b. Key Laboratory of State Forestry Administration on Biodiversity Conservation in Karst Mountainous Areas of Southwestern China, Guizhou Normal University, Guiyang, 550025, China
The species that coexist in an ecological community do so because they are present in the species pool of the region in which the community is located and because they have characteristics that permit them to exist in that community (Webb et al., 2006). Evolutionary and ecological processes interplay to determine the species composition of an ecological community (Ricklefs, 1987). Evolutionary history has informed our view of the world at least since the publication of Charles Darwin (1859) seminal work, On the Origin of Species, but great increases in the availability of molecular data, and advances in computational tools and in the study of diversity gradients and community assembly in the past two decades have simulated the integration of evolutionary history into ecology and vice versa.
The evolutionary history of each clade (e.g., angiosperms) and the relationships among members of the clade at various evolutionary depths or taxonomic levels (e.g., family, genus, species) are imprinted in an evolutionary tree, which is commonly called a phylogeny or phylogenetic tree. A phylogeny can be used in ecological and evolutionary studies either as a backbone for investigation on which ecological information is hung or as a proxy for ecological similarity (Swenson, 2019). Phylogenies can provide important tools to address evolutionary and ecological questions (Baum and Smith, 2012). Since an early attempt of performing a phylogenetic analysis of community structure (Webb, 2000), and a review of the theoretical and empirical roots of methods for phylogenetic analysis (Webb et al., 2002), the use of phylogenies in studies investigating patterns of community structure has gone from an incidental application to a burgeoning subdiscipline (Vamosi et al., 2009), which is often called community phylogenetics (Ndiribe et al., 2013), phylogenetic community ecology (Cavender-Bares et al., 2009; Qian and Jiang, 2014), or phylogenetic ecology (Swenson, 2019).
In community phylogenetics, phylogenies have been commonly used to explore patterns of phylogenetic structure (e.g., phylogenetic diversity and phylogenetic relatedness) of species assemblages across geographical (e.g., latitudinal and elevational) and ecological (e.g., temperature and precipitation) gradients (e.g., Cavender-Bares et al., 2009; Cooper et al., 2008; Li and Sun, 2017; Qian et al., 2014; 2017; 2019; Qian and Ricklefs, 2016; Webb, 2000). Phylogenetic structure of ecological communities is determined by the processes by which species in local communities assemble from regional species pools (Webb et al., 2002). For example, phylogenies have been increasingly used in testing the tropical niche conservatism hypothesis (Algar et al., 2009; Qian et al., 2013, 2019), which predicts that within ecological communities phylogenetic relatedness of species increases from warm, humid environments to cold, dry environments, because most clades of extant species originated during the time when the planet was predominately under tropical environments (Behrensmeyer et al., 1992; Graham, 1999) and many ecological traits (e.g., cold tolerance) are phylogenetically conserved (Donoghue, 2008; Hawkins et al., 2014). Phylogenies can illuminate our knowledge of evolutionary histories of ecological communities at multiple temporal and spatial scales (Pennington et al., 2006), and can provide a historical framework to quantify evolutionary and ecological patterns and to infer evolutionary and ecological processes (Emerson and Gillespie, 2008). Since the publication of Cam Webb's seminal work on community phylogenetics (Webb, 2000), which introduced phylogenetic branch lengths to calculate the degree of relatedness among species in ecological communities, the number of articles published each year on studies of community phylogenetics increases continuously (Fig. 1). Due to the recent availability of tools for building large phylogenies and to conducting sophisticated phylogenetic analyses, a great number of phylogeny-based ecological studies have been published in the past decade (Fig. 1). In particular, recent years have seen an explosion of interest in using phylogenetic relationships to achieve ecological and evolutionary insights.

|
| Fig. 1 The number of publications indexed in Thompson Reuters ISI Web of Science based on searches using a combination of the key words 'phylogen*' and 'community' (gray plus black bars) or a combination of the key words 'phylogen*' and 'community ecology' (black bars). |
A phylogenetic study may involve several thousands or tens of thousands of species (e.g., Brunbjerg et al., 2014; Qian et al., 2013; 2019; Sandel et al., 2020). Because phylogenies that are resolved at the species level are generally not available at broad scales, particularly for plants, ecologists have been frequently using phylogenies resolved at the family level, and in some cases resolved at the genus level, with genera and species being attached to the phylogeny as polytomies (e.g., Chalmandrier et al., 2015; Munguía-Rosas et al., 2011; Giehl and Jarenkow, 2012; Hardy et al., 2012; Qian et al., 2013). For phylogenetic studies involving flowering plants, many recently published studies (e.g., Chen et al., 2020; Lancaster, 2020; Qian et al., 2019) used the megaphylogeny reported by Smith and Brown (2018) and implemented in the software V.PhyloMaker (Jin and Qian, 2019) as a backbone to generate phylogenies. This megaphylogeny, which is the largest dated megaphylogeny for seed plants (Spermatophyta), is resolved completely at the family level and mostly at the genus level for the entire flora of seed plants in the world. It includes 10, 449 genera of seed plants. According to Mabberley (2008), there are 13, 064 genera of seed plants worldwide. Thus, about 80% of the genera of seed plants in the world have been included in Smith and Brown's megaphylogeny. When the megaphylogeny is used as a backbone to generate a phylogeny for a regional or local flora, the vast majority of the plant genera in the regional or local flora can be found in Smith and Brown's megaphylogeny. For example, 87% of the 2919 genera of seed plants in China (Qian et al., 2019), 89% of the 1912 genera of angiosperms in the Himalaya (Rana et al., 2019), 94% of the 339 genera of angiosperms in the Arctic (Elven, 2011), and 98% of the 217 genera of trees in North America (north of Mexico) (Little, 1971–78) are present in Smith and Brown's megaphylogeny. However, the percentage of the species of a regional and local flora that are included in Smith and Brown's megaphylogeny is generally much lower, compared to the percentage for the genera in the flora. For example, for the above-mentioned four cases, the percentages of the species that are present in Smith and Brown's megaphylogeny are 39, 40, 56, and 72%, respectively. When the megaphylogeny is used as a backbone to generate a phylogeny for the species list of a study, additional genera and species (i.e., those that are absent from the megaphylogeny) are commonly attached to the backbone as polytomies within families (in the case of attaching genera) and genera (in the case of attaching species). The phylogeny generated in this way is generally resolved at the genus level. Although phylogenies resolved only at the genus level have been frequently used in ecological studies (e.g., Culmsee and Leuschner, 2013; Eiserhardt et al., 2013; Molina-Venegas et al., 2015), whether using phylogenies resolved at the genus level in ecological studies would bias the results of the studies, compared to those based on phylogenies resolved at the species level, has not been investigated.
In this study, we use the angiosperm tree flora of North America north of Mexico (hereafter North America) as a model system to assess whether the relationships between measures of phylogenetic structure (e.g., phylogenetic diversity and relatedness) and environmental variables based on phylogenies resolved at the genus level with species being attached to the phylogenies as polytomies would significantly differ from those based on a phylogeny completely resolved at the species level. The angiosperm tree flora of North America is an ideal system for addressing this question for several reasons. First, geographical distributions of tree species in North America have been well documented (e.g., Little, 1971–78) and have been commonly used in ecological studies (e.g., Currie and Paquin, 1987; Morin and Lechowicz, 2011), including phylogenetic studies (e.g., Qian et al., 2013; Qian et al., 2015). Second, a time-calibrated phylogeny that includes nearly all genera and the vast majority of species of angiosperm trees in North America can be extracted from the megaphylogeny reported by Smith and Brown (2018). Third, environment (e.g., temperature and precipitation) varies greatly across North America, which is ideal for investigating the relationships between measures of phylogenetic structure and environment.
2. Materials and methodsNorth America was divided into equal area quadrats of 12, 100 km2 (110 km × 110 km; Fig. 2). We determined the presence or absence of each angiosperm tree species in each quadrat by superimposing range maps on the grid system, and then generated species lists for each quadrat. Range maps of tree species distributions were obtained from a USGS website (https://www.sciencebase.gov/catalog/item/4fc518d1e4b00e9c12d8c362). We excluded those quadrats that contain land < 75% of a full-sized quadrat. We only included those angiosperm species that are present in Smith and Brown's (2018) time-calibrated megaphylogeny (GBOTB). In addition, because species-poor assemblages may have extreme values for some metrics of phylogenetic structure (Fritz and Rahbek, 2012) and assemblages with few species may produce results that are unreliable due to a large number of ties (Kamilar and Guidi, 2010), we excluded quadrats with fewer than five species to avoid spurious effects of low sample size. As a result, our final dataset included 1093 quadrats with 388 species (Appendix S1) and 174 genera of angiosperms, which are 75% and 87% of all species (519) and genera (201), respectively, of the angiosperm trees in the 1093 quadrats.

|
| Fig. 2 Geographical variation in tree species richness in North America north of Mexico. Each quadrat is 110 km by 110 km. Species richness in quadrats with land area less than 75% of a full quadrat is not shown. |
We generated four phylogenies for this study. First, we extracted a phylogeny from Smith and Brown's megaphylogeny that included only the 388 species in our final dataset. This species-level phylogeny, which was called PHYLOsp.resolved, is fully resolved for the species in our dataset. Second, we extracted a genus-level phylogeny from Smith and Brown's megaphylogeny by retaining one species per genus. For those genera that have two or more species in the megaphylogeny, we randomly selected one species for each genus. The resulting phylogeny represents a genus-level phylogeny. We attached the 388 species in our dataset to the genus-level phylogeny using two approaches. With one approach, we attached all species of a given genus to the genus-level phylogeny as basal polytomies of the genus (we called this phylogeny PHYLOsp.basal-polytomies); with the other approach, we attached all species of a given genus to the genus-level phylogeny as polytomies at the middle point of the branch length of the genus (we called this phylogeny PHYLOsp.middle-polytomies). Accordingly, the two resulting phylogenies are both species-level phylogenies but resolved at the genus level. We pruned the two phylogenies to retain the 388 species in our dataset. We also extracted a genus-level phylogeny for the 174 genera in our dataset, which was called PHYLOgen.resolved, from the above-mentioned 'global' genus-level phylogeny.
Faith's (1992) phylogenetic diversity (PD) and Webb et al.'s (2008) mean pairwise distance (MPD) and mean nearest taxon distance (MNTD) are among the most commonly used metrics of phylogenetic diversity (Carvallo et al., 2014; Eme et al., 2020). They focus on different depths of evolutionary history. PD represents the sum of the branch lengths of the phylogenetic tree linking all species of a particular assemblage; MPD is the mean phylogenetic distance (i.e., branch length) among all pairs of species within the assemblage; and MNTD is the mean distance between each species within the assemblage and its closest relative. In addition, the standardized effect sizes (ses) of these three metrics (i.e., PDses, MPDses and MNTDses), which account for species richness, are commonly used in studies on phylogenetic structure (Webb et al., 2002; Cadotte and Davies, 2016). They are calculated as: Xses = (Xobs − Xnull)/sd (Xnull), where Xses represents standardized effect size for X (i.e., one of the three metrics under consideration), Xobs is the observed X of an assemblage, Xnull is the expected (i.e., average) X of the randomized assemblages, and sd (Xnull) is the standard deviation of X for the randomized assemblages. PDses is commonly called PDI (e.g., Sandel and Tsirogiannis, 2016; Qian et al., 2019) while MPDses and MNTDses are commonly used as NRI, which is −MPDses, and NTI, which is −MNTDses, respectively (Cadotte and Davies, 2016; Webb et al., 2008). In this study, we examined all the six phylogenetic metrics (i.e., PD, MPD, MNTD, PDI, NRI and NTI).
For each of the 1093 angiosperm assemblages and each of the six phylogenetic metrics, five values were calculated based on different phylogenies as follows: (ⅰ) one value was based on the phylogeny resolved at the species level (i.e., PHYLOsp.resolved); (ⅱ) one value was based on the phylogeny resolved at the genus level with species being attached to their respective genera as basal polytomies (i.e., PHYLO sp.basal-polytomies); (ⅲ) one value was based on the phylogeny resolved at the genus level with species of a genus being attached to the genus as polytomies at the middle point of the branch length of the genus (i.e., PHYLOsp.middle-polytomies); (ⅳ) one value was based on the phylogeny resolved at the genus level (i.e., PHYLOgen.resolved) and the presence or absence of each genus in each assemblage was used in calculation; and (ⅴ) one value was based on the phylogeny resolved at the genus level (i.e., PHYLOgen.resolved) and the species abundance (i.e., species richness) of each genus in each assemblage was used in calculation. In other words, the number of tips in each phylogeny was 388 in the first three cases, and was 174 in the last two cases. We used the software PhyloMeasures (Tsirogiannis and Sandel, 2016) to calculate phylogenetic metrics.
2.2. Environmental dataMean annual temperature, annual precipitation, minimum temperature of the coldest month, precipitation during the driest month, temperature seasonality, and precipitation seasonality were among the most commonly used climate variables in previous studies on phylogenetic structure to represent general environmental conditions for broad-scale sampling units (e.g., Kamilar et al., 2015; Qian et al., 2017; Weigelt et al., 2015). These variables represent the means, extremes and variability of temperature and precipitation. Accordingly, we used these six variables to characterize the climate of each quadrat. We obtained data for these variables from the WorldClim database (worldclim.org/version 2; corresponding to variables BIO1, BIO12, BIO6, BIO14, BIO4, and BIO15, respectively). The mean value of each of the six climate variables was calculated for each quadrat using 30-arc-second resolution data.
2.3. Data analysisWe took several steps to analyze data for each of the six phylogenetic metrics. First, we conducted Pearson correlation analysis to relate scores derived from the phylogeny PHYLOsp.resolved to those derived from the other three phylogenies (i.e., PHYLOsp.basal-polytomies, PHYLOsp.middle-polytomies and PHYLOgen.resolved). We considered a correlation to be strong for |r| > 0.66, moderate for 0.66 ≥ |r| > 0.33, and weak for |r| ≤ 0.33 (Qian et al., 2019). Second, we regressed scores derived from the phylogeny PHYLOsp.resolved simultaneously on the six climate variables using the ordinary least squares model, determined which climate variables were significant (P < 0.05) in each regression, and considered the regression without non-significant climate variables as the structure of a final regression model. Third, we regressed scores derived from each of the four phylogenies on the climate variables retained in each final regression model, and compared coefficients of determination and standardized regression coefficients of climate variables between the model based on scores derived from the phylogeny PHYLOsp.resolved and each of the other models based on scores derived from the other phylogenies. In each regression, every variable was standardized to have mean of zero and standard deviation of one. We used the packages SYSTAT (Wilkinson et al., 1992) for statistical analyses.
3. ResultsThe scores of phylogenetic metrics derived from the phylogeny PHYLOsp.resolved were strongly or perfectly correlated with those derived from the phylogeny PHYLOsp.basal-polytomies (Fig. 3). The average of the correlation coefficients for the six phylogenetic metrics was 0.955 (ranging from 0.837 to 1.000;Fig. 3). The scores of phylogenetic metrics derived from the phylogeny PHYLOsp.resolved were perfectly or nearly perfectly correlated with those derived from the phylogeny PHYLOsp.middle-polytomies (Fig. 3), with the average of the correlation coefficients for the six phylogenetic metrics being 0.991 (ranging from 0.966 to 1.000; Fig. 3). When the scores of phylogenetic metrics derived from the genus-level phylogeny (i.e., PHYLOgen.resolved, with each tip representing a genus) were considered, correlations in the scores of phylogenetic metrics were perfect or nearly perfect between the two sets of analyses (i.e., one set based on the presence or absence of a genus, the other set based on the species abundance of each genus); the average of correlation coefficients for the six phylogenetic metrics was 0.999. When the scores of phylogenetic metrics derived from the phylogeny PHYLOsp.resolved were correlated with the scores phylogenetic metrics derived from the genus-level phylogeny, correlation coefficients for PD remained very high regardless of whether the presence-absence data or the species abundance data were analyzed (r = 0.998 in both cases; Fig. 3); however, correlation coefficients for the other five phylogenetic metrics were much lower than those based on the scores derived from the two species-level phylogenies resolved at the genus level (i.e., PHYLOsp.basal-polytomies and PHYLOsp.middle-polytomies; Fig. 3), with the average of the 10 correlation coefficients being 0.655 (ranging from 0.472 to 0.811; Fig. 3). As a result, we did not conduct regression analyses for the scores of phylogenetic metrics derived from the genus-level phylogeny.
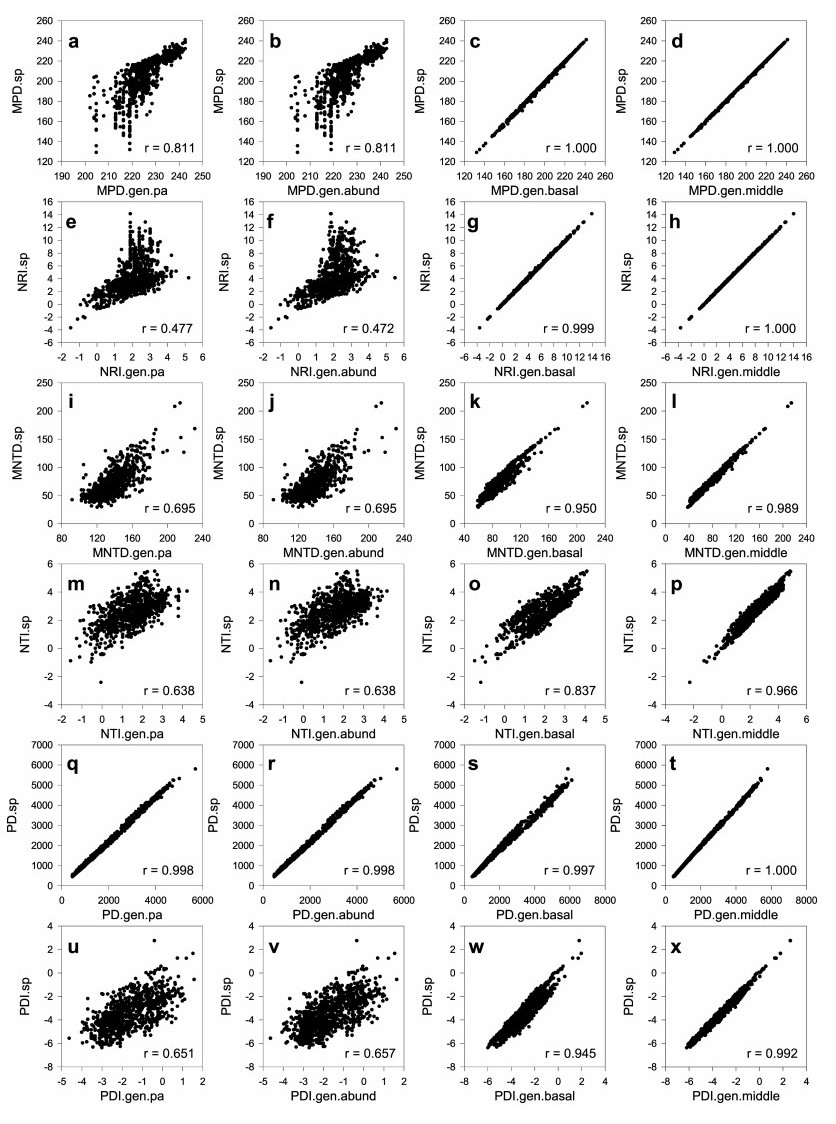
|
| Fig. 3 Relationships between values of phylogenetic metrics based on phylogenies resolved at the species level (METRIC.sp, where METRIC represents MPD, NRI, MNTD, NTI, PD or PDI) and those based on phylogenies resolved at the genus level. Values of METRIC.gen.pa were based on a phylogeny resolved at the genus level with each tip being a genus and genus presence/absence data were used in calculating the phylogenetic metric. Values of METRIC.gen.abund were based on a phylogeny resolved at the genus level with each tip being a genus and genus abundance data (i.e., the number of species in each genus) were used in calculating the phylogenetic metric. Values of METRIC.gen.basal were based on a phylogeny resolved at the genus level with species of each genus being attached to the genus as basal polytomies and each tip representing a species. Values of METRIC.gen.middle were based on a phylogeny resolved at the genus level with species of each genus being attached to the genus at the middle point of its branch and each tip representing a species. See Methods for full names of MPD, NRI, MNTD, NTI, PD, and PDI. |
The final regression models of the six phylogenetic metrics on the six climate variables retained a total of 27 model terms (Table 1). When the models based on the scores of phylogenetic metrics derived from the phylogeny PHYLOsp.basal-polytomies were considered, the rank order of standardized regression coefficients of different climate variables within each model was consistent with that derived from the phylogeny PHYLOsp.resolved for 24 (89%) of the 27 model terms (Table 1). The three inconsistent cases are BIO12 in the model for MNTD, BIO14 in the model for NTI, and BIO12 in the model for PDI (Table 1). Of the 27 terms in the six models, 24 (89%) were significant (P < 0.05 in 22 cases) or marginally significant (P < 0.07 in 2 cases) (Table 1). When the models based on the scores of phylogenetic metrics derived from the phylogeny PHYLOsp.middle-polytomies were considered, the rankings of standardized region coefficients for different climate variables within each model was consistent with that derived from the phylogeny PHYLOsp.resolved for all the 27 model terms, 25 (93%) of which were significant (P < 0.05;Table 1). Coefficients of determination for the regressions based on the scores of phylogenetic metrics derived from the phylogeny PHYLOsp.middle-polytomies were, in general, more similar to those based on the scores of phylogenetic metrics derived from the phylogenyPHYLOsp.resolved, compared to those derived from the phylogeny PHYLOsp.basal-polytomies (Table 1).
| Metric | Climate | PHYLOsp.resolved | PHYLOsp.basal-polytomies | PHYLOsp.middle-polytomies | |||||
| Coeff. | P value | Coeff. | P value | Coeff. | P value | ||||
| MPD | BIO14 | −0.239 | < 0.001 | −0.238 | < 0.001 | −0.238 | < 0.001 | ||
| BIO6 | −0.219 | < 0.001 | −0.198 | 0.001 | −0.215 | < 0.001 | |||
| BIO15 | −0.180 | < 0.001 | −0.179 | < 0.001 | −0.181 | < 0.001 | |||
| BIO12 | 0.335 | < 0.001 | 0.342 | < 0.001 | 0.336 | < 0.001 | |||
| BIO1 | 0.939 | < 0.001 | 0.910 | < 0.001 | 0.932 | < 0.001 | |||
| R2 | 0.702 | 0.697 | 0.700 | ||||||
| NRI | BIO1 | −0.652 | < 0.001 | −0.641 | < 0.001 | −0.647 | < 0.001 | ||
| BIO12 | −0.148 | < 0.001 | −0.166 | < 0.001 | −0.152 | < 0.001 | |||
| R2 | 0.515 | 0.514 | 0.511 | ||||||
| MNTD | BIO6 | −0.861 | < 0.001 | −0.280 | 0.288 | −0.762 | 0.002 | ||
| BIO4 | −0.474 | < 0.001 | −0.231 | 0.060 | −0.358 | 0.002 | |||
| BIO12 | −0.431 | < 0.001 | −0.338 | < 0.001 | −0.400 | < 0.001 | |||
| BIO15 | 0.113 | < 0.001 | 0.080 | 0.020 | 0.073 | 0.025 | |||
| BIO1 | 0.807 | < 0.001 | 0.340 | 0.043 | 0.803 | < 0.001 | |||
| R2 | 0.270 | 0.146 | 0.229 | ||||||
| NTI | BIO1 | −0.662 | < 0.001 | −0.656 | < 0.001 | −0.701 | < 0.001 | ||
| BIO14 | −0.461 | < 0.001 | −0.712 | < 0.001 | −0.596 | < 0.001 | |||
| BIO15 | −0.273 | < 0.001 | −0.245 | < 0.001 | −0.257 | < 0.001 | |||
| BIO12 | 0.505 | < 0.001 | 0.372 | < 0.001 | 0.497 | < 0.001 | |||
| R2 | 0.412 | 0.547 | 0.482 | ||||||
| PD | BIO12 | 0.148 | < 0.001 | 0.151 | < 0.001 | 0.146 | < 0.001 | ||
| BIO1 | 0.246 | 0.002 | 0.157 | 0.051 | 0.233 | 0.003 | |||
| BIO14 | 0.382 | < 0.001 | 0.413 | < 0.001 | 0.396 | < 0.001 | |||
| BIO4 | 0.601 | < 0.001 | 0.648 | < 0.001 | 0.619 | < 0.001 | |||
| BIO6 | 0.835 | < 0.001 | 0.921 | < 0.001 | 0.850 | < 0.001 | |||
| R2 | 0.817 | 0.804 | 0.816 | ||||||
| PDI | BIO6 | −0.471 | 0.025 | 0.105 | 0.580 | −0.347 | 0.089 | ||
| BIO12 | −0.233 | < 0.001 | −0.131 | 0.007 | −0.209 | < 0.001 | |||
| BIO4 | −0.206 | 0.036 | 0.123 | 0.167 | −0.084 | 0.380 | |||
| BIO15 | 0.195 | < 0.001 | 0.142 | < 0.001 | 0.164 | < 0.001 | |||
| BIO14 | 0.243 | 0.001 | 0.336 | < 0.001 | 0.278 | < 0.001 | |||
| BIO1 | 0.986 | < 0.001 | 0.695 | < 0.001 | 0.972 | < 0.001 | |||
| R2 | 0.456 | 0.555 | 0.490 | ||||||
A previous study (Qian and Zhang, 2016) compared the scores of phylogenetic metrics derived from a phylogeny fully resolved at the species level with those derived from a phylogeny resolved at the family level with genera and species being attached to their respective families as polytomies. The present study extends their study by comparing the scores of phylogenetic metrics derived from a phylogeny fully resolved at the species level with those derived from phylogenies resolved at the genus level but not resolved at the species level. We found that the scores of phylogenetic metrics derived from a genus-level phylogeny (i.e., each tip represents a genus, rather than a species, in the phylogeny) were perfectly or nearly perfectly correlated between the two sets of analyses using the genus-level phylogeny (i.e., one set using data with genus presence or absence, the other set using data with species richness (abundance) per genus) for the six phylogenetic metrics. They were also nearly perfectly correlated with the scores of PD derived from the other three phylogenies (Fig. 3q through t). However, they were not strongly correlated with the scores of the other phylogenetic metrics derived from the phylogeny resolved at the species level. Thus, a genus-level phylogeny may be used as a proxy of a species-level phylogeny to explore geographical and ecological patterns for PD, but may not be used as a proxy of a species-level phylogeny to explore such patterns for other phylogenetic metrics, at least those examined in this study. Interestingly, Lehtonen et al. (2015) show that in their study on phylogenetic structure of fern communities, the scores of MPD derived from a phylogeny resolved at the species level is nearly perfectly correlated with the scores of MPD derived from a genus-level phylogeny (r = 0.996), which is much higher than the correlation observed in our study for MPD (r = 0.811; Fig. 3a).
Our study showed that the scores of phylogenetic metrics derived from a phylogeny resolved at the species level were strongly or perfectly correlated with those derived from a phylogeny resolved at the genus level with species being attached to them as polytomies of their respective genera, and were more strongly correlated with those derived from a phylogeny resolved at the genus level with species being attached to their genera as polytomies at the middle points of genus branch lengths than those as basal polytomies of their genera (Fig. 3). Our study also showed that when scores of phylogenetic metrics were regressed on climate variables, the rankings of climate variables in each regression model was generally congruent among models derived from the three above-mentioned species-level phylogenies, with the congruence being stronger between models derived from the phylogeny resolved at the species level and models derived from the phylogeny with species being treated as polytomies at the middle point of the branch of each genus, compared to that between models derived from the phylogeny resolved at the species level and models derived from the phylogeny with species being treated as basal polytomies within each genus (Table 1). In particular, the rankings of climate variables were completely congruent and coefficients of determination were very similar between the model based on the phylogeny resolved at the species level and the model derived from the phylogeny with species being treated as polytomies at the middle point of the branch of each genus for each of the six phylogenetic metrics, suggesting that using a phylogeny resolved at the genus level with species being attached to their genera as polytomies at the middle points of genus branch lengths in a study on community phylogenetics is equivalent to using a phylogeny resolved at the species level in the study.
Ideally, each study on community phylogenetics uses phylogenies well resolved at the species level. Although phylogenies well resolved at the species level are available for some major groups (e.g., Fritz et al., 2009 for mammals; Jetz et al., 2012 for birds), such phylogenies are lacking for many other major groups of organisms such as vascular plants, because many species in these groups have not been sequenced. For example, only ~20% of the vascular plant species worldwide have been sequenced, according to gene sequence data in GenBank (Jin and Qian, 2019). A study may include thousands or tens of thousands of species (e.g., Qian et al., 2013; 2019; Brunbjerg et al., 2014). Because phylogenies that are resolved at the species level are generally not available, ecologists have frequently used a phylogeny more or less resolved at the family or genus level as a backbone and attached species to the backbone phylogeny as polytomies (e.g., Munguía-Rosas et al., 2011; Giehl and Jarenkow, 2012; Hardy et al., 2012; Qian et al., 2013).
Phylogenies that are well resolved at the species level are available for some animal groups, but they do not exist for vascular plants, which include ~391, 000 species worldwide (Royal Botanic Gardens at Kew, 2016). For vascular plants, the time-calibrated megaphylogeny implemented in the software V.PhyloMaker (i.e., GBOTB.extended.tre), which includes Smith and Brown's (2018) phylogeny for seed plants (i.e., GBOTB) and Zanne et al.'s (2014) phylogeny for pteridophytes, is the largest dated phylogeny. Although this megaphylogeny includes only about 20% of vascular plant species worldwide, it includes about 80% of vascular plant genera worldwide. Based on our empirical studies as noted above, 87–98% of seed plant genera in a regional or local species list can be found in the megaphylogeny. Although about 5–15% of plant genera in a study species list may be absent from the megaphylogeny, because about 40–70% of the species in a study species list may be present in the megaphylogeny, a phylogeny generated using the megaphylogeny with V.PhyloMaker as a backbone may be better (more completely resolved) than a species-level phylogeny resolved at the genus level with all species being polytomies, such as those tested in this study. Plant phylogenies generated with V.PhyloMaker have been commonly used in the literature. According to Google Scholar (https://scholar.google.com/; accessed on October 24, 2020), since V.PhyloMaker was made available to the public in 2019, 55 published studies have used V.PhyloMaker as a tool and the megaphylogeny implemented in it as a backbone to generate phylogenies (e.g., Abrahamczyk, 2019; Chen et al., 2020; Gamba and Muchhala, 2020; Lancaster, 2020; Qian et al., 2019; Yue and Li, 2020; Zhang et al., 2020). A phylogeny generated by Scenario 1 of V.PhyloMaker is similar to a species-level phylogeny resolved at the genus level with species of a genus being attached to the phylogeny as basal polytomies of the genus whereas a phylogeny generated by Scenario 3 of V.PhyloMaker is similar to a species-level phylogeny resolved at the genus level with species of a genus being attached to the phylogeny as polytomies at the middle point of the branch length of the genus. Based on the finding of the present study, using a phylogeny generated by V.PhyloMaker under Scenario 3 is likely better than using a phylogeny generated by V.PhyloMaker under Scenario 1 in a study on community phylogenetics. It appears that using a phylogeny generated by V.PhyloMaker under Scenario 3 in a study on community phylogenetics is also better than using a phylogeny resolved at the genus level with species being attached to their genera as polytomies at the middle points of genus branch lengths, because a large number of the species in the former would likely be resolved. The key finding of the present study, i.e., the result of an analysis based on a phylogeny resolved at the species level is qualitatively the same as that based on a phylogeny resolved at the genus level with species being treated as polytomies within genera, indicates that the result of a study based on a phylogeny generated using the megaphylogeny implemented in V.PhyloMaker as a backbone is robust.
5. ConclusionsOur study showed that the scores of phylogenetic metrics derived from a fully resolved genus-level phylogeny are not strongly correlated with those derived from a fully resolved species-level phylogeny, even in the case that the species richness of each genus in each assemblage is accounted for. However, when species are treated as polytomies within their respective genera in a species-level phylogeny resolved at the genus-level, the scores of phylogenetic metrics derived from the phylogeny are strongly or perfectly correlated with those derived from a fully resolved species-level phylogeny, and the relationships between scores of phylogenetic metrics and environmental variables are highly consistent between analyses based on the two types of phylogenies. Furthermore, our study showed that the result of an analysis based on the scores of phylogenetic metrics derived from a phylogeny resolved at the genus level with species being attached to their genera as polytomies at the middle points of the branch lengths of the genera is more robust than that based on the scores of phylogenetic metrics derived from a phylogeny resolved at the genus level with species being attached to their genera as basal polytomies. Although our study focused on tree species, we anticipate that our finding is robust with regard to organismal group. The finding of our study suggests that phylogenies generated based on the megaphylogeny included in V.PhyloMaker as a backbone are appropriate for studies on community phylogenetics, particularly for phylogenies generated by V.PhyloMaker under Scenario 3.
Authors' contributionsH.Q. designed research, analyzed data, and wrote the paper; Y.J. generated phylogenies and calculated phylogenetic metrics; both authors participated in revising the paper.
Declaration of competing interestThe authors declare no conflict of interest.
AcknowledgementsWe thank Yangjian Zhang for compiling tree distribution data, and two reviewers for helpful comments.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.11.005.
Abrahamczyk, S., 2019. Comparison of the ecology and evolution of plants with a generalist bird pollination system between continents and islands worldwide. Biol. Rev., 94: 1658-1671. DOI:10.1111/brv.12520 |
Algar, A.C., Kerr, J.T., Currie, D.J., 2009. Evolutionary constraints on regional faunas: whom, but not how many. Ecol. Lett., 12: 57-65. DOI:10.1111/j.1461-0248.2008.01260.x |
Baum, D.A., Smith., S.D. 2012. Tree thinking: An introduction to phylogenetic biology. Roberts Co., Greenwood Village, Colorado.
|
Behrensmeyer, A.K., Damuth, J.D., DiMichele, W.A., et al., 1992. Terrestrial ecosystems through time: evolutionary paleoecology of terrestrial plants and animals. University of Chicago Press, Chicago.
|
Brunbjerg, A.K., Cavender-Bares, J., Eiserhardt, W.L., et al., 2014. Multi-scale phylogenetic structure in coastal dune plant communities across the globe. J. Plant Ecol., 7: 101-114. DOI:10.1093/jpe/rtt069 |
Cadotte, M.W., Davies, T.J. 2016. Phylogenies in Ecology: A Guide to Concepts and Methods. Princeton University Press, Princeton and Oxford.
|
Carvallo, G.O., Teillier, S., Castro, S.A., et al., 2014. The phylogenetic properties of native- and exotic-dominated plant communities. Austral Ecol., 39: 304-312. DOI:10.1111/aec.12079 |
Cavender-Bares, J., Kozak, K.H., Fine, P.V.A., et al., 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett., 12: 693-715. DOI:10.1111/j.1461-0248.2009.01314.x |
Chalmandrier, L., Munkemuller, T., ünkemüller, S., et al., 2015. Effects of species' similarity and dominance on the functional and phylogenetic structure of a plant meta-community. Ecology, 96: 143-153. DOI:10.1890/13-2153.1 |
Chen, S.-C., Wu, L.-M., Wang, B., et al., 2020. Macroevolutionary patterns in seed component mass and different evolutionary trajectories across seed desiccation responses. New Phytol., 228: 770-777. DOI:10.1111/nph.16706 |
Cooper, N., íguez, J., Purvis, A., 2008. A common tendency for phylogenetic overdispersion in mammalian assemblages. Proc. Roy. Soc.: Biol. Sci., 275: 2031-2037. DOI:10.1098/rspb.2008.0420 |
Culmsee, H., Leuschner, C., 2013. Consistent patterns of elevational change in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr., 40: 1997-2010. DOI:10.1111/jbi.12138 |
Currie, D.J., Paquin, V., 1987. Large-scale biogeographical patterns of species richness of trees. Nature, 329: 326-327. DOI:10.1038/329326a0 |
Darwin, C. 1859. The Origin of Species. Reprinted by Penguin Books, London.
|
Donoghue, M.J., 2008. A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U. S. A., 105: 11549-11555. DOI:10.1073/pnas.0801962105 |
Eiserhardt, W.L., Svenning, J.-C., Borchsenius, F., et al., 2013. Separating environmental and geographical determinants of phylogenetic community structure in Amazonian palms (Arecaceae). Bot. J. Linn. Soc., 171: 244-259. DOI:10.1111/j.1095-8339.2012.01276.x |
Elven, R. 2011. Annotated checklist of the panarctic flora (PAF): vascular plants. National Centre of Biosystematics, Natural History Museum, University of Oslo.
|
Eme, D., Anderson, M.J., Myers, et al., 2020. Phylogenetic measures reveal eco-evolutionary drivers of biodiversity along a depth gradient. Ecography, 43: 689-702. DOI:10.1111/ecog.04836 |
Emerson, B.C., Gillespie, R.G., 2008. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol., 23: 619-630. DOI:10.1016/j.tree.2008.07.005 |
Faith, D.P., 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv., 61: 1-10. DOI:10.1016/0006-3207(92)91201-3 |
Fritz, S.A., Bininda-Emonds, O.R.P., Purvis, A., 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett., 12: 538-554. DOI:10.1111/j.1461-0248.2009.01307.x |
Fritz, S.A., Rahbek, C., 2012. Global patterns of amphibian phylogenetic diversity. J. Biogeogr., 39: 1373-1382. DOI:10.1111/j.1365-2699.2012.02757.x |
Gamba, D., Muchhala, N., 2020. Global patterns of population genetic differentiation in seed plants. Mol. Ecol., 29: 3413-3428. DOI:10.1111/mec.15575 |
Giehl, E.L.H., Jarenkow, J.A., 2012. Niche conservatism and the differences in species richness at the transition of tropical and subtropical climates in South America. Ecography, 35: 933-943. DOI:10.1111/j.1600-0587.2011.07430.x |
Graham, A. 1999. Late Cretaceous and Cenozoic history of North American vegetation north of Mexico. Oxford University Press, New York.
|
Hardy, O.J., Couteron, P., Munoz, F., et al., 2012. Phylogenetic turnover in tropical tree communities: impact of environmental filtering, biogeography and mesoclimatic niche conservatism. Global Ecol. Biogeogr., 21: 1007-1016. DOI:10.1111/j.1466-8238.2011.00742.x |
Hawkins, B.A., Rueda, M., Rangel, T.F., et al., 2014. Community phylogenetics at the biogeographical scale: cold tolerance, niche conservatism and the structure of North American forests. J. Biogeogr., 41: 23-38. DOI:10.1111/jbi.12171 |
Jetz, W., Thomas, G.H., Joy, J.B., et al., 2012. The global diversity of birds in space and time. Nature, 491: 444-448. DOI:10.1038/nature11631 |
Jin, Y., Qian, H., 2019. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography, 42: 1353-1359. DOI:10.1111/ecog.04434 |
Kamilar, J.M., Guidi, L.M., 2010. The phylogenetic structure of primate communities: variation within and across continents. J. Biogeogr., 37: 801-813. DOI:10.1111/j.1365-2699.2009.02267.x |
Kamilar, J.M., Beaudrot, L., Reed, K.E., Climate and species richness predict the phylogenetic structure of African mammal communities. PloS One: e0121808. DOI:10.1371/journal.pone.0121808 |
Lancaster, L.T., 2020. Host use diversification during range shifts shapes global variation in Lepidopteran dietary breadth. Nat. Ecol. Evol., 4: 963-969. DOI:10.1038/s41559-020-1199-1 |
Lehtonen, S., Jones, M.M., Zuquim, G., et al., 2015. Phylogenetic relatedness within Neotropical fern communities increases with soil fertility. Global Ecol. Biogeogr., 24: 695-705. DOI:10.1111/geb.12294 |
Li, X., Sun, H., 2017. Phylogenetic pattern of alpine plants along latitude and longitude in Hengduan Mountains Region. Plant Divers., 39: 37-43. DOI:10.1016/j.pld.2016.11.007 |
Little, E.L., Jr. 1971-78. Atlas of United States trees. Vols. 1, 3, 4, and 5. U.S. Department of Agriculture Miscellaneous Publication, Washington, DC.
|
Mabberley, D.J. 2008. Mabberley's Plant-book A Portable Dictionary of Plants, their Classifications, and Uses, 3rd edn. Cambridge University Press, Cambridge.
|
Molina-Venegas, R., Aparicio, A., Slingsby, J.A., et al., 2015. Investigating the evolutionary assembly of a Mediterranean biodiversity hotspot: deep phylogenetic signal in the distribution of eudicots across elevational belts. J. Biogeogr., 42: 507-518. DOI:10.1111/jbi.12398 |
Morin, X., Lechowicz, M.J., 2011. Geographical and ecological patterns of range size in North American trees. Ecography, 34: 738-750. DOI:10.1111/j.1600-0587.2010.06854.x |
Munguí-Rosas, M.A., Ollerton, J., Parra-Tabla, V., et al., 2011. Meta-analysis of phenotypic selection on flowering phenology suggests that early flowering plants are favoured. Ecol. Lett., 14: 511-521. DOI:10.1111/j.1461-0248.2011.01601.x |
Ndiribe, C., Salamin, N., Guisan, A., 2013. Understanding the concepts of community phylogenetics. Evol. Ecol. Res., 15: 853-868. |
Pennington, R.T., Richardson, J.E., Lavin, M., 2006. Insights into the historical construction of species-rich biomes from dated plant phylogenies, neutral ecological theory and phylogenetic community structure. New Phytol., 172: 605-616. DOI:10.1111/j.1469-8137.2006.01902.x |
Qian, H., Jiang, L., 2014. Phylogenetic community ecology: integrating community ecology and evolutionary biology. J. Plant Ecol., 7: 97-100. DOI:10.1093/jpe/rtt077 |
Qian, H., Ricklefs, R.E., 2016. Out of the tropical lowlands: latitude versus elevation. Trends Ecol. Evol., 31: 738-741. DOI:10.1016/j.tree.2016.07.012 |
Qian, H., Wiens, J.J., Zhang, J., et al., 2015. Evolutionary and ecological causes of species richness patterns in North American angiosperm trees. Ecography, 38: 241-250. DOI:10.1111/ecog.00952 |
Qian, H., Deng, T., Jin, Y., et al., 2019. Phylogenetic dispersion and diversity in regional assemblages of seed plants in China. Proc. Natl. Acad. Sci. U. S. A., 116: 23192-23201. DOI:10.1073/pnas.1822153116 |
Qian, H., Jin, Y., Ricklefs, R.E., 2017. Phylogenetic diversity anomaly in angiosperms between eastern Asia and eastern North America. Proc. Natl. Acad. Sci. U. S. A., 114: 11452-11457. DOI:10.1073/pnas.1703985114 |
Qian, H., Hao, Z., Zhang, J., 2014. Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J. Plant Ecol., 7: 154-165. DOI:10.1093/jpe/rtt072 |
Qian, H., Zhang, J., 2016. Are phylogenies derived from family-level supertrees robust for studies on macroecological patterns along environmental gradients?. J. Systemat. Evol., 54: 29-36. DOI:10.1111/jse.12161 |
Qian, H., Zhang, Y., Zhang, J., et al., 2013. Latitudinal gradients in phylogenetic relatedness of angiosperm trees in North America. Global Ecol. Biogeogr., 22: 1183-1191. DOI:10.1111/geb.12069 |
Rana, S.K., Price, T.D., Qian., H. 2019. Plant species richness across the Himalaya driven by evolutionary history and current climate. Ecosphere 10, Article e02945.
|
Ricklefs, R.E., 1987. Community diversity: relative roles of local and regional processes. Science, 235: 167-171. DOI:10.1126/science.235.4785.167 |
Royal Botanic Gardens at Kew 2016. The state of the World's Plants 2016. Royal Botanic Gardens, Kew.
|
Sandel, B., Tsirogiannis, C., 2016. Species introductions and the phylogenetic and functional structure of California's grasses. Ecology, 97: 472-483. DOI:10.1890/15-0220.1 |
Sandel, B., Weigelt, P., Kreft, H., et al., 2020. Current climate, isolation and history drive global patterns of tree phylogenetic endemism. Global Ecol. Biogeogr., 29: 4-15. DOI:10.1111/geb.13001 |
Smith, S.A., Brown, J.W., 2018. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot., 105: 302-314. DOI:10.1002/ajb2.1019 |
Swenson, N.G. 2019. Phylogenetic Ecology: A History, Critique, and Remodeling. University of Chicago Press.
|
Tsirogiannis, C., Sandel, B., 2016. PhyloMeasures: a package for computing phylogenetic biodiversity measures and their statistical moments. Ecography, 39: 709-714. DOI:10.1111/ecog.01814 |
Vamosi, S.M., Heard, S.B., Vamosi, J.C., et al., 2009. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol., 18: 572-592. DOI:10.1111/j.1365-294X.2008.04001.x |
Webb, C.O., 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat., 156: 145-155. DOI:10.1086/303378 |
Webb, C.O., Ackerly, D.D., Kembel, S.W., 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics, 24: 2098-2100. DOI:10.1093/bioinformatics/btn358 |
Webb, C.O., Ackerly, D.D., McPeek, M.A., et al., 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Systemat., 33: 475-505. DOI:10.1146/annurev.ecolsys.33.010802.150448 |
Webb, C.O., Losos, J.B., Agrawal, A.A., 2006. Integrating phylogenies into community ecology. Ecology, 87: S1-S2. DOI:10.1890/0012-9658(2006)87[1:IPICE]2.0.CO;2 |
Weigelt, P., Kissling, W.D., Kisel, Y., et al., 2015. Global patterns and drivers of phylogenetic structure in island floras. Sci. Rep., 5: 12213. DOI:10.1038/srep12213 |
Wilkinson, L., Hill, M., Welna, J.P., et al., 1992. SYSTAT for Windows: statistics. SYSTAT Inc., Evanston.
|
Yue, J., Li, R., Phylogenetic relatedness of woody angiosperm assemblages and its environmental determinants along a subtropical elevational gradient in China. Plant Divers.: 10.1016/j.pld.2020.08.003. DOI:10.1016/j.pld.2020.08.003 |
Zanne, A.E., Tank, D.C., Cornwell, W.K., et al., 2014. Three keys to the radiation of angiosperms into freezing environments. Nature, 506: 89-92. DOI:10.1038/nature12872 |
Zhang, Y., Qian, L., Spalink, D., et al., Spatial phylogenetics of two topographic extremes of the Hengduan Mountains in southwestern China and its implications for biodiversity conservation. Plant Divers.: 10.1016/j.pld.2020.09.001. DOI:10.1016/j.pld.2020.09.001 |



