis Fenzl | FJ404840 | JQ933385 | KP149043 | FJ404989 | KP148941 |
| Moenchia erecta (L.) G. Gaertn., B. Mey. & Scherb. | JN895271 | JN892479 | FJ404926 | FJ405002 | JN589103 |
| Myosoton aquaticum (L.) Moench | JN894058 | KM360890 | MH243547 | FJ405004 | AY594303 |
| Odontostemma barbatum (Franch.) Sadeghian & Zarre | – | – | – | – | KP148852 |
| Odontostemma fridericae (Hand.-Mazz.) Sadeghian & Zarre | – | – | – | – | AY936332 |
| Odontostemma roseiflorum (Sprague) Sadeghian & Zarre | FJ404825 | – | FJ404895 | FJ404971 | AY936244 |
| Odontostemma trichophorum (Franch.) Sadeghian & Zarre | – | – | – | – | AY936243 |
| Pseudocerastium stellarioides X. H. Guo et X. P. Zhang | MT507771 | MT507771 | MT507771 | MT507771 | MT791125 |
| Pseudostellaria heterophylla (Miq.) Pax | KX158371 | KX158408 | KX158445 | EU785992 | KX158334 |
| Pseudostellaria jamesiana (Torr.) W.A. Weber & R.L. Hartm. | KX158343 | KX158380 | KX158417 | FJ405010 | KX158306 |
| Pseudostellaria japonica (Korsh.) Pax | KX158344 | KX158381 | KX158418 | – | KX158307 |
| Pseudostellaria maximowicziana (Franch. & Sav.) Pax | KX158346 | KX158383 | KX158420 | – | KX158309 |
| Pseudostellaria tianmushanensis G.H. Xia & G.Y. Li | KX158355 | KX158392 | KX158429 | – | KX158318 |
| Pseudostellaria tibetica Ohwi | KX158354 | KX158391 | KX158428 | – | KX158317 |
| Rabelera holostea (L.) M.T.Sharples & E.Tripp | KX183916 | FJ395575 | MH243549 | JN589664 | KX183997 |
| Shivparvatia ciliolata (Edgew.) Pusalkar & D. K. Singh | – | – | – | – | KP148859 |
| Shivparvatia glanduligera (Edgew.) Pusalkar & D. K. Singh | – | – | – | – | KP148867 |
| Shivparvatia stracheyi (Edgew.) Pusalkar & D. K. Singh | – | – | – | – | KP148898 |
| Stellaria alsine Grimm | HM850778 | HM850385 | – | EU785987 | AY438312 |
| Stellaria americana (Porter ex B.L. Rob.) Standl. | KX158372 | KX158409 | KX158446 | JN589675 | KX158335 |
| Stellaria borealis Bigelow | JN589285 | MG247728 | – | JN589713 | JN589064 |
| Stellaria chinensis Regel | JN589241 | – | – | EU785990 | JN589133 |
| Stellaria corei Shinners | JN589300 | – | – | JN589715 | JN589046 |
| Stellaria crassifolia Ehrh. | KC475924 | KC484145 | – | JN589701 | JN589071 |
| Stellaria cuspidata Willd. ex D.F.K. Schltdl. | JN589268 | – | FJ404952 | JN589641 | JN589099 |
| Stellaria graminea L. | MK520714 | KM360998 | MH243548 | JN589687 | AY594304 |
| Stellaria longifolia Muhl. ex Willd. | MK520715 | JX848448 | – | GQ245567 | JN589146 |
| Stellaria longipes Goldie | KC475949 | JX848449 | – | JN589672 | JN589086 |
| Stellaria media (L.) Vill. | HM850779 | AF206823 | Z83152 | EU785989 | MK044722 |
| Stellaria nemorum L. | AY936298 | JN893484 | – | HM590349 | AY936246 |
| Stellaria palustris Ehrh. ex Retz. | MK520716 | KX158401 | KX158438 | – | JN589080 |
| Stellaria pubera Michx. | FJ404878 | KP643834 | – | FJ405027 | JN589127 |
| Stellaria soongorica Roshev. | MF158660 | KX158402 | KX158439 | – | KX158328 |
| Stellaria umbellata Turcz. | JN589254 | MG246195 | – | JN589737 | JN589109 |
| Stellaria vestita Kurz | MH116882 | MH116433 | – | EU785988 | MH117776 |
| Outgroup | |||||
| Arenaria serpyllifolia L. | KX158357 | KX158394 | KX158431 | FJ404972 | KX158320 |
In the Caryophyllaceae-wide analysis, 83 coding regions (79 protein-coding genes and four rRNA genes) were extracted from the plastid genomes of all species sampled, and then concatenated. A maximum likelihood (ML) tree was reconstructed based on the concatenated data set using RAxML-HPC2 (8.1.2) (Stamatakis, 2006) on the CIPRES cluster (Miller et al., 2010) under the GTR + Γ model, with remaining parameters left at default values. A rapid bootstrap (BS) analysis using the same model with 1000 pseudoreplicates was conducted to obtain the support values.
In the Alsineae-wide analyses, three data sets (i.e., cpDNA data set including matK, rbcL, rps16 intron and trnL-F; ITS data set; the combined cpDNA-ITS data set including all five regions) were constructed and used in phylogenetic analyses through two approaches: ML and Bayesian inference (BI). In these three data sets, a few gene sequences of some individuals were absent and thus coded as missing data. ML analyses of the three data sets were similar to Caryophyllaceae-wide analysis. BI analysis was conducted using MrBayes v.3.2.6 (Ronquist and Huelsenbeck, 2003) on the CIPRES cluster (Miller et al., 2010) with default parameters. Models of nucleotide substitution were selected under the Akaike Information Criterion (AIC) using jModelTest v. 3.7 (Posada, 2008). Selected models included the TPM1uf + Γ for matK, HKY + I + Γ for rbcL, TPM1uf + Γfor rps16 intron, TVM + Γ for trnL-F, and SYM + I + Γ for ITS. Each Markov chain Monte Carlo (MCMC) analysis was sampled once every 100 generations for 2, 000, 000 generations, and the chain convergence was assessed by confirming that the average standard deviation (SD) of the split frequencies fell below 0.01. Tracer v.1.6 (Rambaut et al., 2014) was used to determine whether the parameter samples were drawn from a stationary, unimodal distribution, and whether adequate effective sample sizes (ESS) for each parameter (ESS > 200) were reached. Posterior probabilities (PP) were determined from the posterior distribution after, discarding the first 25% trees of each run as burn-in.
2.3. Morphological character evolutionTwo morphological characters, viz. the number of styles, and the number of lobes at the apex of the capsule, have been suggested to be important in distinguishing different genera within the tribe Alsineae (Lu et al., 2001; Zhang et al., 2017). These two morphological characters were selected and analyzed using Mesquite v.3.01 (Maddison and Maddison, 2014) under the phylogenetic framework of Alsineae derived from the ML analysis of the combined cpDNA-ITS data set. Characters were unordered and equally weighted. Morphological characters and their state were coded as follows: (a) the number of styles is two (coded as 0), three (1), four (2), five (3), two or three (4); (b) the number of lobes at the apex of capsule is two (0), three (1), four (2), six (3), eight (4), ten (5), or unknown (?).
3. Results 3.1. Phylogenetic analysesThe Caryophyllaceae-wide concatenated data set of 83 plastid genes contained 74, 334 characters. ML analysis using this data set strongly suggested the monophyly of Caryophyllaceae (MLBS = 100%), resolved all relationships among the six tribes sampled, and provided strong support for all phylogenetic nodes within the family (MLBSs = 100%) (Fig. 1). The tribe Paronychieae diverged first within the Caryophyllaceae, and the two tribes Sagineae and Sperguleae were successively sister to a large clade comprised of other members sampled in family. A well-supported relationship between the tribe Alsineae and the Caryophylleae-Sileneae clade was recovered. Additionally, the genusPseudocerastium clustered within Alsineae and was strongly supported (MLBS = 100%) to be sister to the genus Cerastium (Cerastium arvense L. represented here).
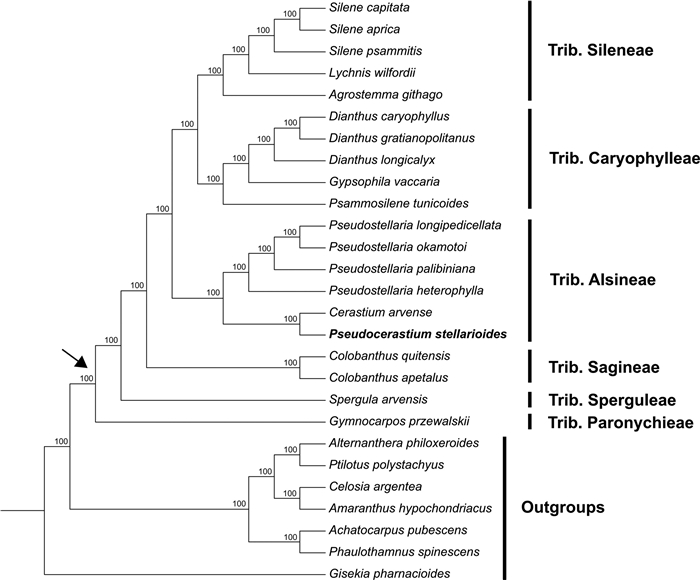
|
| Fig. 1 Maximum likelihood (ML) tree of Caryophyllaceae inferred from concatenated data set of 83 plastid genes. Numbers associated with nodes are ML bootstrap support. The crown node of Caryophyllaceae is shown by the arrowhead. |
Both BI and ML analyses of the three data sets focused on the Alsineae (cpDNA, ITS, combined cpDNA-ITS data sets) yielded largely consistent topologies (Fig. 2, Fig. 3). Results from the combined cpDNA-ITS analyses recovered two main, well-supported clades (MLBSs = 100%, PPs = 1.00) within the tribe (Fig. 3): one contains four genera (viz. Lepyrodiclis Fenzl, Odontostemma, Pseudostellaria, Shivparvatia) and the species Stellaria americana (Porter ex B.L. Rob.) Standl; the other clade comprises nine genera, viz. Dichodon (Bartl. ex Rchb.) Rchb., Moenchia Ehrh., Holosteum L., Cerastium, Pseudocerastium, Stellaria (except S. americana), Myosoton, Hartmaniella and Rabelera. Relationships among all of the genera sampled in Alsineae were resolved with high support except several nodes that were not well supported, such as the positions of Holosteum and Moenchia, and the sister relationship between Lepyrodiclis and Shivparvatia (Fig. 3).
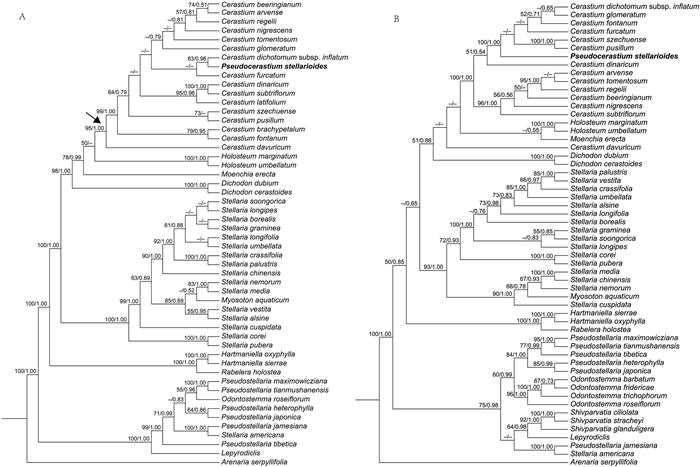
|
| Fig. 2 Maximum likelihood (ML) trees of Alsineae inferred from the cpDNA data set (A; including four plastid regions: matK, rbcL, rps16 intron and trnL-F) and ITS data set (B). Bootstrap (BS) value ≥ 50% in ML analysis and posterior probability (PP) ≥ 0.50 in Bayesian inference (BI) is indicated on the left and right of slanting bar associated with phylogenetic node, respectively. Dashes denote that the phylogenetic node associated was not supported or the BS value is < 50% in ML analyses or PP < 0.50 in BI. |
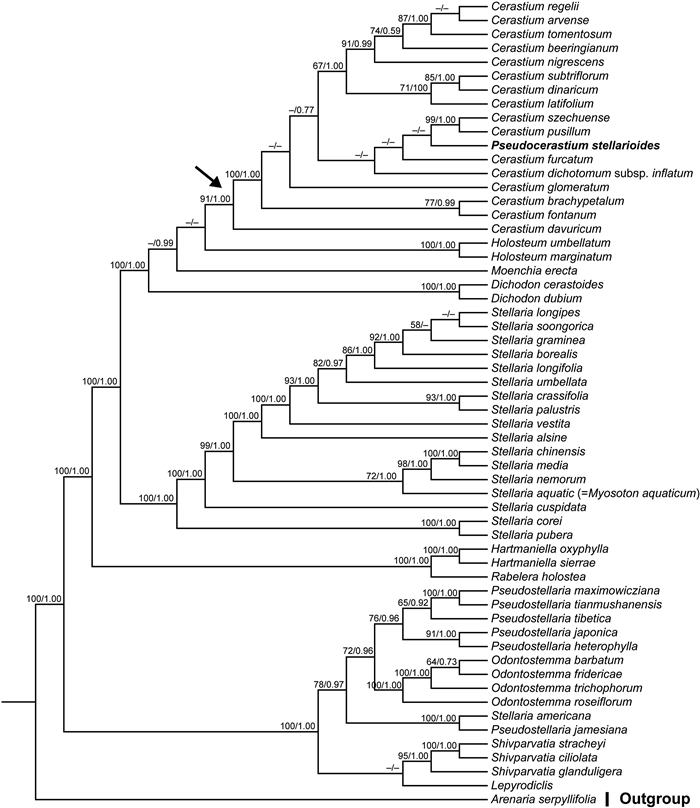
|
| Fig. 3 Maximum likelihood (ML) trees of Alsineae inferred from the combined cpDNA-ITS data set (including matK, rbcL, rps16 intron, trnL-F and ITS). Bootstrap (BS) value ≥ 50% in ML analysis and posterior probability (PP) ≥ 0.50 in Bayesian inference (BI) is indicated on the left and right of slanting bar associated with phylogenetic node, respectively. Dashes denote that the phylogenetic node associated was not supported or the BS value is < 50% in ML analyses or PP < 0.50 in BI. The crown node of Cerastium is shown by the arrowhead. |
Phylogenetic trees of Alsineae generated using BI and ML methods based on the three data sets all showed that the species P. stellarioides was nested within the large genus Cerastium with strong support, although the relationships among its closest relatives were not well supported (Fig. 2, Fig. 3). Based solely on the combined cpDNA-ITS data set, P. stellarioides clustered in a clade containing four Cerastium species (viz. C. szechuense F.N. Williams, C. pusillum Ser., C. furcatum Cham. & Schltdl. and C. dichotomum L. subsp. inflatum (Link) Cullen), all with low support. Additionally, the monophyly of the large genus Stellaria was not supported due to the nesting of the monotypic genus Myosoton within Stellaria and separation of S. americana from the core Stellaria. A similar situation also existed in Pseudostellaria, with the species Pseudostellaria jamesiana (Torr.) W.A. Weber & R.L. Hartm. recovered as sister to Stellaria americana and isolated from the core Pseudostellaria.
3.2. Morphological character evolutionOur analysis of morphological character evolution in Alsineae indicated that three styles and six lobes at the apex of the capsule may be ancestral characters, while other characters (e.g., two, four, or five styles, and two, three, four, eight, or ten lobes at the apex of the capsule) might be derived (Fig. 4). The genus Pseudocerastium has five styles and ten lobes at the apex of the capsule, traits that it shares with the large genus Cerastium and the monotypic genus Myosoton (Fig. 4).
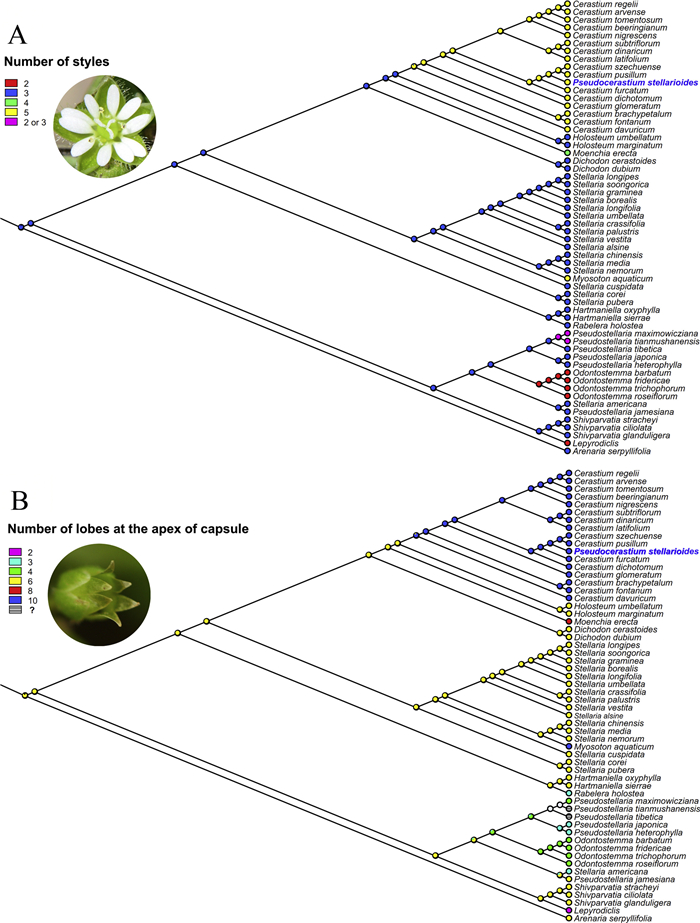
|
| Fig. 4 Selected morphological characters (A: number of styles; B: number of lobes at the apex of capsule) optimized onto the Maximum likelihood tree of Fig. 3. |
Molecular evidence has been used to circumscribe that large family Caryophyllaceae into 11 tribes; however, the phylogenetic relationships among some tribes are weakly supported or even in conflict in previous studies (Harbaugh et al., 2010; Greenberg and Donoghue, 2011). For example, previous phylogenetic analyses using three cpDNA regions (matK, trnL-F, rps16) showed that the Alsineae-Arenarieae clade and Sclerantheae-Sagineae clade were successively sister to the (Caryophylleae-Eremogoneae)-Sileneae clade, although some nodes were not well-supported (Harbaugh et al., 2010). In contrast, phylogenetic analysis using six DNA regions (matK, ndhF, trnL-F, trnQ-rps16, trnS-trnfM, ITS) suggested that the sister relationship between the Alsineae-Arenarieae clade and the Sclerantheae-Sagineae clade were weakly supported, as was the large clade consisted of these four tribes being sister to the clade of (Caryophylleae-Eremogoneae)-Sileneae (Greenberg and Donoghue, 2011).
In the present study, plastid phylogenomic analysis revealed that Alsineae was strongly supported (MLBS = 100%) to be sister to the clade consisting of Caryophylleae and Sileneae, while Sagineae was sister to the clade containing the former three tribes with high support (MLBS = 100%) (Fig. 1). The phylogenetic relationships recovered here are largely consistent with that reported in Harbaugh et al. (2010), but support values of relevant nodes increased markedly (Fig. 1). This result indicates that the plastid phylogenomic approach is helpful in clarifying the relationships of Caryophyllaceae members. Increased taxon sampling of the family, especially members of the five tribes not sampled in the current phylogenomic analysis, is important for improving our understanding of the evolutionary history of the family.
Our understating of the relationships among members of Alsineae was changed dramatically by a series of recent molecular phylogenetic studies (Greenberg and Donoghue, 2011; Zhang et al., 2017; Sharples and Tripp, 2019). As a result, the description of some new genera (i.e., Hartmaniella, Nubelaria, Rabelera and Shivparvatia), the recircumscription of Cerastium, Pseudostellaria and Stellaria, as well as the reinstatement of Dichodon and Odontostemma, have been suggested (Pusalkar and Singh, 2015; Sadeghian et al., 2015; Zhang et al., 2017; Sharples and Tripp, 2019). These taxonomic suggestions are further supported in the present study, although some nodes were weakly supported, such as the phylogenetic positions of Holosteum and Moenchia, and also the sister relationship between Lepyrodiclis and Shivparvatia (Fig. 3). Additionally, the inclusion of Myosoton within the core Stellaria was also reported in previous phylogenetic studies (Greenberg and Donoghue, 2011; Sharples and Tripp, 2019), although a series of morphological traits between the two genera seem to be quite different between the two genera, such as the characters of the style and capsule (Fig. 4). A monotypic genus nested within a large morphologically diverse genus might be a common phenomenon in angiosperms, and the different morphological features of the monotypic genus might indicate that it may have undergone different evolutionary history compared with its close relatives, such as those reported in the monotypic genera Guihaiothamnus Lo (Xie et al., 2014) and Parapteropyrum A.J. Li (Tian et al., 2011; Yang et al., 2020). Thus, the specific name Stellaria aquatic (L.) Scop [ = Myosoton aquaticum (L.). Moench.] should be adopted based on phylogenetic results. Furthermore, the present study also revealed the polyphyly of bothPseudostellaria and Stellaria (Fig. 2, Fig. 3), due to the separation of P. jamesiana and Stellaria americana from the core members of both Pseudostellaria and Stellaria, respectively. The strongly supported sister relationship between the two North American species P. jamesiana and S. americana as well as their independent phylogenetic position within Alsineae, were also reported in Greenberg and Donoghue (2011) and Zhang et al. (2017). However, a recent study based on ITS sequences provided strong support for the close relationship between P. jamesiana and the genus Cerastium (Xu et al., 2019). Although the two North American species have three styles, similar to other members of the corePseudostellaria and Stellaria, S. americana has three lobes at the apex of the capsule, which is very different from that of the core Stellaria (six lobes). Also, P. jamesiana has six lobes at the apex of the capsule, which differs from that of the other Pseudostellaria species (three or four lobes). Therefore, phylogenetic and morphological evidence suggests an independent taxonomic status of the clade comprising P. jamesiana and S. americana, but an enlarged taxon sampling of the tribe will be necessary for further study.
4.2. Phylogenetic position and taxonomic status of PseudocerastiumThe genus Pseudocerastium has been widely accepted in taxonomic literature since it was published (Lu et al., 2001; Wu et al., 2005; Hernández-Ledesma et al., 2015; Wang et al., 2015; Yu et al., 2016). Zhang and Guo (1998) noted that a series of morphological characters observed in Pseudocerastium differ from those of the genera Cerastium, Myosoton and Stellaria; specifically, Pseudocerastium has a short cylindric capsule, 10 lobes at the apex of the capsule, 10 stamens, the lower part of filaments complanate and villous, 5 styles opposite sepals, seeds small and tuberculate on the surface, and 5 deeply bifid petals. However, the present phylogenetic analyses based on both plastid (Fig. 2-A) and nuclear (Fig. 2-B) sequence data sets as well as the combined cpDNA-ITS data set (Fig. 3) all indicated that Pseudocerastium is a member of Cerastium, and the close relationship between the two genera was further supported by two morphological characters (i.e., five styles and ten lobes at the apex of the capsule), although these two characters were also shared by Myosoton (Fig. 4). Other morphological characters were also shared by the genera Cerastium and Pseudocerastium, such as the cylindric capsule, 10 stamens, 5 styles with opposite sepals, and a tuberculate seed surface. Additionally, the complanation at the lower part of filaments is described in Cerastium furcatum and C. fontanum subsp. vulgare (Hartman) Greuter, and the villous filaments are also characterized by C. furcatum (Ke, 1996). The deeply 2-lobed petal was suggested to be an important character to distinguish Pseudocerastium from Cerastium; however, several species, such as C. arvense L. subsp. strictum Gaudin and C. wilsonii Takeda, all have a similar or even larger ratio of the length of petal lobes and the petals (Ke, 1996). Thus, the morphological characters used to distinguish Pseudocerastium from Cerastium seem to be unreliable.
Morphologically, the species Pseudocerastium stellarioides is mostly similar to Cerastium wilsonii, but differs from the latter by its pyriform ovary (Fig. 5-F) (vs. subglobose; Fig. 5-K), villous indumentum on the lower part of the filaments (Fig. 5-B) (vs. glabrous, Fig. 5-I) and compressed globose seeds (Fig. 5-G) (vs. subtriangular-globose; Fig. 5-M). Moreover, P. stellarioides is also morphologically different from C. dichotomum subsp. inflatum, which is the sister of P. stellarioides revealed by the phylogenetic analysis of cpDNA data set (Fig. 2-A). P. stellarioides is a perennial herb that is 5–25 cm tall, with leaves 1–2 cm wide, sepals that are ovate-oblong and 4–5 mm long, and apex of petals that are deeply 2-lobed. In contrast, C. dichotomum subsp. inflatum is an annual herb that is 10–15 cm tall, leaves much narrower and 0.2–0.5 cm wide, sepals ovate and 10–12 mm long, and apex of petals that are retuse. Therefore, based on results of molecular and morphological analyses, the reduction of Pseudocerastium to a new synonym of Cerastium and the transfer of P. stellarioides to the latter genus are proposed here.

|
| Fig. 5 Morphological comparisons between Pseudocerastium stellarioides X.H. Guo et X.P. Zhang (A–G) and Cerastium wilsonii Takeda (H–M). A & H, habit; B–C & I‒J, flower; F & K, ovary; D‒E & L, capsule; G & M, seed. |
Cerastium L., Sp. Pl. 1: 437. 1753. Type: C. arvense L.
Pseudocerastium C.Y. Wu, X.H. Guo et X.P. Zhang, in Acta Bot. Yunnan. 20 (4): 395. 1998. syn. nov. Type: P. stellarioides X.H. Guo et X.P. Zhang, in Acta Bot. Yunnan. 20 (4): 396.
Cerastium jiuhuashanense Gang Yao et J.W. Zhai, nom. nov.
Basionym: P. stellarioides X.H. Guo et X.P. Zhang, in Acta Bot. Yunnan. 20 (4): 396. Type: China. Anhui province, Jiuhuashan, alt. 800–1000 m, 29 July 1995, X.H. Guo 951054 (holotype: ANUB-13023624!; isotypes: ANUB-13023625!, ANUB-13023626!, KUN!).
Distribution: China. Anhui and Hubei.
Note: A new specific epithet is proposed here for the species studied because there is an earlier and validly published name Cerastium stellarioides Moç. ex Ser. in the genus Cerastium, and the epithet proposed here commemorates the type locality of the species studied. Additionally, it is worth noting that in the protologue of P. stellarioides, capsule of the species was described as shortly cylindric and included in sepals, however, the mature capsule of the species (Fig. 5-E) observed in the field is similar to that of many other Cerastium species (Fig. 5-L) and exceeded evidently from sepals.
Additional specimens examined: China. Hubei province, Sui-zhou, Guangshui, 15 April 2018, ZHUXX426 (CSH, KUN).
Author contributionsGang Yao and Junwen Zhai designed the research; Kun Liu prepared the DNA material of Cerastium jiuhuashanense; Gang Yao, Jiuxiang Huang and Yuling Li performed the research; Bine Xue and Yuling Li analyzed the data; Gang Yao, Bine Xue and Junwen Zhai wrote the paper.
Declaration of competing interestThe author declares no conflict of interest.
AcknowledgementsThe authors are indebted to Dr. Xinxin Zhu from Xinyang Normal University, China, for providing field images of Cerastium jiuhuashanense and C. wilsonii, for Dr. Jacob B. Landis from Cornell University, USA, for revising the English writing. This study was financially supported by grant awards from the Natural Science Foundation of Guangdong Province, China (2019A151501 1695) and the National Natural Science Foundation of China (31500180).
Dillenberger M.S., Kadereit J.W., 2014. Maximum polyphyly: Multiple origins and delimitation with plesiomorphic characters require a new circumscription of Minuartia (Caryophyllaceae). Taxon, 63: 64-88. DOI:10.12705/631.5 |
Greenberg A.K., Donoghue M.J., 2011. Molecular systematics and character evolution in Caryophyllaceae. Taxon, 60: 1637-1652. DOI:10.1002/tax.606009 |
Harbaugh D.T., Nepokroeff M., Rabeler R.K., et al, 2010. A new lineage-based tribal classification of the family Caryophyllaceae. Int. J. Plant Sci., 171: 185-198. DOI:10.1086/648993 |
Hernández-Ledesma P., Berendsohn W.G., Borsch T., et al, 2015. A taxonomic backbone for the global synthesis of species diversity in the angiosperm order Caryophyllales. Willdenowia, 45: 281-383. DOI:10.3372/wi.45.45301 |
Jin J.J., Yu W.B., Yang J.B., et al, 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 241. DOI:10.1186/s13059-020-02154-5 |
Ke, P., 1996. Cerastium. In: Tang CL (ed. ) Flora Reipublicae Popularis Sinicae 26. Science Press, Beijing, 76-93.
|
Li Y.L., Huang J.X., Yao G., 2019. Characterization of the complete plastid genome of Psammosilene tunicoides (Caryophyllaceae), and endangered medical herb endemic to south-western China. Mitochondrial DNA B., 4: 2798-2799. DOI:10.1080/23802359.2019.1659120 |
Lu, D.Q., Wu, Z.Y., Zhou, L.H., et al., 2001. Caryophyllaceae. In: Wu ZY, Raven PH (eds) Flora of China 6. Science Press & Missouri Botanical Garden Press, Beijing & St. Louis, 1-113.
|
Maddison, W.P., Maddison, D.R., 2014. Mesquite: Amodular System for Evolutionary Analysis. Version 3.01. http://mesquiteproject.org.
|
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetics trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, 1-8.
|
Posada D., 2008. JModelTest: phylogenetic model averaging. Mol. Biol. Evol., 25: 1253-1256. DOI:10.1093/molbev/msn083 |
Pusalkar P.K., Singh D.K., 2015. Taxonomic rearrangement of Arenaria (Caryophyllaceae) in Indian Western Himalaya. J. Jpn. Bot., 90: 77-91. |
Qu X.J., Moore M.J., Li D.Z., et al, 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. DOI:10.1186/s13007-019-0435-7 |
Rambaut, A., Suchard, M.A., Drummond, A.J., 2014. Tracer v1.6. Available at: http://beast.bio.ed.ac.uk/Tracer.
|
Ronquist F., Huelsenbeck J.P., 2003. MrBayes 3:Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
Sadeghian S., Zarre S., Rabeler R.K., et al, 2015. Molecular phylogenetic analysis of Arenaria (Caryophyllaceae: tribe Arenarieae) and its allies inferred from nuclear DNA internal transcribed spacer and plastid DNA rps16 sequences. Bot. J. Linn. Soc., 178: 648-669. DOI:10.1111/boj.12293 |
Sharples M.T., Tripp E.A., 2019. Phylogenetic relationships within and delimitation of the cosmopolitan flowering plant genus Stellaria L. (Caryophyllaceae): core stars and fallen stars. Syst. Bot., 44: 857-876. DOI:10.1600/036364419X15710776741440 |
Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
Tian X., Luo J., Wang A., et al, 2011. On the origin of the woody buckwheat Fagopyrum tibeticum (=Parapteropyrum tibeticum) in the Qinghai-Tibetan Plateau. Mol. Phylogent. Evol., 61: 515-520. DOI:10.1016/j.ympev.2011.07.001 |
Wang L.S., Jia Y., Zhang X.C., Qin H.N., 2015. Overview of higher plant diversity in China. Biodiv. Sci., 23: 217-224. DOI:10.17520/biods.2015049 |
Wu Z.Y., Sun H., Zhou Z.K., et al, 2005. Origin and differentiation of endemism in the flora of China. Acta Bot. Yunnan., 27: 577-604. DOI:10.1007%2Fs11515-007-0020-8 |
Xie P., Tu T., Razafimandimbison S.G., et al, 2014. Phylogenetic position of Guihaiothamnus (Rabiaceae): Its evolutionary and ecological implications. Mol. Phylogent. Evol., 78: 375-385. DOI:10.1016/j.ympev.2014.05.022 |
Xu B., Luo D., Li Z.M., et al, 2019. Evolutionary radiations of cushion plants on the Qinghai-Tibet Plateau: Insights from molecular phylogenetic analysis of two subgenera of Arenaria and Thylacospermum (Caryophyllaceae). Taxon, 68: 1003-1020. DOI:10.1002/tax.12127 |
Yao G., Jin J.J., Li H.T., et al, 2019. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogent. Evol., 134: 74-86. DOI:10.1016/j.ympev.2018.12.023 |
Yang, B., Li, L., Liu, J., et al., 2020. Plastome and phylogenetic relationship of the woody buckwheat Fagopyrum tibeticum in the Qinghai-Tibet Plateau. Plant Divers. https://doi.org/10.1016/j.pld.2020.10.001
|
Yu, S.X., Hao, G., Jin, X.F., 2016. Caryophyllaceae. In: Species Catalogue of China Vol. 1 Plants, Spermatophytes (Ⅶ), Angiosperms (Caryophyllaceae-Ericaceae). Science Press, Beijing, 1-33.
|
Zeng C.X., Hollingsworth P.M., Yang J., et al, 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods, 14: 43. DOI:10.1186/s13007-018-0300-0 |
Zhang, M.L., Zeng, X.Q., Li, C., et al., 2017. Molecular phylogenetic analysis and character evolution in Pseudostellaria (Caryophyllaceae) and description of a new genus, Hartmaniella, In: North America. Bot. J. Linn. Soc. 184, 444-456.
|
Zhang X.P., Guo X.H., 1998. A new genus of Caryophyllaceae from China. Acta Bot. Yunnan., 20: 395-398. |



