b. Yunnan Key Laboratory for Integrative Conservation of Plant Species with Extremely Small Populations, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China;
c. University of Chinese Academy of Sciences, Beijing, 100049, China;
d. Sichuan Panzhihua Cycas National Nature Reserver Bureau, Panzhihua, 617000, China;
e. Department of Botany and Biodiversity Research, University of Vienna, Rennweg 14, A-1030, Vienna, Austria;
f. CAS Key Laboratory for Plant Biodiversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China
The perennial herb Aristolochia delavayi Franch. (Aristolochiaceae) is an endemic species in the dry-warm valleys along the Jinsha River in southwestern China (Chen et al., 2018; Yu et al., 2020). The plant species has a short single-flower flowering period (~3 days), but the duration of flowering within a population is approximate three months (mostly from June to September). It mainly reproduces in a sexual way, which relies on cross-pollination by pollinators, but the plant itself tends to increase breeding opportunities through tillering (unpublished data). The fruits of A. delavayi are capsules, and its seeds are lacking elaiosomes. A previous study showed that seed dispersal mainly depends on gravity (Chen et al., 2015). Leaves of this plant species are locally used as a spice (Zhou et al., 1995; Sun et al., 2008) and also for medicinal purposes e.g. to strengthen the stomach, to increase the appetite and treat flatulence and malaria (Zhou and Yang, 1995). This plant species is also an important food source for the endangered butterfly species Byasa daemonius Alphéraky (Chen et al., 2015).
Aristolochia delavayi grows mainly in sparse vegetation in river valleys at an altitude of 1220–2250 m (Yang et al., 2014; He et al., 2017). The habitat in this area is fragile with high erosion rate, which makes vegetation restoration difficult (Zhong, 2000; Guan et al., 2013). Due to the habitat destruction by various human activities (e.g. hydropower construction, agricultural activities), and also the long-term overharvesting of this plant species, the populations of A. delavayi decreased gradually, so the species is on the verge of extinction (Yang et al., 2014; Chen et al., 2015). At present, A. delavayi is listed as a wild endangered species (EN) on the red list of higher plants in China and the red list of IUCN (Chen et al., 2015; Qin et al., 2017). It is also treated as a plant species with extremely small populations (PSESP) (Ma et al., 2013). To protect this species and to prevent extinction, protection measures are required.
Generally, before implementation of conservation, studying the genetic structure and diversity of a given species can help to develop scientific conservation strategies (Guan et al., 2013). The precipitous terrain and severe habitat fragmentation in the studied area may hinder gene flow among plant populations. Relevant studies revealed, the endemic taxa in this area generally show high genetic differentiation among populations, but the level of genetic diversity within populations is either high or low. This for example was shown on Cycas panzhihuaensis L. Zhou & S.Y. Yang (Xiao et al., 2020), Musella lasiocarpa (Franch.) C.Y. Wu (Ma et al., 2019), Trailliaedoxa gracilis W.W. Sm. & G. Forrest (Jia et al., 2016) and Munronia delavayi Franch. (Jia et al., 2014). By comparing the genetic differences between A. delavayi and other endemic plant species in this region, it is helpful to clarify the population history, its dynamics, and the effect of external environmental factors on the genetic pattern of the existing populations. Recent genetic analyses on A. delavayi was undertaken by Yang et al. (2014). They analyzed four populations using eight ISSR markers, which showed a high genetic diversity (PPB = 84.71%). However, during our comprehensive field surveys in recent years, we found that the above-mentioned four populations are only located in the upstream part of the distribution area of A. delavayi, whilst the populations in the middle and lower part of the distribution area were not involved. Considering that A. delavayi is vulnerable, it is necessary to use a higher number of molecular markers to re-study and evaluate its population genetics comprehensively.
At present, molecular genetic marker methodologies have been well developed which now allow performing sophisticated analyses. In particular, microsatellite markers have widely been used in the genetic research of wild natural populations and endangered species (Balloux and Lugon-Moulin, 2002; Chen et al., 2009; Tang et al., 2008; Wang et al., 2006; Yang et al., 2018). This kind of markers are rich in polymorphism, good in repeatability, mostly co-dominant, widely distributed in the genome, and reveal a large amount of information even within a small sample size (Jarne and Lagoda, 1996; Zhang and Hou, 2004; Habel et al., 2010; Zhang et al., 2019). In this study, SSR primers for A. delavayi were developed for the first time by using genome skimming technology. Based on these primers, we were able to analyze the genetic structure and its diversity of existing wild populations of this species. The obtained results allowed us to analyze the internal causes of the species' endangerment and propose reasonable conservation strategies.
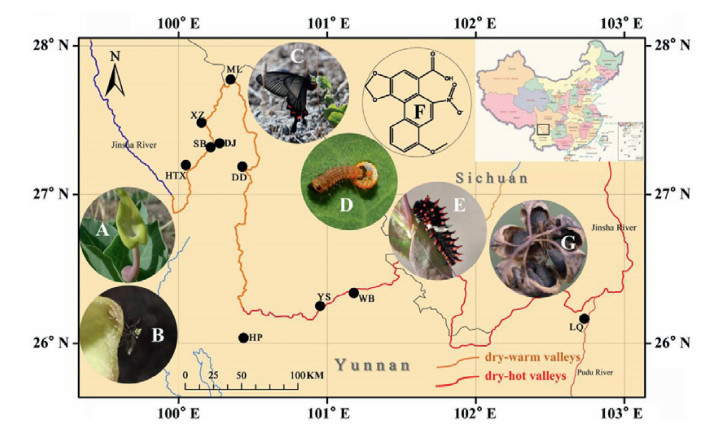
2. Materials and methods 2.1. Plant materialsDuring the population and habitat surveys of the past few years, molecular samples of 193 individuals from ten populations of Aristolochia delavayi were collected along the Jinsha River Basin in July 2018 and August 2019. Detailed information about the plant material is provided in Table 1. Most of the samples were collected in Yunnan Province, except the samples belonging to the population ML, which was collected from Sichuan province (Fig. 1). From each population were more than 20 individuals collected, with a few exceptions, such as DD (only two individuals) and WB (seven individuals) due to the rare occurrence of this plant species. Fresh and healthy leaves were dried with silica gel immediately after collection and stored at −20 ℃ prior DNA extraction. Voucher specimens of partial populations were deposited in the Herbarium of Kunming Institute of Botany (KUN), Chinese Academy of Sciences.
| Population | Location | Longitude (E) | Latitude (N) | Altitude (m) | Sample size |
| HTX | Tiger Leaping Gorge Town | 100.051503 | 27.186242 | 1860 | 21 |
| SB | Jiangbian Village, Sanba Town | 100.228188 | 27.316460 | 1858 | 29 |
| DJ | Daju Town | 100.237005 | 27.334241 | 1732 | 24 |
| XZ | Xiazhuen Village, Sanba Town | 100.159363 | 27.486931 | 1931 | 24 |
| ML | Eya Town | 100.358434 | 27.767007 | 1832 | 20 |
| DD | Dadong Town | 100.449564 | 27.163259 | 1584 | 2 |
| HP | Huangping Town | 100.443849 | 26.027998 | 2002 | 21 |
| YS | Dongshan Town | 100.969334 | 26.241000 | 1322 | 21 |
| WB | Wanbi Town | 101.179270 | 26.342586 | 1222 | 7 |
| LQ | Zehei Town | 102.755256 | 26.158288 | 1228 | 24 |
|
| Fig. 1 Distribution of collected populations of Aristolochia delavayi. (A) A blooming flower (typical pollinator captive strategy in Aristolochiaceae, female flowering stage attracts pollinators and male flowering stage releases pollinators); (B) A pollinator carrying pollen crawls out of the split utricle; (C) A Byasa daemonius individual locates its spawning site based on the smell of Aristolochia delavayi leaves; (D) A newborn larvae leaving out of its shell (Byasa daemonius); (E) A larvae is feeding on Aristolochia delavayi leaf; (F) Aristolochic acid derivates can be found in leaves and Byasa daemonius body parts (accumulated these compounds to repel its potential natural enemies); (G) Seed dispersal from dehiscent capsule by gravity. A, G pictures were taken by Gao Chen; B, D pictures were taken by Zhi Chen and C, E pictures were taken by Yang Niu. |
Genomic DNA was extracted from the dried leaves using a modified CTAB method (Doyle and Doyle, 1987), and the quality and concentration of DNA were measured by using a spectrophotometer (Nanodrop - 1000). Using the genome skimming technology, the genomic DNA from two individuals in the DJ and LQ populations were used for genome skimming. The specific process is as follows: The sequencing was performed on the Miseq Benchtop Sequencer (Illumina X-Ten) using the 2 × 250 bp read mode. The obtained data were assembled using software SPAdes and the microsatellite sequences of two samples were compared by using the software QDD 2.1 Beta (Meglecz et al., 2010). MISA software was used to design SSR primers based on the flanking sequence of the SSR loci.
The overall PCR reaction system volume was 20 μL, including 1 μL DNA template of 50–60 ng/μL; 2 ×Taq PCR MasterMix 10 μL, 0.3 μL of each primer at a concentration of 10 μmol/L; and 8.4 μL ddH2O. The amplification procedure was set as follows: Pre denaturation at 95 ℃ for 3 min; denaturation at 95 ℃ for 30 s, annealing at appropriate temperature for 30 s, extension at 72 ℃ for 30 s, a total of 32 cycles; and the final extension step at 72 ℃ for 5 min. The annealing temperature (Tm) was determined according to the reference value of primer synthesis and optimized in the experiment. The PCR reaction was performed on the DNA Thermal Cycler (Applied Biosystems).
For the primer screening, the genomic DNA of four individuals from four populations were selected for PCR amplification. The PCR products were detected using 8% non-denaturing polyacrylamide gel electrophoresis. The gel was stained with silver and developed after electrophoresis, and the amplification was observed. The amplification effects of primers were compared, and polymorphic primers were screened for genetic analysis of all samples. The PCR products were separated and visualized using QIAxcel of capillary gel electrophoresis system (ABI PRISM 3730 XL, USA). The final banding data were read by GeneMarker v.2.2.
2.3. Data analysisTo detect linkage disequilibrium (LD) between the loci, we used the program GENEPOP v.4.7 (Rousset, 2008). The following parameters of genetic diversity at population and species level were calculated by the GenAlEx v.6.41 (Peakall and Smouse, 2006): Population sample sizes (N), number of alleles (Na), effective number of alleles (Ne), Shannon's information index (I), observed (Ho) and expected (He) frequency of heterozygotes, percentage of polymorphic loci (PPL), and fixation index (F). The GenAIEx program was also used to calculate the inbreeding coefficient (FIS) and gene flow (Nm) at population level.
The geographic distances between populations were calculated using the Franson CoordTrans Program v.2.3 Meanwhile, POPGENE v.1.32 was used to calculate the Nei's genetic distance between populations of Aristolochia delavayi. We established a geographic distance as well as a genetic distance matrix, and used Mantel test in GenAlEx v.6.41 to detect correlation between geographic and genetic distance (IBD) (Mantel, 1967; Smouse et al., 1986; Peakall and Smouse, 2006). The recent bottleneck effects were detected by using BOTTLENECK v.1.2.2. For this purpose, two evolutionary models, infinite alleles model (IAM) and two-phase model (TPM) (Wang et al., 2010), were selected, and 1000 iterations statistics were carried out by performing the Sign test and Wilcoxon sign-rank test, respectively.
Concerning the genetic structure, hierarchical analyses of molecular variance (AMOVA) were performed to assess the genetic structure within and between populations by using Arlequin v.3.11 (Excoffier et al., 2005). Principal coordinate analysis (PCoA) was conducted using GenAlEx v.6.41 to test the genetic similarity of all the individuals in the included population (Peakall and Smouse, 2006). UPGMA cluster analysis based on the Nei's genetic distance between populations was performed by using MEGA v.7.0.14. Bayesian clustering method in Structure v.2.3.4 was used to cluster all individuals individually and determine subsequently the number of genetic groups (Pritchard et al., 2000). The parameters for the calculation of the ΔK value and analyze possible genetic structure were set as follows: The K value was set to 10, 10 replicates, using the admixture model and assumed allele frequencies correlated, followed by a burn-in of 1 × 106 iterations (Evanno et al., 2005).
3. Results 3.1. Development and screening of SSR primersIn this study, 3689 pairs of SSR primers were successfully discovered and designed based on the genome skimming technique, and 100 pairs of primers were randomly selected for polymorphism primer screening. Finally, 15 pairs of SSR primers with good amplification effects and high polymorphism were selected and used for genetic analysis. The details of these primers are shown in Table 2.
| Primer No. | Primer sequence (5′–3′) | Repeat motif | Fragment size (bp) | Tm (℃) |
| YYL-40 | F: AAGCAGGATGTGGGTAATGG R: CCTACCCAACAAGGAAACGA |
(CTT)10 | 304–325 | 55.4 |
| YYL-42 | F: CAAGGGAACTGCACACATTG R: GGCTGACCTTAGCCTGAATG |
(TAA)11 | 254–284 | 56.4 |
| YYL-43 | F: TTGCGATGCTAGAGAACACG R: ACGTACCCAAGATGGCACTC |
(AGA)11 | 175–205 | 56.4 |
| YYL-45 | F: AGAGGGGTAAGAGAAAGCGG R: GAATCCTGTACCAGCGGAGA |
(GAA)12 | 246–267 | 57.4 |
| YYL-47 | F: CATGCATCAGGAGTTGTGCT R: AGTTGCAGAGAAGGGAACGA |
(AAG)12 | 140–164 | 55.4 |
| YYL-48 | F: GTATTACCACCATGGGGACG R: CGGAGCTCCATCTTCATCTC |
(GAA)13 | 252–279 | 57.4 |
| YYL-49 | F: TAATCACCTGCTTCCTGCTG R: CTCTTGAGTACTGGGGCGAC |
(GAA)14 | 306–330 | 57.4 |
| YYL-54 | F: CAGATTCGACGACGTCATGT R: GCCTACGTACTGATGCCCTT |
(CGAT)5 | 246–278 | 56.4 |
| YYL-59 | F: ACACCCGTTTCGATTTGAAG R: CTTGCTTTTGTGGTTGGGTT |
(AGAA)5 | 193–205 | 53.8 |
| YYL-68 | F: CACGATCGGATCATCAACAC R: CTTTGTCGTCCTCCAGCTTC |
(TGGA)6 | 215–247 | 56.4 |
| YYL-69 | F: AGATACATCGAATTTGGGCG R: GGGATGGATTGGCTTAAGTTT |
(TGTT)7 | 133–161 | 53.8 |
| YYL-70 | F: TCCACAGCCACCTAAATTCC R: TCGACGATCTCAAAATTCCC |
(ATCT)7 | 163–199 | 54.6 |
| YYL-78 | F: TCGTCGAAGAACCCAATTTC R: CACGCCATGGAACACTACAG |
(AATC)8 | 238–294 | 55.4 |
| YYL-81 | F: TAACGGGCAAAACTGGAATC R: ATAGAAACCCGCAATCATGC |
(TTTAT)5 | 195–215 | 53.8 |
| YYL-86 | F: CCTCACAAGGCCACAAGAAT R: CAATTCTCAAACCGTCCCAT |
(AAATA)5 | 249–274 | 54.6 |
The GENEPOP test showed that only a few detectable loci were linked in a few populations, but none of them were linked with other loci in more than three populations. Due to this result, the above-mentioned 15 pairs of SSR primers were used for the genetic analysis of A. delavayi (Wang et al., 2013) and the results of these analyses are presented in Table 3, Table 4. Overall, a total of 78.50 alleles (Na) and 44.45 effective alleles (Ne) were detectable. Within these 15 pairs of SSR primers, 3–10 alleles and 2–6 effective alleles with an average of 5.23 alleles and 2.96 effective alleles could be detected (Table 3). The calculated observed heterozygosity (Ho) varied from 0.333 (DD) to 0.705 (SB), whilst the expected heterozygosity (He) varied from 0.233 (DD) to 0.677 (XZ). The calculated Shannon information index (I) ranges from 0.335 (DD) to 1.485 (SB). At population level, except the populations DD and WB, the percentages of polymorphic loci (PPL) in all other populations was 100% (Table 4). The calculated inbreeding coefficient (FIS) value varied from −0.336 to 0.319, with an average value of 0.056. The average values of the genetic differentiation coefficient (FST) and the gene flow (Nm) among populations are 0.328 and 0.591, respectively.
| Locus | Sample size | Na | Ne | FIS | FIT | FST | Nm |
| YYL40 | 193 | 5.200 | 2.239 | 0.253 | 0.640 | 0.527 | 0.224 |
| YYL42 | 193 | 6.300 | 3.664 | −0.150 | 0.027 | 0.205 | 0.967 |
| YYL43 | 193 | 7.400 | 4.146 | 0.004 | 0.143 | 0.217 | 0.902 |
| YYL45 | 193 | 4.500 | 2.406 | 0.046 | 0.387 | 0.307 | 0.566 |
| YYL47 | 193 | 4.800 | 2.556 | −0.010 | 0.245 | 0.316 | 0.540 |
| YYL48 | 193 | 5.100 | 3.032 | 0.046 | 0.204 | 0.258 | 0.718 |
| YYL49 | 193 | 7.400 | 3.931 | 0.207 | 0.401 | 0.283 | 0.633 |
| YYL54 | 193 | 4.600 | 2.248 | 0.199 | 0.435 | 0.364 | 0.437 |
| YYL59 | 193 | 3.300 | 2.069 | −0.083 | 0.391 | 0.429 | 0.333 |
| YYL68 | 193 | 3.800 | 2.478 | −0.336 | −0.028 | 0.239 | 0.794 |
| YYL69 | 193 | 4.700 | 2.338 | 0.319 | 0.616 | 0.446 | 0.311 |
| YYL70 | 193 | 5.100 | 3.171 | 0.129 | 0.403 | 0.316 | 0.542 |
| YYL78 | 193 | 9.600 | 6.391 | 0.048 | 0.164 | 0.172 | 1.206 |
| YYL81 | 193 | 3.500 | 1.64 | 0.111 | 0.457 | 0.469 | 0.283 |
| YYL86 | 193 | 3.200 | 2.139 | 0.058 | 0.382 | 0.378 | 0.412 |
| Mean | 5.233 | 2.963 | 0.056 | 0.324 | 0.328 | 0.591 |
| Pop | N | Na | Ne | Ho | He | I | F | PPL |
| HTX | 21 | 5.600 | 3.252 | 0.644 | 0.585 | 1.197 | −0.056 | 100.00% |
| SB | 29 | 7.733 | 4.071 | 0.705 | 0.673 | 1.485 | −0.067 | 100.00% |
| DJ | 24 | 6.200 | 3.309 | 0.608 | 0.613 | 1.289 | 0.014 | 100.00% |
| XZ | 24 | 7.133 | 3.840 | 0.579 | 0.677 | 1.467 | 0.121 | 100.00% |
| ML | 20 | 5.067 | 3.072 | 0.494 | 0.582 | 1.169 | 0.129 | 100.00% |
| DD | 2 | 1.533 | 1.427 | 0.333 | 0.233 | 0.335 | −0.417 | 53.33% |
| HP | 21 | 4.200 | 2.080 | 0.386 | 0.433 | 0.829 | 0.151 | 100.00% |
| YS | 21 | 4.933 | 2.948 | 0.581 | 0.579 | 1.127 | 0.012 | 100.00% |
| WB | 7 | 4.000 | 2.736 | 0.667 | 0.556 | 1.046 | −0.193 | 93.33% |
| LQ | 24 | 5.933 | 2.897 | 0.530 | 0.566 | 1.180 | 0.144 | 100.00% |
| Mean | 5.233 | 2.963 | 0.553 | 0.550 | 1.112 | 0.005 | 94.67% |
In the performed Bottleneck test, under the IAM hypothesis, both methods exhibited, that the population YS deviated from the mutation drift equilibrium, showing a significant heterozygosity excess. Under the TPM hypothesis, the sign test revealed that the population LQ deviated from the mutation drift equilibrium, exhibiting a significant heterozygosity excess (Table 5). This test indicated, that the above-mentioned two populations have recently experienced a bottleneck effect.
| Population | Mutation-drift equilibrium test model | ||||||
| Infinite allele model (IAM) | Two-phase model (TPM) | ||||||
| Sign test | Wilcoxon test | Sign test | Wilcoxon test | ||||
| HTX | 0.28867 | 0.33026 | 0.51241 | 0.89038 | |||
| SB | 0.38354 | 0.15143 | 0.21489 | 0.56140 | |||
| DJ | 0.54858 | 0.52448 | 0.58928 | 0.48871 | |||
| XZ | 0.58743 | 0.10699 | 0.42055 | 0.35913 | |||
| ML | 0.05394 | 0.05536 | 0.53515 | 0.38940 | |||
| DD | 0.12492 | 0.31250 | 0.18390 | 0.31250 | |||
| HP | 0.56735 | 0.63867 | 0.28262 | 0.27686 | |||
| YS | 0.04278a | 0.02155a | 0.16179 | 0.30280 | |||
| WB | 0.16491 | 0.24121 | 0.49945 | 0.76086 | |||
| LQ | 0.55134 | 0.89038 | 0.01079a | 0.08325 | |||
| Note: P-value is the test of heterozygosity excess. asignificant with P < 0.05. |
|||||||
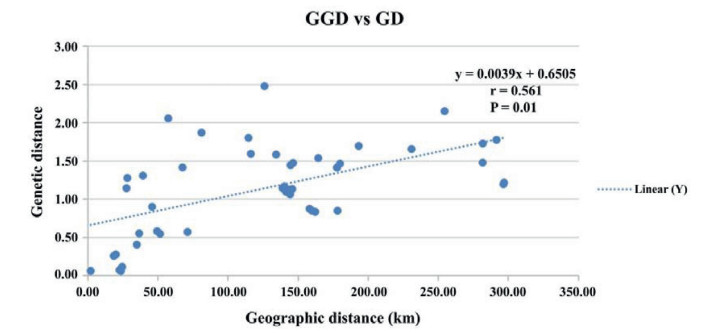
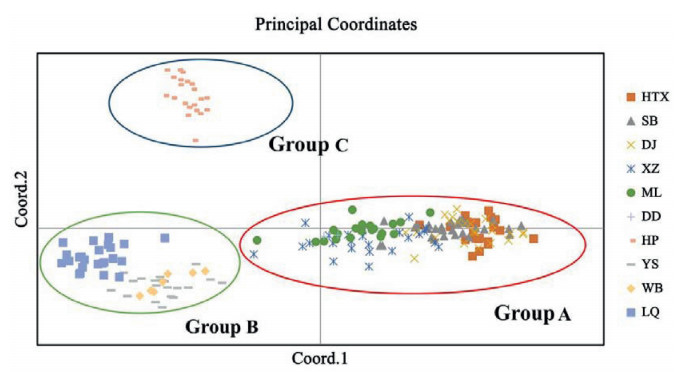
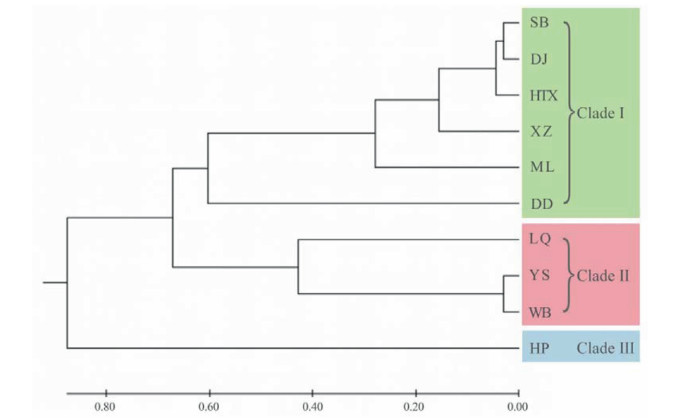
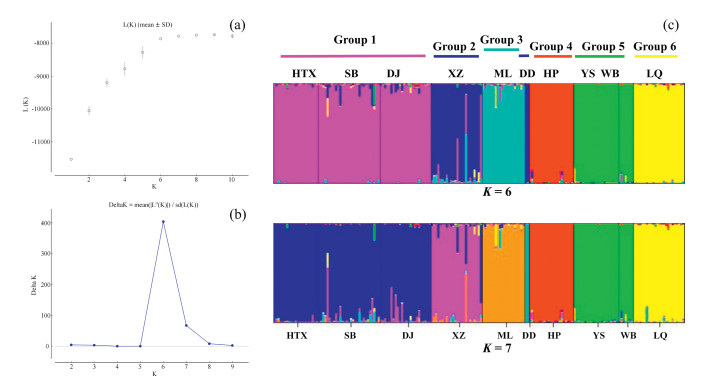
The AMOVA results (Table 6) indicated significant differences among populations of Aristolochia delavayi, because 31.62% of the variation existed among populations and 68.38% occurred within populations. The genetic differentiation coefficient (FST) at species level is 0.316, which is considered as significant, i.e. 0.25 < FST < 1. This indicates a significant genetic differentiation among populations (Wright, 1978). The performed Mantel test resulted in a significant positive correlation between the genetic distances of the studied populations and its geographic distance (P = 0.01 < 0.05 and r = 0.561 > 0.5) (Fig. 2), which also indicates the presence of IBD in the population structure. The result of principal coordinate analysis indicated, that the analyzed ten natural populations can be roughly grouped into three groups (Group A includes populations HTX, SB, DJ, XZ, ML, DD, Group B includes YS, WB, LQ and Group C includes HP) (Fig. 3). This result was supported by the performed UPGMA cluster analysis based on genetic distance (Fig. 4). Structural analysis showed, a ΔK is at the maximum, when K = 6. This indicated that all populations can be assembled in six groups. These results further refine the grouping results of the above-mentioned methods. Among all analyzed samples, six populations collected in Northwestern Yunnan consist of three groups; the three populations in Central Yunnan clustered in two groups, and the Huangping population was still remained separately (Fig. 5).
| Source of variation | df | Sum of squares | Variance components | Percentage of variation [%] | FST |
| Among groups | 1 | 200.41 | 0.78 | 12.28 | |
| Among populations within groups | 8 | 401.98 | 1.23 | 19.34 | |
| Within populations | 376 | 1641.18 | 4.36 | 68.38 | 0.32a |
| Total | 385 | 2243.56 | 6.38 | 100 | |
| Note: df means degree of freedom; grouping of populations: Yunnan northwestern group (populations HTX, SB, DJ, XZ, ML and DD) and Yunnan central group (populations HP, YS, WB and LQ). aextremely significant with P < 0.01. |
|||||
|
| Fig. 2 Correlation analysis of geographic distance (GGD) and genetic distance (GD) of ten populations of Aristolochia delavayi (Mantel test). |
|
| Fig. 3 The result of Principal Coordinates Analysis (PCoA) of Aristolochia delavayi populations. The first and second axis explained 34.13% and 25.23% of the total genetic variance, respectively. |
|
| Fig. 4 UPGMA dendrogram based on Nei's genetic distance. Population codes see Table 1. |
|
| Fig. 5 Genetic structure of Aristolochia delavayi inferred by Bayesian clustering of SSR data. (a) Plot of mean posterior probability Ln(K) values in the range of K from 1 to 10; (b) The corresponding ΔK statistics calculated according to Evanno et al. (2005); (c) Bar plots showing assignment probabilities from structure analysis when K = 6 and K = 7, the same color in the figure represents the same cluster. Population codes see Table 1. |
Genetic variation of species is the premise of local adaptation and evolution. It is also considered as an important parameter to determine the priority of population conservation in the protection of endangered plants (Schaal et al., 1998; Laikre et al., 2010; Zhao and Gong, 2015). So, understanding the genetic status of Aristolochia delavayi provides a rational basis for the evaluation of conservation work and formulating of effective protection measures. Young et al. (1996) showed that genetic variation of population eroded with reduced remnant population size. However, our analysis results showed that the expected and observed heterozygosity of A. delavayi at species level are 0.550 and 0.553, respectively. These values are higher than reported values from other endemic or endangered taxa in this region, such as Nouelia insignis Franch. (He = 0.149, Ho = 0.216) (Luan et al., 2006), Buddleja crispa Benth. (H = 0.314, I = 0.485) (Zhang et al., 2015), Cycas hongheensis S.Y. Yang & S.L. Yang ex D.Y. Wang (He = 0.435, Ho = 0.403) (Zhao and Gong, 2015), T. gracilis W.W. Smith et Forrest (Hs = 0.489) (Jia et al., 2016), Amorphophallus albus P.Y. Liu & J.F. Chen (He = 0.504, Ho = 0.528) (Tang et al., 2020), C. panzhihuaensis L. Zhou & S.Y. Yang (He = 0.328, Ho = 0.189) (Xiao et al., 2020). Our calculated result is consistent with the result reported by Yang et al. (2014), indicating that the species possess relatively high genetic diversity. Moreover, this indicates that the recent population size reduction only affects little the genetic variation of the species. According to Loveless and Hamrick (1984) and Nybom and Bartish (2000), the factors which affect the genetic variation of species generally include population history, population size, reproduction pattern, breeding system, genetic drift, gene flow, natural selection, geographical distribution, etc. The reduction of the population size cause genetic drift effects, but drift has only a little effect on genetic variation in limited generations (Young et al., 1996). The recent habitat fragmentation event may be below the fragmentation threshold, which probably not cause the loss of genetic variation (Prober and Brown, 1994). We assume that in the past few decades, with the rapid reduction of the population size, the genetic drift effects are not yet accumulated, and the species may retain its genetic variation by its sexual as well as asexual reproduction.
4.2. Genetic differentiation and structureThe results of AMOVA analysis showed that the genetic variation of Aristolochia delavayi occurred mainly within the population, but there was a significant genetic differentiation among the populations observable. The high percentage of genetic diversity within the population may be due to the retention of genetic resources prior the population reduction and to the outcrossing of sexual reproduction within the population. For genetic differentiation among populations, it is generally considered to be caused by restricted gene flow (Loveless and Hamrick, 1984; Slatkin, 1985). The results of this study showed, that the level of gene flow among the population is generally low (Nm = 0.591 < 1). This reduced gene flow may be related to the limited reproductive characteristics of the plant itself.
During the performed field work, it was observed that the sexual propagation strategy of Aristolochia delavayi relies on the participation of pollinators. The main pollinators are some small flies belonging to the families Ceratopogonidae and Chironomidae, which basically have weak flying abilities and low pollination efficiency. The seed dispersal only depends on gravity with a limited dispersal distance, and the seeds are prone to dry under this climatic circumstance. In addition, the active geological structure and erosion in the area caused major rivers in the region which formed valleys with a steep and rugged topographies. This became an obstacle for gene exchange among most plant species in the region as reported previously (Yue et al., 2012; Zhang et al., 2015; Tang et al., 2020). At the same time, the fragmentation of habitat caused by human activities became another obstacle for the gene flow of the studied plant species. Presumably, the restricted pollen and seed dispersal mechanisms of A. delavayi and its relative geographical isolation may be the main reasons for the observed genetic differentiation among populations of this species. Though the limited number of populations, the present outcrossing reproductive strategy may enable sufficient gene exchange among individuals, which maintain a high genetic diversity within the population.
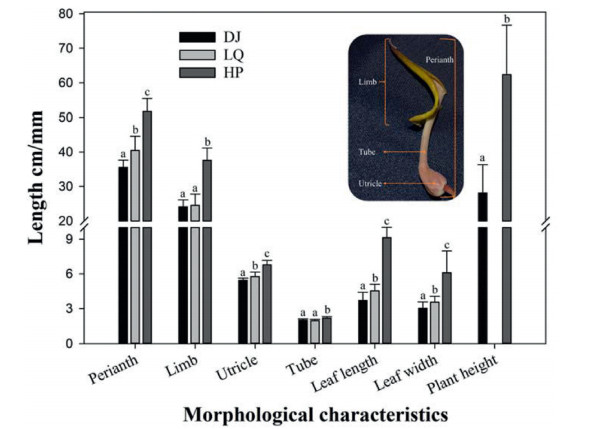
The performed Mantel test detected isolation by distance (IBD) in the population structure of the studied species, which indicated that geographic barriers played an important role in the formation of the present genetic structure of populations, but there may be potential biases in further cluster analysis (Perez et al., 2018). Therefore, we have performed principal coordinate analysis. In this study, the results of principal coordinate analysis and UPGMA clustering are consistent, showing a clear geographic regionality. The ten populations of Aristolochia delavayi are assembled in three groups, which are clustered in Northwest Yunnan, Central Yunnan, and Huangping areas, respectively. Bayes cluster analysis further refined the above grouping results. The genetic clustering results of the three methods all disclosed the separation of the Huangping population. This indicates a distant genetic relationship with the other studied populations. The geographical distribution of this population is rather isolated, far away from the Jinsha River and its tributaries. Compared to other populations, there were no obvious differences in the main taxonomic characteristics (flower and leaf) of A. delavayi in Huangping population. But the different plant size (Fig. 6), leaf scent (Yu et al., 2020), and diverged genetic data suggest that this population could be a subspecies or variety of A. delavayi. But further efforts are required to validate this.
|
| Fig. 6 Comparison of flower and leaf morphological characteristics of Aristolochia delavayi from different populations. The two populations of DJ (Daju) and LQ (Luquan) are the farthest apart in geographical distribution, HP (Huangping) population is far away from the Jinsha River, a main distribution area of A. delavayi. Except for the leaf length, width and plant height in cm, the other features are in mm. Sample sizes of each population: flowers (n = 32) and leaves (n = 90). a, b and c represent the significant difference (P < 0.05). The plant height data of LQ is lacking because of the plants have been destroyed before measurement. |
Aristolochia delavayi is an important species in the traditional medicinal and as spice, and its essential oil is essential for the development and application of antibacterial drugs (Li et al., 2013). At the same time, this species is an important host of the endangered butterfly Byasa daemonius. To sustain this plant species may effectively promote the gradual recovery of the B. daemonius, and may also avoid cascading effects (Koh, 2004; Brodie et al., 2014; Chen et al., 2015). Over the past few decades, the habitat destruction and overharvesting by human activities have led to a rapid decline in the population sizes. At present, only ten isolated populations of A. delavayi were found and most of them comprise of only few individuals. Therefore, the wild resource of the plant species needs effective protection urgently.
Because of the rarity of wild populations of Aristolochia delavayi and its significant genetic differentiation among the populations, it is necessary to protect the present populations. In particular, the two populations (YS and LQ), which have recently experienced genetic bottlenecks, would need special attention. For example, intensify the implementation of in situ protection may maintain the existing population size, and improve their population adaptability.
In view of the uniqueness of Huangping population from other populations, it is recommended to perform in situ conservation measures to sustain the current genetic resources for future research and utilization. For the Dadong population, due to the limited number of individuals found within this population, it is recommended to conduct first a thorough survey in order to assess the size of the existing population. Afterwards propagation of these plants, ex situ conservation should be implemented. Additionally, re-introduction of these cultivated individuals should be performed in order to increase the number of individuals within this population. If ex situ conservation measures are implemented, the pollination and seed dispersal processes should be taken into account, otherwise limited pollinators and seed dispersers may impact the reproduction systems negatively (Tang et al., 2019).
According to the genetic differences of the populations of Aristolochia delavayi in different regions, it is recommended to divide them into three protection units. Due to the low gene flow among populations, measures such as artificial pollination and seed dispersal may be taken into account. This would ensure and, as a consequence, increase the gene flow between populations. These measures should be carried out within the same protection unit in order to avoid outbreeding depression (Tallmon et al., 2004; Barmenttlo et al., 2018).
Remarkably, a large number of Aristolochia zhongdianensis J.S. Ma individuals are mixed with A. delavayi in the population SB. Our morphological data indicated that some suspected hybrids occurred in this population (unpublished data). Previous studies exhibited, that in two species showing hybridization phenomenon, gene introgression tends to dilute genetic variation within a small population (Levin et al., 1996; Wolf et al., 2001). Therefore, further research is needed to reveal possible gene introgression between A. delavayi and A. zhongdianensis in the population SB. This would clarify the direction of gene flow, and the trends of genetic variation of the population. Such studies may also provide a scientific guidance for reasonable and successful protection.
Author contributionsYu-Long Yu and Gao Chen collected plant materials, and Zhi-Xiang Yu assisted in the field population survey. Yu-Long Yu independently performed the experiments, analyzed the data and wrote the manuscript. Hui-Chun Wang and Johann Schinnerl provided assistance in the language modification and polish of the manuscript. Rong Tang provided useful suggestions for the writing of the article. Gao Chen and Yu-Peng Geng designed and supervised the study, and also revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interestThe authors declare that there are no conflicts of interest.
AcknowledgmentsWe thank Chencan Liao, Maoyao Yan and Qi Yu for their assistance in collecting samples. We appreciate Zhi Chen for picture optimization. Support for this study was provided by grants from the NSFC (National Natural Science Foundation of China)-Yunnan Joint fund to support key projects (No. U1602264) and Yunnan Ten Thousand Talents Plan Young & Elite Talent Project to G. Chen (YNWR-QNBJ-2018-017); and NSFC (National Natural Science Foundation of China) (No. 31660057) to Y.P. Geng; Science & Technology Basic Resources Investigation Program of China for Survey and Germplasm Conservation of PSESP in Southwest China (2017FY100100) to W.B. Sun.
Balloux F., Lugone-Moulin N., 2002. The estimation of population differentiation with microsatellite markers. Mol. Ecol, 11: 155-165. DOI:10.1046/j.0962-1083.2001.01436.x |
Barmenttlo S.H., Meirmans P.G., Luijten S.H., et al, 2018. Outbreeding depression and breeding system evolution in small, remnant populations of Primula vulgaris: consequences for genetic rescue. Conserv. Genet, 19: 545-554. DOI:10.1007/s10592-017-1031-x |
Brodie J.F., Aslan C.E., Roders H.S., et al, 2014. Secondary extinctions of biodiversity. Trends Ecol. Evol, 29: 664-672. DOI:10.1016/j.tree.2014.09.012 |
Chen G., Ge J., Qin Y., et al, 2018. Research on the volatiles from leaves and roots of Aristolochia delavayi Franch. and their potential safety hazards. China Condiment, 43: 22-27. |
Chen G., Luo S.H., Mei N.S., et al, 2015. Case study of building of conservation coalitions to conserve ecological interactions. Conserv. Biol, 29: 1527-1536. DOI:10.1111/cobi.12583 |
Chen F.J., Wang A.L., Chen K.M., et al, 2009. Genetic diversity and population structure of the endangered and medically important Rheum tanguticum (Polygonaceae) revealed by SSR markers. Biochem. Syst. Ecol, 37: 613-621. DOI:10.1016/j.bse.2009.08.004 |
Doyle J.J., Doyle J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19: 11-15. |
Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol, 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Excoffier L., Laval G., Schneider S., 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinf. Online, 1: 47-50. |
Guan M.M., Ma R., Gong X., 2013. Conservation genetics of an endemic plant, Anemoclema glaucifolium, in the Jinsha River valley. Plant Divers. Resour, 35: 555-562. |
Habel J.C., Schmitt T., Meyer M., et al, 2010. Biogeography meets conservation: the genetic structure of the endangered lycaenid butterfly Lycaena helle (Denis Schiffermüller 1775). Biol. J. Linn. Soc, 101: 155-168. DOI:10.1111/j.1095-8312.2010.01471.x |
He W.J., Li Z.J., Zhang H.X., et al, 2017. Study on chemical components in the volatile oils obtained from aseptic seeding and callus of Aristolochia delavayi. Flavour Fragr. Cosmet, 45: 5-8. |
Jia J., Wu H., Wang J.F., et al, 2014. Genetic diversity and structure of Munronia delavayi Franch. (Meliaceae), an endemic species in the dry-hot valley of Jinsha River, south-western China. Genet. Resour. Crop Evol, 61: 1381-1395. DOI:10.1007/s10722-014-0120-7 |
Jia J., Zeng L.Q., Gong X., 2016. High genetic diversity and population differentiation in the critically endangered plant species Trailliaedoxa gracilis (Rubiaceae). Plant Mol. Biol. Rep, 34: 327-338. DOI:10.1007/s11105-015-0924-4 |
Jarne P., Lagoda P.J.L., 1996. Microsatellites, from molecules to populations and back. Trends Ecol. Evol, 11: 424-429. DOI:10.1016/0169-5347(96)10049-5 |
Koh L.P., 2004. Species coextinctions and the biodiversity crisis. Science, 305: 1632-1634. DOI:10.1126/science.1101101 |
Laikre L., Allendorf F.W., Aroner L.C., et al, 2010. Neglect of genetic diversity in implementation of the convention on biological diversity. Conserv. Biol, 24: 86-88. DOI:10.1111/j.1523-1739.2009.01425.x |
Levin D.A., FranciscoOrtega J., Jansen R.K., 1996. Hybridization and the extinction of rare plant species. Conserv. Biol, 10: 10-16. DOI:10.1046/j.1523-1739.1996.10010010.x |
Li Z.J., Njateng G.S.S., He W.J., et al, 2013. Chemical composition and antimicrobial activity of the essential oil from the edible aromatic plant Aristolochia delavayi. Chem. Biodivers, 10: 2032-2041. DOI:10.1002/cbdv.201300066 |
Luan S.S., Chiang T.Y., Gong X., 2006. High genetic diversity vs. low genetic differentiation in Nouelia insignis (Asteraceae), a narrowly distributed and endemic species in China, revealed by ISSR fingerprinting. Ann. Bot, 98: 583-589. DOI:10.1093/aob/mcl129 |
Loveless M.D., Hamrick J.L., 1984. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst, 15: 65-95. DOI:10.1146/annurev.es.15.110184.000433 |
Ma H., Wang D.X., Li T.Q., et al, 2019. Phylogeographic study of Musella lasiocarpa(Musaceae): providing insight into the historical river capture events. Pakistan J. Bot, 51: 191-199. |
Ma Y.P., Chen G., Grumbine R.E., et al, 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers. Conserv, 22: 803-809. DOI:10.1007/s10531-013-0434-3 |
Mantel N., 1967. The detection of disease clustering and a generalized regression approach. Canc. Res, 27: 209-220. |
Meglecz E., Costedoat C., Dubut V., et al, 2010. QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformation, 26: 403-404. DOI:10.1093/bioinformatics/btp670 |
Nybom H., Bartish L.V., 2000. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect.Plant Ecol. Evol. Syst, 3: 93-114. DOI:10.1078/1433-8319-00006 |
Peakall R., Smouse P.E., 2006. GENALEX 6:genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes, 6: 288-295. DOI:10.1111/j.1471-8286.2005.01155.x |
Perez M.F., Franco F.F., Bombonato J.R., et al, 2018. Assessing population structure in the face of isolation by distance: are we neglecting the problem?. Divers. Distrib, 24: 1883-1889. DOI:10.1111/ddi.12816 |
Pritchard J.K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1093/genetics/155.2.945 |
Prober S.M., Brown A.H.D., 1994. Conservation of the grassy white box woodlands: population genetics and fragmentation of Eucalyptus albens. Conserv. Biol, 8: 1003-1013. DOI:10.1046/j.1523-1739.1994.08041003.x |
Qin H.N., Yang Y., Dong S.Y., et al, 2017. Threatened species list of China's Higher Plants. Biodivers. Sci, 25: 696-744. DOI:10.17520/biods.2017144 |
Rousset F., 2008. Genepop' 007:a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour, 8: 103-106. DOI:10.1111/j.1471-8286.2007.01931.x |
Schaal B.A., Hayworth D.A., Olsen K.M., et al, 1998. Phylogeographic studies in plants: problems and prospects. Mol. Ecol, 7: 465-474. DOI:10.1046/j.1365-294x.1998.00318.x |
Slatkin M., 1985. Gene flow in natural population. Annu. Rev. Ecol. Systemat, 16: 393-430. DOI:10.1146/annurev.es.16.110185.002141 |
Smouse P.E., Long J.C., Sokal R.R., 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool, 35: 627-632. DOI:10.2307/2413122 |
Sun J.M., Qin J.L., Li L.J., et al, 2008. Resources status and protection tactics of Aristolochia delavayi, an endangered and endemic species in China. China New Technolog. Prod, 13: 130. |
Tallmon D.A., Luikart G., Waples R.S., 2004. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol, 19: 489-496. DOI:10.1016/j.tree.2004.07.003 |
Tang S.Q., Dai W.J., Li M.S., et al, 2008. Genetic diversity of relictual and endangered plant Abies ziyuanensis (Pinaceae) revealed by AFLP and SSR markers. Genetica, 133: 21-30. DOI:10.1007/s10709-007-9178-x |
Tang R., Li Y., Xu Y.L., et al, 2019. In situ and ex situ pollination biology of the four threatened plant species and the significance for conservation. Biodivers.Conserv, 29: 381-391. DOI:10.1007/s10531-019-01887-5 |
Tang R., Liu E.X., Zhang Y.Z., et al, 2020. Genetic diversity and population structure of Amorphophallus albus, a plant species with extremely small populations(PSESP) endemic to dry-hot valley of Jinsha River. BMC Genet, 21: 102. |
Wang L., Guo J., Zhao G.F., 2006. Genetic diversity of the endangered and endemic species Psathyrostachys huashanica natural populations using simple sequence repeats (SSRs) markers. Biochem. Systemat. Ecol, 34: 310-318. DOI:10.1016/j.bse.2005.09.009 |
Wang D.Y., Peng J., Chen Y.J., et al, 2013. Genetic diversity and genetic structure of the rare and endangered species, Primula ranunculoides. Biodivers. Sci, 21: 601-609. |
Wang Z.G., Wu J.P., Liu C.S., et al, 2010. Bottleneck effect analysis of Chinese goat breeds using microsatellites. Chin. J. Anim. Vet. Sci, 41: 664-670. |
Wolf D.E., Takebayashi N., Rieseberg L.H., 2001. Predicting the risk of extinction through hybridization. Conserv. Biol, 15: 1039-1053. DOI:10.1046/j.1523-1739.2001.0150041039.x |
Wright, S., 1978. Evolution and the genetics of populations. In: Variability within and Among Natural Populations, ume 4. University of Chicago Press.
|
Xiao S.Y., Ji Y.H., Liu J., et al, 2020. Genetic characterization of the entire range of Cycas panzhihuaensis (Cycadaceae). Plant Divers, 42: 7-18. DOI:10.1016/j.pld.2019.10.001 |
Yang X., Yang Z., Li H., 2018. Genetic diversity, population genetic structure and protection strategies for Houpo#235;a officinalis (Magnoliaceae) an endangered Chinese medical plant. J. Plant Biol, 61: 159-168. DOI:10.1007/s12374-017-0373-8 |
Yang Z.Y., Yi T.S., Zeng L.Q., et al, 2014. The population genetic structure and diversification of Aristolochia delavayi (Aristolochiaceae), an endangered species of the dry hot valleys of the Jinsha River, southwestern China. Botany, 92: 579-587. DOI:10.1139/cjb-2013-0267 |
Young A., Boyle T., Brown T., 1996. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol, 11: 413-418. DOI:10.1016/0169-5347(96)10045-8 |
Yu Y.L., Geng Y.P., Chang N., et al, 2020. Variation of volatile compound composition of leaves from different Aristolochia delavayi Franch. populations and its potential value. Guihaia, 40: 1251-1258. |
Yue L.L., Chen G., Sun W.B., et al, 2012. Phylogeography of Buddleja crispa (Buddlejaceae) and its correlation with drainage system evolution in southwestern China. Am. J. Bot, 99: 1726-1735. DOI:10.3732/ajb.1100506 |
Zhang X., Chen G., Ma Y.P., et al, 2015. Genetic diversity and population structure of Buddleja crispa Bentham in the Himalaya-Hengduan Mountains region revealed by AFLP. Biochem. Systemat. Ecol, 58: 13-20. DOI:10.1016/j.bse.2014.10.015 |
Zhang Z.C., Hou X.L., 2004. Strategies for development of SSR molecular markers. Hereditas, 26: 763-768. |
Zhang X., Zhang L., Schinnerl J., et al, 2019. Genetic diversity and population structure of Hibiscus aridicola, an endangered ornamental species in dry-hot valleys of Jinsha River. Plant Divers, 41: 300-306. DOI:10.1016/j.pld.2019.07.001 |
Zhao Y.J., Gong X., 2015. Diversity and conservation of plant species in dry valleys, southwest China. Biodivers. Conserv, 24: 2611-2623. DOI:10.1007/s10531-015-0952-2 |
Zhong X.H., 2000. Degradation of ecosystem and ways of ITS rehabilitation and reconstruction in dry and hot valley-take representative area of Jinsha River, Yunnan province as an example. Resour. Environ. Yangtze Basin, 9: 376-383. |
Zhou T.S., Yang Q.K., 1995. Research on extraction of essential oil from Aristolochia delavayi in Yunnan. Flavour Fragr. Cosmet, 23: 10-12. |
Zhou T.S., Yang Q.K., Zhang Z.J., et al, 1995. Studies on the chemical constituents and aroma of the essential oil from Aristolochia delavayi of Yunnan. Flavour Fragr. Cosmet, 23: 13-17. |



