b. Key Laboratory for Plant Diversity and Biogeography of East Asia, Chinese Academy of Sciences, Kunming, Yunnan 650201, China;
c. Dobbies Garden Centre, UK-DD2 Perth, Scotland, UK;
d. Flower Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, Yunnan 650051, China
Rhododendron L. is the largest genus in Ericaceae. It contains about 1025 species and can be classified into eight subgenera (Fang and Min, 1995; Chamberlain et al., 1996). Members of this genus are distributed widely in the northern hemisphere, with the Sino-Himalayan as the main center of abundance and diversity (Fang and Min, 1995). Because of its high ornamental value and long history of cultivation, rhododendrons have become popular horticultural plants worldwide. Rhododendron species richness is highest in China, which also contains numerous endemic species (Fang et al., 2005). In fact, an estimated 600 Rhododendron species have been described in China, and large numbers of new taxa are still being discovered since the publication of Flora of China (Gao and Zhang, 2008; Gao and Li, 2009; Chen et al., 2010, 2012; Ma et al., 2013, 2015; Liao et al., 2015; Cai et al., 2016; Liu et al., 2018; Tian et al., 2019).
In recent years, field investigations in Yiliang County (Zhaotong prefecture), within the Wumeng Mountain system in northeastern Yunnan, China, have discovered several new species of Rhododendron (Gao and Zhang, 2008; Gao and Li, 2009; Tian et al., 2019), suggesting that Rhododendron diversity in this region has been underestimated. During field investigations in this region in June 2018, we collected specimens from a Rhododendron population with white flowers that were growing on steep cliffs and rocks. After a detailed examination of relevant specimens and literature, we concluded that these specimens represent a species new to science in subsect.Maddenia (Hutch.) Sleumer in subgen. Rhododendron.
Morphologically, this species is similar to Rhododendron valentinianum Forrest ex Hutch. and Rhododendron changii (W.P. Fang) W.P. Fang: it has a broadly elliptic to obovate leaf blade, rusty-yellow setae on the petiole and funnelform-campanulate corollas. However, the new species can be distinguished from these species by having sparse scales on the abaxial surface of the leaf blade, 1–2 flowers per inflorescence, and a larger, white corolla. Although both the new species and Rhododendron linearilobum R.C. Fang & A.L. Chang have similar corollas, the new species can be distinguished by having a broadly elliptic to obovate leaf blade, dense rusty-yellow setae on the petioles, and wider calyx lobes.
In recent years, as many new species have been described, molecular marker technology has greatly advanced our understanding of Rhododendron phylogeny (Gao et al., 2003; Ma et al., 2013; Yan et al., 2014; Liu et al., 2018; Du et al., 2020). However, these studies have mainly relied on a few common markers, which have limited their resolution. Next Generation Sequencing (NGS) offers a solution to these issues (Hohenlohe et al., 2010). For example, restriction site-associated DNA sequencing (RAD-seq) has been widely used in non-model species (e.g., temperate bamboos) to resolve evolutionary relationships (Heckenhauer et al., 2018; Zhang et al., 2018; Liu et al., 2020; Guo et al., 2020).
Here we confirmed that our newly collected specimen is a hitherto undescribed species using RAD-seq data to reconstruct the phylogenetic relationships of 11 Rhododendron species.
2. Materials and methods 2.1. Morphological analysisDuring field investigations in June 2018 in Xiaocaoba Nature Reserve, Zhaotong prefecture, northeastern Yunnan, China, specimens of the new species were collected. Some living plants were introduced and cultivated at the Kunming Botanical Garden, Kunming Institute of Botany, CAS. The microscopic morphology of leaves was observed through a KEYENCE VHX-6000 stereoscopic microscope (KEYENCE Corp., Osaka, Japan). In addition, three similar species were sampled for morphological comparison. All voucher specimens in this study were deposited in the herbarium of the Kunming Institute of Botany (KUN), CAS.
2.2. Restriction site-associated DNA sequencing and SNP identificationA total of 11 individuals were sampled to construct the phylogram (Appendix A). Because the morphological characters of the new species are very similar to species within subsect. Maddenia in Rhododendron, we sampled nine additional species within the subsection and selected Rhododendron simsii Planch. as the outgroup for subsequent phylogenetic analysis. High quality genomic DNA was extracted from silica gel dried young leaves using a modified CTAB protocol (Doyle, 1991).
The RAD-seq library was prepared using EcoRI to digest DNA, following the adapted protocol of Miller et al. (2007). The de novo assembly of the RAD-seq library and SNP genotyping from short-read sequences was performed using the STACKS 2.5.2 pipeline (Rochette, 2017, 2019). The raw data was demultiplexed and filtered using the process_radtags program (key parameters included -c -q -E -t 135). For each sample, short-reads were merged into loci based on a maximum likelihood framework using the program ustacks, and were then aligned into exactly-matching stacks. The stack–depth parameter was set to three (m = 3) and the within-individual distance parameter was five (M = 5). The catalog of loci from all samples was built using cstacks with a between-individual distance parameter of three (n = 3). Lastly, the loci of each sample were matched against the catalog to confirm alleles via the program sstacks. To reconstruct a phylogenetic tree, we randomly selected one SNP from each locus via the program populations. The SNP obtained from the previous STACKS pipeline was first applied by VCFtools (Danecek, 2011) with the following key parameters: –min-alleles 2 –max-alleles 2 –remove-indels –maf 0.05 –max-missing 0.2, –minGQ 30 –minDP 3.
After filtering with VCFtools, 42, 083 SNPs were obtained and then tested for neutrality using the key parameter Tajima's D with the sliding windows of 2500 and a 95% confidence interval (Tajima, 1989). The VCF file of neutral site was converted to phy format using Tassel 5.0 (Bradbury et al., 2007). Phylogenetic analysis of SNP matrices in phy format was performed using the maximum likelihood method of concatenation using IQTREE (Minh et al., 2020; Kalyaanamoorthy et al., 2017; Hoang et al., 2018) on partition A of the Beijing Supercloud Computing Center, and the best base substitution model selected from 242 DNA base substitution models was TVM+F+ASC, and the bootstrap was set as 1000. Phylogenetic trees were graphically visualized using FigTree (Price et al., 2010).
To capitalize on phylogenetic information from the sequencing data and obtain a robust taxonomic status for the new species, SNP loci were genotyped based on the de novo assembly of the RAD-seq data. After non-linkage filtering and neutral tests, we genotyped 12, 380 loci. By calculating and sorting the BIC scores for these genotyped sites, the TVMe+ASC model was selected as the best-fit model for the maximum likelihood method of the phylogenetic tree.
3. Results 3.1. Morphological charactersThe morphological characters of the new species differ from all species previously described in Rhododendron. Several traits of the new species indicate that it belongs to the subsect. Maddenia; specifically, it has a large corolla, the abaxial surface of the leaf is scaly, and a quarter of the base of the style has scales (Fig. 1, Fig. 2). The new species can be distinguished from the two most similar species (i.e., R. valentinianum and R. changii) by having sparse scales (ca. 1–2 × their own diameter apart) on the abaxial surface of leaf blade (Fig. 3), fewer flowers (1–2) per inflorescence, and a larger, white corolla (3.5–4.5 cm) (Table 1). R. linearilobum has a corolla with similar morphology and color as the new species, but can be easily distinguished from the new species by having a narrowly obovate leaf blade, densely rusty-red-villous petiole, and calyx lobes ca. 2 mm in width (Table 1).
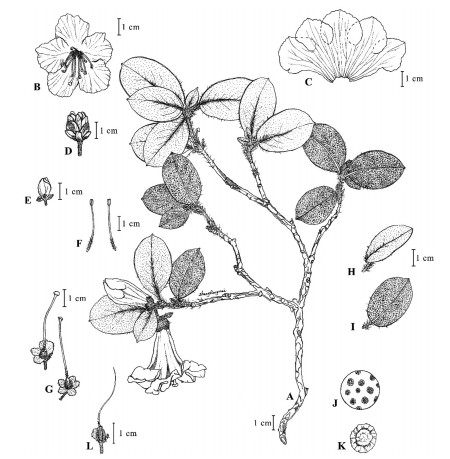
|
| Fig. 1 Illustrations of Rhododendron kuomeianum Y.H. Chang, J. Nielsen & Y.P. Ma sp. nov. Drawn by R.M. Zhang. (A) habit; (B) corolla; (C) dissected corolla; (D) inflorescence bud; (E) flower bud; (F) stamen; (G) pistil and calyx; (H) leaf adaxial surface; (I) leaf abaxial surface; (J) distribution of scales on the adaxial surface of the leaf blade; (K) single scale on the abaxial surface of leaf blade; (L) capsule. |

|
| Fig. 2 Rhododendron kuomeianum Y.H. Chang, J. Nielsen & Y.P. Ma sp. nov. (A–B) plant and habitat; (C–D) leaf; (E) inflorescence bud; (F) corolla anatomy; (G) fruit. Scale bar = 1 cm. |

|
| Fig. 3 The micrograph of the leaf surface of Rhododendron kuomeianum Y.H. Chang, J. Nielsen & Y.P. Ma sp. nov. (A) distribution of scales on the adaxial surface of the leaf blade; (B) distribution of scales on the abaxial surface of leaf blade; (C) single scale on the on the adaxial surface of the leaf blade; (D) single scale on the abaxial surface of leaf blade; (E) indumentum characteristics of the petiole. |
| Characters | R.kuomeianum | R.valentinianum | R.changii | R. linearilobum |
| Geographic distribution | NE Yunnan | SE W Yunnan NW NE Guizhou |
Chongqing | SE Yunnan |
| Elevation | 1800–2000 m | 2400–3000m | 1600–2000 m | 2200m |
| Petiole | setose | setose | setose | villous |
| Shape of the leaf blade | broadly elliptic to obovate | obovate to oblong-elliptic | obovate to oblong-elliptic | elliptic |
| Size of leaf blade | 3.5–5.5×2.5–3.5cm | 3–4×1.5–2cm | 3–4.5×2–2.5cm | 4–7.5×1.5–2.5cm |
| Scale density (as own diameter apart) | 1–2 | 0.5 or contiguous | nearly contiguous | 1–2 |
| Inflorescence | 1 or 2-flowered | 2–4-flowered | 3 or 4-flowered | 2–4-flowered |
| Pedicel | glabrous | setose | glabrous | pubescent |
| Calyx lobes | 4–7×3–5mm, ovate or broadly elliptic | 8×4–5mm, ovoid or oblong-ovate | 10×4–5mm, ovoid or oblong-ovate | 6–12×2mm, linear |
| Color of the corolla | white with pale red margins | light to bright yellow | light to bright yellow | white with pale red margins |
| Corolla size | 3.5–4.5cm long, tube 2–3cm long | 2–3.5cm long, tube 1.8–2cm long | 3.5–4cm long, tube 1.8–2cm long | ca. 4cm long, tube 1.8–2cm long |
| Proportion of scales to style | 1/4 | 1/8 | 1/8 | 2/5 |
| Indumentum characteristics of the style | glabrous | glabrous | glabrous | base pubescent |
We constructed a RAD-seq libraries and genotyped SNPs from 11 species of Rhododendron. A total of 4.6 gigabase pairs (Gb) of data was obtained, with an average of 428.22 megabase pairs (Mb) of data per sample (see Appendix B for details).
Phylogenetic trees based on the de novo assembly of our RAD-seq library reveal that all 10 species within subsect. Maddenia are clustered into one clade, with Rhododendron maddendii subsp. crassum (Franch.) Cullen as the basal species (Fig. 4). One subclade includes R. valentinianum, R. changii, Rhododendron pachypodum Balf. f. & W.W. Smith and Rhododendron kuomeianum Y.H. Chang, J. Nielsen & Y.P. Ma sp. nov., which verifies that R. kuomeianum is genetically distinct and supports its status as a new species. Notably, the topological structure of the ML tree indicates that the phylogenetic position of R. valentinianum is very close to that of R. changii. The remaining five species (Rhododendron liliiflorum Levl., R. chunienii Chun & W.P. Fang, R. maddenii subsp. maddenii (Batalin) H. Hara, R. excellens Hemsl. & Wils. and R. ciliatum Hook. f.) cluster into a separate subclade, althoughR. liliiflorum and R. chunienii cannot be distinguished from each other. In addition, R. maddenii subsp. maddenii is genetically distant from the subspecies R. maddenii subsp. crassum (Fig. 4).
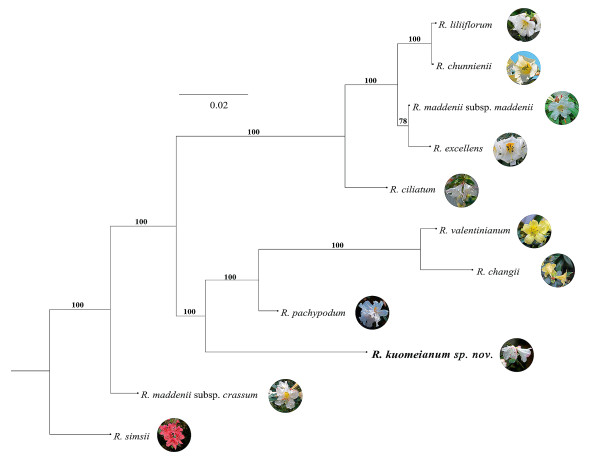
|
| Fig. 4 Maximum likelihood phylogenetic topology based on 12380 unlinked high-quality and neutral SNP sites without a reference genome. The phylogenetic tree bootstrap probabilities ≥75 are shown at branches. |
In the present study, morphological comparisons and phylogenetic analysis based on de novo assembly of a RAD-seq library confirmed that Rhododendron kuomeianum is a new species. R. kuomeianum has similar floral traits as R. linearilobum; however, the new species has an oblong-elliptic leaf blade similar to R. valentinianum and R. changii. Due to unavailability of sequencing materials of R. linearilobum, which was estimated to be < 50 plants and hence evaluated to be critically endangered by IUCN criteria (Gibbs et al., 2011), the most genetically close relative remains to be unclear.
Rhododendron is the largest genus of seed plants in China, and recognized as one of the most taxonomically challenging plants due to recent adaptive radiations and natural hybridization (Yan et al., 2014). One major taxonomic problem for this genus is that many hybrids have been incorrectly identified as new taxa. Previous studies at the Baili Rhododendron Nature Reserve in Guizhou have confirmed that many of the new taxa described in this area are actually different genotypes produced by natural hybridization betweenRhododendron delavayi Franch. and Rhododendron irroratum Franch. (Marczewski et al., 2016; Zhang et al., 2017). Morphological evidence has been used to describe taxa for over a century; however, this approach often leads to uncertainty and debate. For instance, R. changii was initially described as a variety of R. valentinianum; subsequently, it was upgraded to species status because of its larger, scaleless corolla, glabrous style, and the absence of hairs on the pedicel or calyx (Fang, 1983). After comparing holotypes and native plants, Geng (2014) restored R. changii as a variety of R. valentinianum based on morphological similarities and habitats. Our phylogenetic tree clusters these two species together, indicating that R. changii can reasonably be treated as a variety of R. valentinianum. Similarly, Li (1995) and Geng (2004) proposed that the holotype of R. chunienii, which had previously been described as having five stamens (the most unique character in this subsection), had been misdiagnosed because of damage to the corolla. Both living plants and specimens of R. chunienii collected at the type locality have 10 stamens; furthermore, other characteristics of R. chunienii are the same as in R. liliiflorum. Thus, R. chunienii has been treated as a synonym for R. liliiflorum. Our phylogenetic analysis supports this classification.
Rhododendron maddenii contains two subspecies, R. maddenii subsp.maddenii and R. maddenii subsp. crassum. Although these subspecies are morphologically similar, they are geographically isolated from one another. R. maddenii subsp. maddenii is mainly distributed in Cuona County and Naidong District in southern Tibet, China, as well as in Bhutan and northeastern India; in contrast, R. maddenii subsp. crassum is mainly distributed in western Yunnan. In this study, the high-confidence phylogenetic tree reveals that the genetic difference between these two subspecies is very large, which has also been confirmed by DNA barcoding (Yan et al., 2014). One explanation for the genetic differences between these two subspecies is that long-term geographic isolation has allowed mutations to accumulate in their respective populations.
Taxonomic treatment
Rhododendron kuomeianum Y.H. Chang, J. Nielsen & Y.P. Ma sp. nov. (Fig. 1, Fig. 2). 国楣杜鹃【guó méi dù juān】
Type. CHINA. Yunnan: Yiliang County Xiaocaoba Nature Reserve, 25°50′01″N, 104°17′41″E, alt. 2000 m, 15 Apr. 2019, Y.H. Chang, Y.P. Ma & D.T. Liu, Cyh20190402 (holotype KUN! 1498888; isotype KUN! 1498889, 1498890).
Diagnosis. Rhododendron kuomeianum resembles R. valentinianum in having a broadly elliptic to obovate leaf blade, dense rusty-yellow setae on the petiole, and a funnelform-campanulate corolla. It differs from R. valentinianum in having a white corolla with pale red margins (versus light to bright yellow), fewer flowers (1–2) per inflorescence (versus 2–4), and sparser distribution of scales on the abaxial surface of the leaf blade (1–2 × scales diameter apart) (versus 0.5 × scales diameter apart) (Table 1).
Description. Multi-branched shrubs, evergreen, 40–100 cm tall; branches short, old branches deep red to brown, young shoots scaly, hispid. Petiole 5–9 mm, scaly, hispid; leaf blade leathery, broadly elliptic to obovate, 3.5–5.5 × 2.5–3.5 cm; base broadly cuneate or rounded; margin entire and slightly revolute, sparsely setose; apex obtuse or rounded, apiculate; abaxial surface pale green, scales 1–2 × their own diameter apart, about equal in size, golden, seldom contiguous, adaxial surface deep green and shiny, densely scaly, brown hispid along midrib; midveins raised adaxially and lateral veins hardly raised; midrib concave adaxially. Inflorescence terminal, cymose, 1 or 2-flowered. Pedicel stout, 0.6–0.9 cm, scaly, without hairs; calyx 4–5-lobed to the half, lobes pale green, 4–7 × 3–5 mm, ovate or broadly elliptic, persistent in fruit, scaly abaxially and on margin, margin sparsely white-ciliate; corolla funnelform-campanulate, white with pale red margin and without blotch, 3.5–4.5 cm, tube 2–3 cm, outer surface glabrous, lobes orbicular or broadly elliptic, ca. 1.8–2.5 × 2.0–2.5 cm; stamens 10, 1.5–3.7 cm, unequal, shorter than corolla; filaments densely pubescent in lower 1/5; anthers oblong-elliptic, red brown, ca. 2.5–3 mm; ovary 5-locular, ca. 4.5 mm, densely scaly; style slightly arched, as long as corolla or exserted from corolla, 3.5–4.2 cm base densely scaly in lower 1/4. Capsule ovoid, ca. 1 cm, dehiscing from top, densely scaly. Flowering: April–May, Fruiting: after June.
Paratype. China. Yunnan: Yiliang county Xiaocaoba Nature Reserve, 25°50′06″N, 104°17′39″E, alt. 1950 m, 5 Apr. 2019, Y.H. Chang & F.M. Yang, Cyh20190401 (KUN! 1498891). Same locality, alt. 2030 m, 6 June 2018, Y.P. Ma, Y.H. Chang & J. Neilsen, Ma20180603 (KUN! 1498892, 1498893).
Distribution and habitat. To date, Rhododendron kuomeianum has only been found in the type locality at Xiaocaoba Nature Reserve, Zhaotong City, NE Yunnan, China, where it is represented by one population at elevations of 1800–2000 m (Fig. 5). In addition, we found all plants growing on rocks and cliffs, which are steep and barren (Fig. 2A). Based on the known occurrences, we conclude that R. kuomeianum is well-adapted to cold and wet environments.
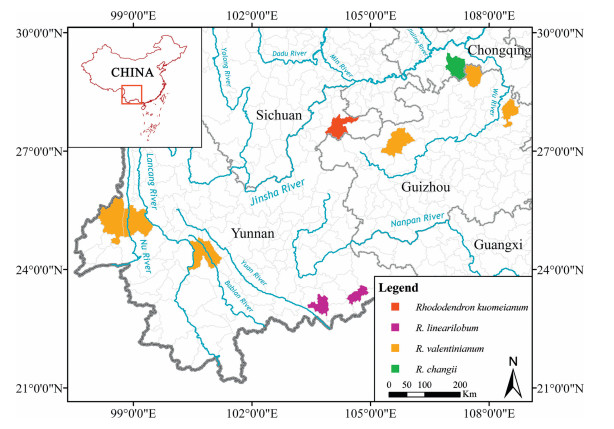
|
| Fig. 5 Distribution of Rhododendron kuomeianum Y.H. Chang, J. Nielsen & Y.P. Ma sp. nov. and other morphologically similar species in China. |
Conservation status. During many field investigations since 2018, we have only found this species in the type locality, and there are about 200 mature individuals in this population. Detailed examination of relevant specimens and literature available for Rhododendron species collected from Xiaocaoba Nature Reserve and adjacent regions revealed no additional information about the distribution of this species; therefore, this species is likely to be endemic to this region. Based on the limited information currently available, we thus tentatively assign this species to an IUCN Red List status of Data Deficient (DD) (IUCN, 2019). Further field investigation is urgently needed.
Etymology. The new species Rhododendron kuomeianum was named after Professor Kuo Mei Feng, a botanist and horticulturalist from the Kunming Institute of Botany, Chinese Academy of Sciences, to honor his great contribution to research on the genusRhododendron in China. In pinyin, the Chinese name is "Guó méi dù juān".
Author contributionsY.P. Ma conceived and designed experiments. Y.H. Chang, Y.P. Ma, D.T. Liu, and J. Neilsen performed field investigations and collected the specimens. G. Yao and L. Zhang analyzed the molecular data. Y.H. Chang wrote the manuscript. Y.H. Chang, G. Yao and Y.P. Ma revised the manuscript.
Declaration of competing interestWe declare that the named authors have no conflicts of interest, financial or otherwise.
AcknowledgmentsWe are grateful to Mrs. Rongmei Zhang for the botanical line drawing, Mr. Lianyi Li for the microscopic scanning of the leaf surface, Ms. Songting Du, Mrs. Wei Wei, and Mr. Yechun Xu for providing photos and Mr. Fengmao Yang for help during fieldwork. We also thank Mr. Shiwei Guo for his constructive comments on the manuscript. This study was financially supported by the National Natural Science Foundation of China (Grant No. 31770418 and No. 31760231) the Youth Innovation Promotion Association, Chinese Academy of Sciences (Grant No. 2018428), and the Reserve Talents for Academic and Technical Leaders of Middle-aged and Young People in Yunnan Province (Grant No. 2018HB066).
Appendix A and B. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.04.003.
Bradbury, P.J., Zhang, Z.W., Kroon, D.E., et al., 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 23: 2633-2635. DOI:10.1093/bioinformatics/btm308 |
Cai, L., Neilsen, J., Dao, Z.L., et al., 2016. Rhododendron longipedicellatum (Ericaceae), a new species from Southeastern Yunnan, China. Phytotaxa, 282: 296-300. DOI:10.11646/phytotaxa.282.4.7 |
Chamberlain, D.F., Hyam, R., Argent, G., et al., 1996. The genus Rhododendron: its classification and synonymy. Royal Botanic Garden Edinburgh, Edinburgh, pp. 181.
|
Chen, X., Huang, J.Y., Consaul, L., et al., 2010. Two new species of Rhododendron (Ericaceae) from Guizhou, China. Novon, 20: 386-391. DOI:10.3417/2008102 |
Chen, X., Huang, J.Y., Xie, H., et al., 2012. Rhododendron cochlearifolium (Ericaceae), a new species from China. Ann. Bot. Fenn., 49: 397-402. DOI:10.5735/085.049.0614 |
Danecek, P., Auton, A., Abecasis, G., et al., 2011. The variant call format and VCFtools. Bioinformatics, 15: 2156-2158. DOI:10.1093/bioinformatics/btr330 |
Doyle, J., 1991. DNA protocols for plants-CTAB total DNA isolation. In: Hewitt GM, Johnston A eds. Molecular techniques in taxonomy. Berlin: Springer. pp. 283-293.
|
Du, C., Liao S., Boufford D.E., et al., 2020. Twenty years of Chinese vascular plant novelties, 2000 through 2019. Plant Divers., 42: 393-398. DOI:10.1016/j.pld.2020.08.004 |
Fang, W.P., 1983. New taxa of the genus Rhododendron from China. Acta Phytotaxon. Sin., 21: 457-469. |
Fang, R.Z., Min, T.L., 1995. The floristic study on the genus Rhododendron. Acta Bot. Yunnanica, 17: 359-379. |
Fang, M.Y., Fang, R.C., He, M.Y., et al., 2005. Ericaceae. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds. ), Flora of China, vol. 14. Science Press, Beijing & Missouri Botanical garden Press, St. louis, pp. 260-455.
|
Gao, L.M., Li, D.Z., 2009. Rhododendron qiaojiaense (Ericaceae), a new species from Yunnan, China. Ann. Bot. Fenn., 46: 67-70. DOI:10.5735/085.046.0108 |
Gao, L.M., Zhang, S.D., 2008. Rhododendron yaoshanense (Ericaceae), a new species from NE Yunnan, China. Ann. Bot. Fenn., 45: 204-206. DOI:10.5735/085.045.0305 |
Gao, L.M., Li, D.Z., Zhang, C.Q., 2003. Phylogenetic relationships of Rhododendron section Azaleastrum (Ericaceae) based on ITS sequences. Acta Phytotaxon. Sin., 41: 173-179. |
Geng, Y.Y., 2004. New synonymies in the genus Rhododendron from China. Acta Phytotaxon. Sin., 42: 566-570. |
Geng, Y.Y., 2014. The genus Rhododendron of China. Shanghai Scientific and Technical Publishers, Shanghai, pp. 328-347.
|
Gibbs, D., Chamberlain, D., Argent, G., et al., 2011. The red list of rhododendrons. Botanic Gardens Conservation International, Richmond, UK, p. 57.
|
Guo, C., Ma, P.F., Yang, G.Q., et al., 2020. Parallel ddRAD and genome skimming analyses reveal a radiative and reticulate evolutionary history of the temperate bamboos. Syst. Biol. syaa076.
|
Heckenhauer, J., Samuel, R., Aston, P.S., et al., 2018. Phylogenomics resolves evolutionary relationships and provides insights into floral evolution in the tribe Shoreeae (Dipterocarpaceae). Mol. Phylogenet. Evol., 127: 1-13. DOI:10.1016/j.ympev.2018.05.010 |
Hoang, D.T., Chernomor, O., Von, Haeseler. A., et al., 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol., 2: 518-522. |
Hohenlohe, P., A., Bassham, S., Etter, P., D., et al., 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6, e1000862.
|
IUCN Standards and Petitions Committee., 2019. Guidelines for using the IUCN red list categories and criteria. Version 14. Prepared by the Standards and petitions committee of the IUCN species survival commission. http://www.iucnredlist.org/documents/RedListGuidelines.pdf.
|
Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K., et al., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods, 6: 587-589. DOI:10.1038/nmeth.4285 |
Li, G.Z., 1995. A revison and geographical distribution of the genus Rhododendron in Guangxi. Guihaia, 15: 193-208. |
Liao, R.L., Xue, D., Neilsen, J., et al., 2015. A new species of Rhododendron (Ericaceae) from Shangri-La, NW Yunnan, China. Phytotaxa, 238: 293-297. DOI:10.11646/phytotaxa.238.3.10 |
Liu, R.L., Zhong, B.T., Gao, L.M., 2018. A new species of Rhododendron (Ericaceae) from Jiangxi of China based on morphological and molecular evidences. Phytotaxa, 4: 267-275. |
Liu, D.T., Zhang, L., Wang, J.H., et al., 2020. Conservation genomics of a threatened Rhododendron: contrasting patterns of population structure revealed from neutral and selected SNPs. Front. Genet., 11: 757. DOI:10.1201/9781351187435-95 |
Ma, Y.P., Wu, Z.K., Xue, R.J., et al., 2013. A new species of Rhododendron (Ericaceae) from the Gaoligong Mountains, Yunnan, China, supported by morphological and DNA barcoding data. Phytotaxa, 114: 42-50. DOI:10.11646/phytotaxa.114.1.4 |
Ma, Y.P., Chamberlain, D.F., Sun, W.B., et al., 2015. A new species of Rhododendron (Ericaceae) from Baili Rhododendron nature reserve, NW Guizhou, China. Phytotaxa, 195: 197-200. DOI:10.11646/phytotaxa.195.2.11 |
Marczewski, T., Ma, Y.P., Zhang, X.M., et al., 2016. Why is population information crucial for taxonomy? A case study involving a hybrid swarm and related varieties. AoB Plants, 8: 1-17. |
Miller, M.R., Dunham, J.P., Amores, A., et al., 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA(RAD) markers. Genome Res., 2: 240-248. DOI:10.1101/gr.5681207 |
Minh, B.Q., Schmidt, H.A., Chernomor, O., et al., 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomicera. Mol. Biol. Evol., 37: 1530-1534. DOI:10.1093/molbev/msaa015 |
Price, M.N., Dehal, P.S., Arkin, A.P., 2010. FastTree 2-Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One 3, e9490.
|
Rochette, N.C., Catchen, J.M., 2017. Deriving genotypes from Rad-seq short-read data using stacks. Nat. Protoc., 12: 2640-2659. DOI:10.1038/nprot.2017.123 |
Rochette, N.C., Rivera-Colón, A.G., Catchen, J. M., 2019. Stacks 2: analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol., 21: 4737-4754. DOI:10.1111/mec.15253 |
Tajima, T., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123: 585-595. DOI:10.1093/genetics/123.3.585 |
Tian, X.L., Chang, Y.H., Neilsen, J., et al., 2019. A new species of Rhododendron (Ericaceae) from northeastern Yunnan, China. Phytotaxa, 395: 66-70. DOI:10.11646/phytotaxa.395.2.2 |
Yan, L.J., Liu, J., öller, M., et al., 2014. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. Mol. Ecol. Res., 15: 932-944. |
Zhang, J.L., Ma, Y.P., Wu, Z.K., et al., 2017. Natural hybridization and introgression among sympatrically distributed Rhododendron species in Guizhou, China. Biochem. Systemat. Ecol., 70: 268-273. DOI:10.1016/j.bse.2016.12.014 |
Zhang N.N., Ma Y.P., Folk. R.A., et al., 2018. Maintenance of species boundaries in three sympatric Ligularia (Senecioneae, Asteraceae) species. J. Integr. Plant Biol., 60: 986-999. DOI:10.1111/jipb.12674 |



