b. University of the Chinese Academy of Sciences, Beijing, 100049, China
Stem cells are critical for the growth and development of all multicellular organisms. Accordingly, a fundamental goal of developmental biology is understanding the regulatory mechanisms that maintain the balance between stem cell division and differentiation. In animals, stem cell research has attracted considerable attention because of the potential for drug discovery, regenerative medicine, and cancer therapy. Consequently, various chemicals have been identified that promote and/or inhibit animal stem cell division (Ahmed et al., 2017; Hou et al., 2013; Chanda et al., 2013). Although both genetic and chemical approaches for regulating plant meristem have been extensively investigated, to date no chemical elicitor/promoter for root meristem maintenance has been reported.
In plants, the root meristem consists of a layer of stem cells or initials surrounding the mitotically less active quiescent centre (QC) cells. Outside of the stem cells the tissues of the root consist of the stele, pericycle, endodermis, cortex, epidermis, lateral cap, and columella root cap (Fig. 1a). Stem cells such as the cortex/endodermal initials (CEI) undergo asymmetric cell division and differentiate into the cells of cortex and endodermis (Fig. 1a). The number of cells in the cortex and endodermis determines meristem size (Casamitjana-Martinez et al., 2003) and is regulated by the SHORT ROOT (SHR)-mediated transcriptional pathway (Wu et al., 2014; Cui et al., 2007) (Fig. 1b). In Arabidopsis thaliana, loss of SHR results in short roots (Helariutta et al., 2000; Levesque et al., 2006). The SHR pathway includes a series of transcription factors, including its binding partner SCARECROW (SCR) and RETINOBLASTOMA RELATED (RBR) (Fig. 1b), and downstream factors NUTCRACKER (NUC) and Magpie (MGP) (Levesque et al., 2006; Cui et al., 2007; Cruz-Ramirez et al., 2012). SHR is expressed in stele cells and is able to move into QC cells or CEI daughter (CED) cells. In the CEI (Fig. 1b), interactions between SHR and SCR, or between SCR and RBR, form a circuit that controls the expression of NUC and MGP in during the process that signals of asymmetric cell division. The circuit is a core mediator of asymmetric cell division signalling that coordinates periclinal division of CED cells with anticlinal division of CEI cells to regulate the growth of the cortex/endodermis lineage (Wu et al., 2014; Moreno-Risueno et al., 2015; Cruz-Ramirez et al., 2013).

|
| Fig. 1 (a) Schematic of Arabidopsis thaliana root meristem with each cell type. QC, quiescent centre; CEI, cortex/endodermal initials; VI, Vascular initials; LRC, Lateral root cap; Ep, Epidermis Initials; CSC, Columella Stem Cells; CC, Columella Cells. (b) Schematic of regulatory pathway for asymmetric cell division (ACD). RBR, RETINOBLASTOMA RELATED; SCR, SCARECROW; SHR, SHORT ROOT. (c) Structures of the flavonoids tested in this study. |
Flavonoids are widespread secondary metabolites that have strong biological activity in root development (Buer et al., 2010; Peer et al., 2011; Peer and Murphy, 2007; Jacobs and Rubery, 1988). Exogenous application of flavonoids is known to cause primary root shortening (Buer et al., 2010; Laffont et al., 2010; Peer et al., 2011). All previously tested flavonoids (e.g., naringenin, quercetin, taxifolin, tamarixetin, kaempferol, apigenin, fisetin, and galangin) inhibit root growth (Yin et al., 2014; Jacobs and Rubery, 1988). In the present study, however, we found that scutellarin (Fig. 1c), a flavnoid with a 6-hydroxyl and a 7-glucoside, causes root lengthening through the NUC pathway. In contrast, naringenin, a flavonoid with same chemical backbone as scutellarin but without the 6-hydroxyl and with a 7-hydroxyl, has the opposite or no effect on root length. Our transcriptional, genetic, and cellular analyses show that flavonoids with and without a 6-hydroxyl and 7-glucoside have opposite effects on the NUC pathway. Our results reveal a novel function for flavonoids and provide a pharmacological approach to regulate root meristem.
2. Materials and methods 2.1. ChemicalsScutellarin (purity > 98%, HPLC) was provided by Dr. Qinshi Zhao, State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences. Naringenin was purchased from the Aladdin Industrial Corporation. Chemicals were dissolved in 0.5‰ DMSO (dimethylsulfoxide, as mock) at the indicated concentration.
2.2. Plant material and growth conditionsArabidopsis thaliana (Columbia ecotype) nuc-1 mutants (SALK_124222C, NUC-knockout line from Columbia ecotype) were ordered from the Arabidopsis Biological Resource Center, and the homozygous insertion site of the mutant was confirmed by PCR (http://signal.salk.edu/tdnaprimers.2.html) with T-DNA–specific primer LBb1 and NUC-specific primers NUC-LP and NUC-RP.
We constructed the overexpression transgenic plant NUC-OE under the Col background. The full-length cDNA of NUC was cloned into the binary vector pEGAD behind the CaMV 35S promoter. Thenuc-1 recovery lines 35S::nuc/nuc-1 and nucpro::nuc/nuc-1 were rescued by nuc CDs subcloned into pEGAD vectors separately under the 35S promoter and nuc promoter, respectively. The transgenic seedlings were screened by BASTA and PCR, and the overexpression of NUC was detected by reverse transcription PCR.
Surface-sterilized A. thaliana seeds were soaked in 0.5‰ DMSO (mock), 50 μM scutellarin in 0.5‰ DMSO, or 50 μM naringenin 0.5‰ DMSO at 4 ℃ in the dark for 2 days for homogeneous germination, and then were placed on filter paper (for microarray analysis) or petri dishes with agar supplemented with 1/4 MS and 0.5‰ DMSO (mock) or with flavonoids in 1/4 MS and 0.5‰ DMSO. Nitrogen sources antagonized scutellarin-induced root lengthening; therefore, in these experiments, we used an MS medium without a nitrogen source. Seedlings were grown vertically in a growth chamber for 12 days at 25 ℃, 120 μmol m−2 s−1 fluorescent lighting, 60% relative humidity, and a 12/12-h photoperiod.
Elongation zone size was measured as the length from the first elongated cell adjacent to meristem to the first root hair. Data were subjected to one-way analysis of variance (ANOVA) using SPSS 19.0 software.
2.3. RNA extraction and microarray analysisSeedlings treated with 0.5‰ DMSO or 100 μM scutellarin were harvested 10 days after treatment. Total RNA of 10-day-old seedlings was extracted using RNAiso Plus reagent (TAKARA). For each treatment, three independent experiments were conducted, each containing 100 whole seedlings. cDNA preparation and hybridization were performed with an A. thaliana ATH1 Genome Array (Affymetrix). Data were analyzed using GeneSpring v.7.2.
2.4. Gene expression quantificationRNA was extracted from the roots of mock and treated seedlings, and used as template for cDNA synthesis using PrimeScript 1st Strand cDNA Synthesis Kit. Reale-time PCR was performed using a FastStart Universal SYBR Green Master (ROX) kit (Roche) and gene-specific primers (Table S1) in an ABI 7500 q-RT PCR System (Applied Biosystems). The resulting data were normalized against the housekeeping gene actin2 and relative quantification analysis was performed by the comparative CT method (2−ΔΔCT) (Yuan et al., 2006). RT-PCR was performed using Tiangen reagents and gene-specific primers (Table S1) over 28 cycles, and three independent replicates were performed for each gene.
2.5. Histological analyses and microscopyThe fluorescence of GFP was examined using a confocal laser scanning microscope (Olympus Fluvial FV1000). PI staining (Fernandez-Marcos et al., 2011) and FDA staining (Toda et al., 1999) were performed as described previously and observed using a confocal laser scanning microscope.
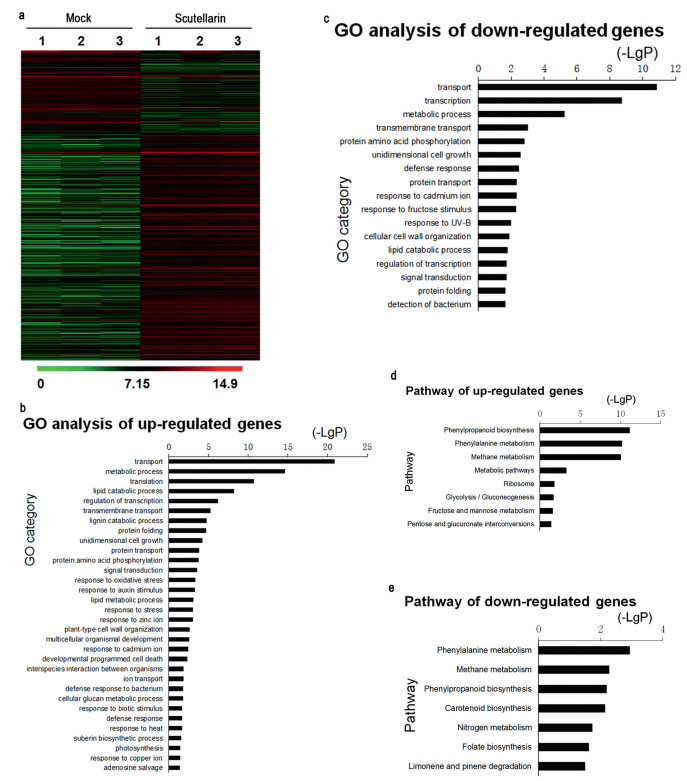
3. Results 3.1. Scutellarin alters gene transcription in Arabidopsis thaliana seedlingsTo evaluate the effects of scutellarin on plants, we compared the transcriptome profiles between mock- and scutellarin-treated Arabidopsis thaliana seedlings. Of the 22, 500 genes on the A. thaliana microarray, scutellarin induced a > 2-fold change in expression in 1, 253 genes: 802 were up-regulated and 451 were down-regulated compared to the mock treatment (Fig. 2). The results of the microarray analysis were confirmed by RT-PCR (Fig. S1). The percentage of genes that were significantly influenced by scutellarin treatment was about 5.6%. Given that auxin, a major hormone for plants, affects 5.5% of gene expression during root development (Lewis et al., 2013), these transcriptional data suggest that scutellain plays a significant role in plant development.
|
| Fig. 2 (a) Comparison of the effects of mock and scutellarin at the transcriptional level. Genome-wide identification of > 2-fold changes in gene expression between mock and 100 μM scutellarin treatment in 10-day seedlings. Three biological replicates were used for each treatment. GO analysis of differentially expressed genes, up-regulated (b) and down-regulated (c). Significant pathways over-represented among differentially expressed genes, up-regulated (d) and down-regulated (e). |
Gene ontology (GO) analysis of the differentially expressed genes revealed that response to auxin stimulus was one of the biological processes most markedly affected by scutellarin treatment (Fig. 2b). By querying the Kyoto Encyclopedia of Genes and Genomes pathway database, we found that phenylpropanoid biosynthesis and phenylalanine metabolism were the two pathways most enriched by scutellarin treatment (Fig. 2d). The products of these major pathways for secondary metabolite synthesis are the precursors of flavonoid synthesis (Laffont et al., 2010) (Fig. S2a). Among the differentially expressed genes, we found that flavonol synthase (FLS) and dihydroflavonol 4-reductase (DFR), which control whether intermediates in flavonoid biosynthesis are converted to flavonols or anthocyanins (Buer et al., 2010) (Fig. S2a), were respectively induced and suppressed (Fig. S2b). Given that auxin signaling and flavonol concentration are both key factors for root growth and development (Peer et al., 2011; Ubeda-Tomas et al., 2012; Jacobs and Rubery, 1988), these results suggest that scutellarin may affect root growth and development.
3.3. Scutellarin induced expression of NUC in rootsWe analyzed coexpression networks of global transcriptomes to identify genes that may play important roles in root development. Among the differentially expressed genes, 228 genes were coexpressed with at least one other gene under mock treatment (Fig. S3a). After scutellarin treatment, coexpression patterns of 209/228 genes changed (Fig. S3b), indicating that scutellarin altered their regulation network. For 97 of these genes, the number of coexpression neighbors changed by > 50%. One such gene is the transcription factor NUC, whose expression increased 3.8-fold. Given that NUC is located at the apical meristerm of primary root and is a key component of the regulatory pathway in root development (Levesque et al., 2006; Cui et al., 2007; Cruz-Ramirez et al., 2012), these data indicate that scutellarin may affect NUC during root development.
To test whether scutellarin induces NUC expression during root development, we compared the expression ofNUC in roots after scutellarin and naringenin treatments. Naringenin is a flavonoid inhibitor of root development that has the same chemical backbone as scutellarin but with 7-hydroxyl and without 6-hydroxyl (Fig. 1c) (Buer et al., 2010; Laffont et al., 2010; Peer et al., 2011). We found that NUC expression was significantly induced by scutellarin but not affected by naringenin (Fig. 3). These results suggest that scutellarin positively affects NUC in roots and this effect may be related to its 6-hydroxyl and 7-glucuronide.

|
| Fig. 3 Expression of NUC after treatment with mock, 100 μM scutellarin, or 100 μM naringenin in the roots of Arabidopsis thaliana seedlings for 7 and 10 days. ** indicates significant difference from that of mock or naringenin (P < 0.01, n = 3). |
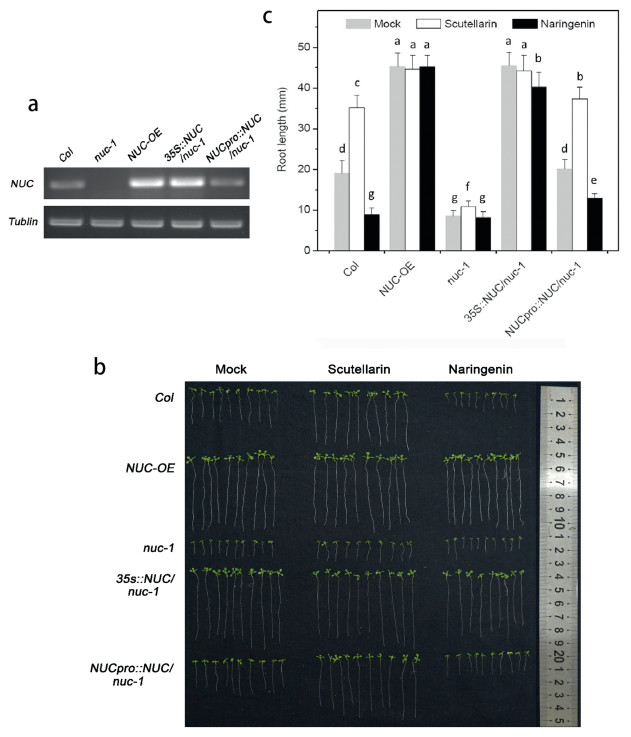
To test the effects of scutellarin on NUC and examine possible physiological consequences on roots, we isolated a NUC-knockout A. thaliana line nuc-1 and generated a NUC-overexpression (35::NUC, NUC-OE) line and two NUC-rescue lines (35S::NUC/nuc-1 and NUCpro::NUC/nuc-1) (Fig. 4a). We treated these lines with mock, scutellarin, or naringenin (Fig. 4b and c). Under control conditions (mock), the roots ofnuc-1 seedlings were significantly shorter, whereas the roots of NUC-OE seedlings were significantly longer (Fig. 4b and c); the roots of 35S::NUC/nuc-1 seedlings were longer than Col seedlings, whereas and the roots of NUCpro::NUC/nuc-1 seedlings were approximately the same length of Col seedlings (Fig. 4b and c, Table S2). In addition, we measured the meristem size of these seedlings by counting the cortex cell numbers in meristem and found the meristem sizes corresponded to their root-lengths (Fig. S4). These phenotypes verify previous speculation based on transcriptional evidence that NUC postively regulates root development (Levesque et al., 2006; Cui et al., 2007; Cruz-Ramirez et al., 2012). Furthermore, these findings suggest that scutellarin-induced expression of NUC (Fig. 3) might increase root length by increasing mersitem size.
|
| Fig. 4 (a) Expression of NUC in Col, nuc-knockout line nuc-1, nuc-overexpression line NUC-OE, and NUC-rescue lines 35S::NUC/nuc-1 and NUCpro::NUC/nuc-1. (b) Phenotypes of Col, NUC-OE, nuc-1, 35S::NUC/nuc-1 and NUCpro::NUC/nuc-1 under mock, 100 μM scutellarin, or 100 μM naringenin in 1/4 MS vertical plate for 12 days. (c) Root length ofCol, NUC-OE, nuc-1, 35S::NUC/nuc-1 and NUCpro::NUC/nuc-1 under mock, 100 μM scutellarin, or 100 μM naringenin in 1/4 MS vertical plate for 12 days. Values are means ± SD (n = 30). Bars with different letters differ significantly at P < 0.05. |
We indeed found that scutellarin lengthened roots in Col; in contrast, naringenin shortened roots (Fig. 4b and c). Furthermore, scutellarin-induced root lengthening almost disappeared in nuc-1 but reappeared in NUCpro::NUC/nuc-1. Note that in nuc-1, the roots treated with scutellarin were significantly but slightly longer than those given the mock treatment (Fig. 4c). These results indicate that loss of NUC may block the major effects of scutellarin or naringenin on roots. In addition, neither scutellarin nor naringenin affected root length in NUC-OE or 35S::NUC/nuc-1 (Fig. 4b and c, Table S2). This means that excess NUC may limit the effects of scutellarin or naringenin on roots. These results demonstrate that scutellarin promotes root length via a NUC-mediated pathway. The opposite effects of scutellarin and naringenin on root length may result from the structural differences of these compounds (i.e., 6-hydroxyl and 7-glucuronide).
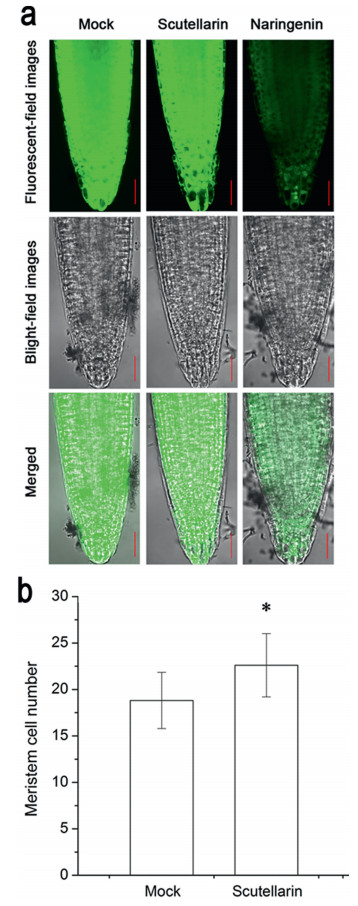
3.5. Scutellarin promotes the division of CEIBecause NUC expression was positively related to mersitem size (Fig. S4) and the increase of mersitem size might result from the asymmetric division of CEI cells into cortex/endodermis cells (Kinoshita et al., 2015), we speculated that scutellarin might promote the division of CEI. We first examined the cellular viability of root meristem and found that the scutellarin-treated roots retained the same viability as that of mock-treated roots. By contrast, naringenin treatment strongly decreased viability in the whole root tip (Fig. 5a). We further examined whether 6-hydroxyl group flavonoids promote meristem size of seedlings following mock and scutellarin treatments. The number of cortex cells in the meristem of plants treated with 100 μM scutellarin was significantly greater than that of plants given the mock treatment (Fig. 5b). As the consequence of viability loss in the root tip, the structure of meristem treated by naringein could not be clearly observed and thus meristem size could not be measured. These results indicate that scutellarin promotes meristem size and suggest that this promotion may be the cellular cause of root lengthening. These results also suggest that flavonoids with and without a 6-hydroxyl and 7-glucuronide have opposite effects on the cell division of root meristem.
|
| Fig. 5 (a) Root tip viability of Arabidopsis thaliana Col grown on mock, 100 μM scutellarin, 100 μM naringenin for 5 days. The viable cells showed green fluorescence due to Fluorescein Diacetate (FDA). Bar, 30 μm. (b) Root cortex meristematic cell number of Arabidopsis thaliana Col grown on mock, 100 μM scutellarin for 5 days. The number of cortex cells in a single file extending from the QC up to elongated cell was counted. Values are means ± SD (n = 10–11). * indicates that the value is significantly different from that of mock (P < 0.05). |
We found that scutellarin promotes lengthening of roots through NUC-mediated regulation of root meristem. Scutellarin increased expression of NUC and increased root length. Its effect on root length depended on the transcription factor NUC. Scutellarin increased cortex meristematic cell number. In contrast, naringenin had no effect on NUC expression and decreased cellular activity in the meristem. Taken together, these results show that flavonoids with a 6-hydroxyl and 7-glucoside and those without a 6-hydroxyl and 7-glucoside were able to regulate root meristem in opposite ways through a NUC-mediated pathway.
Cell fate and differentiation in the root meristem is regulated by a circuit of key components that include SHR, SCR, and RBR (Menke and Scheres, 2009; Petricka et al., 2009). Loss of SHR or SCR inhibits asymmetric cell division in the CEI and the formation of endodermis and cortex, ultimately decreasing root length (Helariutta et al., 2000; Di Laurenzio et al., 1996; Sabatini et al., 2003). NUC is a down-stream factor in the genetic pathway that regulates asymmetric cell division. In our study, loss of NUC led to short roots (Fig. 4b, c and S4). Scutellarin treatment increased NUC expression, which in turn increased the number of cortex cells, as shown in previous studies (Sozzani et al., 2010; Cui et al., 2007) (Fig. 3, Fig. 5). Increase in meristem size eventually lengthens roots. This signalling pathway is supported by our observation that the overexpression ofNUC increases root length (Fig. 4b and c). This pathway might be the mechanism by which scutellarin increases root length (Fig. 4b and Fig. S4). In contrast, naringenin treatment strongly decreased the viability of the whole root tip (Fig. 5a), which may decrease root length. Roots were slightly but significantly lengthened by scutellarin in nuc-1 (Fig. 4c). Given that NUC and MGP are both down-stream components of the pathway the regulates asymmetric cell division and that the expression of MGP can be induced by scutellarin (Fig. 5), we speculate that this slight increase in root length may result from the increase in MGP expression when NUC is blocked. In summary, the opposite effects that flavonoids with and without a 6-hydroxyl group have on root elongation appear to be due to their different effects on the NUC-mediated pathway that regulates asymmetric cell division.
Root development can be regulated by both polar auxin transport and asymmetric cell division pathways. The mechanisms by which flavonoids modulate the polar auxin transport pathway are well documented (Buer et al., 2010; Laffont et al., 2010; Peer et al., 2011). To the best of our knowledge, our study is the first to demonstrate that flavonoids play a role in the regulation of asymmetric cell division. Flavonoids with or without 6-hydroxyl and 7-glucoside can positive or negatively regulate the NUC-mediated pathway of asymmetric cell division, respectively. This finding provides a chemical approach to regulate root meristem.
Author contributionsInvestigation, X.H. and X.Z.; conceptualization, W.L.; writing, X.Z. and W.Li; funding acquisition, W.L. and X.Z.; supervision, W.L. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interestNone.
AcknowledgementsThis research was funded by Yunnan Applied Basic Research Project (2017FB057 and 2017AB001), as well as National Natural Science Foundation of China (31700235 and 31770375). We thank Dr. Qinshi Zhao for donations of scutellarin, Dr. Yanxia Jia for assistance with confocal laser scanning microscope analysis, and Liyan Peng, Yaowen Chang, Dr. Buzhu Yu, and Dr. Peiji Zhao for assistance with the flavonoid assay, and Dr. Buzhu Yu for the chemical structure diagram.
Appendix A. Supplementary dataSupplementary data to this article can be found online at _aaaaaa_paichu__.
Ahmed M., Chaudhari K., Babaei-Jadidi R., et al, 2017. Concise review: emerging drugs targeting epithelial cancer stem-like cells. Stem Cell, 35: 839-850. DOI:10.1002/stem.2579 |
Buer C.S., Imin N., Djordjevic M.A., 2010. Flavonoids: new roles for old molecules. J. Integr. Plant Biol, 52: 98-111. DOI:10.1111/j.1744-7909.2010.00905.x |
Casamitjana-Martinez E., Hofhuis H.F., Xu J., et al, 2003. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol, 13: 1435-1441. DOI:10.1016/S0960-9822(03)00533-5 |
Chanda B., Ditadi A., Iscove N.N., Keller G., 2013. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell, 155: 215-227. DOI:10.1016/j.cell.2013.08.055 |
Cruz-Ramirez A., Diaz-Trivino S., Blilou I., et al, 2012. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell, 150: 1002-1015. DOI:10.1016/j.cell.2012.07.017 |
Cruz-Ramirez A., Diaz-Trivino S., Wachsman G., et al, 2013. A SCARECROWRETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol, 11: e1001724. DOI:10.1371/journal.pbio.1001724 |
Cui H., Levesque M.P., Vernoux T., et al, 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science, 316: 421-425. DOI:10.1126/science.1139531 |
Di Laurenzio L., Wysocka-Diller J., Malamy J.E., et al, 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell, 86: 423-433. DOI:10.1016/S0092-8674(00)80115-4 |
Fernandez-Marcos M., Sanz L., Lewis D.R., et al, 2011. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1(PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U.S.A, 108: 18506-18511. DOI:10.1073/pnas.1108644108 |
Helariutta Y., Fukaki H., Wysocka-Diller J., et al, 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell, 101: 555-567. DOI:10.1016/S0092-8674(00)80865-X |
Hou P., Li Y., Zhang X., et al, 2013. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science, 341: 651-654. DOI:10.1126/science.1239278 |
Jacobs M., Rubery P.H., 1988. Naturally occurring auxin transport regulators. Science, 241: 346-349. DOI:10.1126/science.241.4863.346 |
Kinoshita A., Ten Hove C.A., Tabata R., et al, 2015. A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development, 142: 444-453. DOI:10.1242/dev.113167 |
Laffont C., Blanchet S., Lapierre C., et al, 2010. The compact root architecture1 gene regulates lignification, flavonoid production, and polar auxin transport in Medicago truncatula. Plant Physiol, 153: 1597-1607. DOI:10.1104/pp.110.156620 |
Levesque M.P., Vernoux T., Busch W., et al, 2006. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol, 4: e143. DOI:10.1371/journal.pbio.0040143 |
Lewis D.R., Olex A.L., Lundy S.R., et al, 2013. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell, 25: 3329-3346. DOI:10.1105/tpc.113.114868 |
Menke F.L., Scheres B., 2009. Plant asymmetric cell division, vive la difference!. Cell, 137: 1189-1192. DOI:10.1016/j.cell.2009.06.007 |
Moreno-Risueno M.A., Sozzani R., Yardimci G.G., et al, 2015. Transcriptional control of tissue formation throughout root development. Science, 350: 426-430. DOI:10.1126/science.aad1171 |
Peer W.A., Blakeslee J.J., Yang H., Murphy A.S., 2011. Seven things we think we know about auxin transport. Mol. Plant, 4: 487-504. DOI:10.1093/mp/ssr034 |
Peer W.A., Murphy A.S., 2007. Flavonoids and auxin transport: modulators or regulators?. Trends Plant Sci., 12: 556-563. DOI:10.1016/j.tplants.2007.10.003 |
Petricka J.J., Van Norman J.M., Benfey P.N., 2009. Symmetry breaking in plants: molecular mechanisms regulating asymmetric cell divisions in Arabidopsis. Cold Spring Harb. Perspect Biol, 1: a000497. |
Sabatini S., Heidstra R., Wildwater M., et al, 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev, 17: 354-358. DOI:10.1101/gad.252503 |
Sozzani R., Cui H., Moreno-Risueno M.A., et al, 2010. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature, 466: 128-132. DOI:10.1038/nature09143 |
Toda T., Koyama H., Hara T., 1999. A simple hydroponic culture method for the development of a highly viable root system in Arabidopsis thaliana. Biosc.Biotech. Biochem, 63: 210-212. DOI:10.1271/bbb.63.210 |
Ubeda-Tomas S., Beemster G.T.S., Bennett M.J., 2012. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci, 17: 326-331. DOI:10.1016/j.tplants.2012.02.002 |
Wu S., Lee C.M., Hayashi T., et al, 2014. A plausible mechanism, based upon ShortRoot movement, for regulating the number of cortex cell layers in roots. Proc.Natl. Acad. Sci. U.S.A, 111: 16184-16189. DOI:10.1073/pnas.1407371111 |
Yin R., Han K., Heller W., et al, 2014. Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol, 201: 466-475. DOI:10.1111/nph.12558 |
Yuan J.S., Reed A., Chen F., Stewart Jr., C.N., 2006. Statistical analysis of real-time PCR data. BMC Bioinf, 7: 85. DOI:10.1186/1471-2105-7-85 |



