The Atraphaxideae is a woody tribe in the family Polygonaceae, which initially included three genera, Calligonum L., Atraphaxis L., and Pteropyrum Jaub. & Spach. These genera are shrubs or small trees, widely distributed in arid and semi-arid regions in the interior of Eurasia (Bao and Li, 1993; Li 1998). A new shrub genus, Parapteropyrum A.J. Li, has since been described and added to this tribe (Li, 1981). This is the only genus of tribe Atraphaxideae that contains an endangered species, Parapteropyrum tibeticum A.J. Li, which occurs in the dry and hot valley along Yarlung Zangbo in southeastern part of the Qinghai-Tibet Plateau. Morphological (Ronse De Craene and Akeroyd, 2008) and pollen traits (Hong, 1995) support the placement of Parapteropyrum in the Atraphaxideae. However, although the chromosome base number of Parapteropyrum and Fagopyrum Mill. are the same, Parapteropyrum is a polyploid (Tian et al., 2009). Phylogenetic analyses based on sequences of the nuclear ITS and two or three chloroplast genome (plastome) genes sequences have similarly suggested thatParapteropyrum is more closely related to some herbal Fagopyrum species (the tribe Polygoneae) than to the woody genera of the Atraphaxideae (Sanchez et al., 2009; Tavakkoli et al., 2010). Comprehensive species sampling indicated that P. tibeticum nests within the buckwheat genus (Tian et al., 2011; Sun et al., 2014). However, these inferences have relatively low statistical support, mainly because of the low number of phylogenetically informative sites in short DNA fragments, and require further confirmation.
Plastomes contain 110-130 genes with a relatively conserved genome structure (Jansen and Ruhlman, 2012). Furthermore, plastomes lack paralogues and gene recombination is extremely rare. Therefore, whole plastomes provide sufficient informative variation for high resolution phylogenetic analyses with strong support (Raubeson and Jansen, 2005; Yang et al., 2019). In most previous plastome studies, whole-genome Illumina reads from a second-generation sequencer are used to assemble unknown plastomes of targeted species through a randomly chosen reference plastome of a closely related species (Carbonell-Caballero et al., 2015; Dierckxsens et al., 2017; Guo et al., 2017; Zhang et al., 2018b, Zhang et al., 2018a; Yang et al., 2018). Such a reference plastome is crucial for assembling and improving the plastome sequence of the targeted species based on different algorithms (Dierckxsens et al., 2017). The conserved nature of plastome structure allows for convenient selection of a reference plastome, including plastome sequences from relatively distantly related species (Dierckxsens et al., 2017). However, it remains unknown whether the choice of reference plastome affects the assembly of target species plastomes. If plastome sequences vary in response to the choice of reference plastome, does this variation affect plastome-based phylogenetic reconstructions?
Polygonaceae plastomes have been reported for several genera (Fagopyrum, Rheum L., Rumex L., Oxyria Hill and Muehlenbeckia Meisn.). However, no studies have reported plastome data for the genera of the Atraphaxideae. In this study, we used plastome data to analyze the phylogeny of family Polygonaceae with the specific aim of determining the phylogenetic position of Parapteropyrum and whether the woody genera of the Atraphaxideae are polyphyletic, as suggested previously (Tian et al., 2009, 2011; Sanchez et al., 2009; Tavakkoli et al., 2010; Sun et al., 2014). For this purpose, we first assembled plastomes for the monotypic Parapteropyrum and two additional genera of the tribe Atraphaxideae. To determine whether the choice of reference plastome affects plastome assembly and phylogenetic analyses, we assembled new plastomes using reference plastomes from different species. We also summarized plastome characters based on these available plastomes.
2. Material and methods 2.1. DNA extraction and complete chloroplast genome assemblyWe sampled three species from the Atraphaxideae in our study including Calligonum ebinuricum N.A. Ivanova ex Soskov (sampled from the Turpan Eremophyte Botanical Garden, Chinese Academy of Sciences, Xinjiang, China), Atraphaxis bracteata Losinsk. (sampled from Minqin Desert Botanical Garden, Gansu, China) and P. tibeticum (sampled from Shannan, Xizang, China, 93.1728°E, 29.0128°N). We used a modified CTAB method (Doyle and Doyle, 1987) to extract the total DNA of the silica-dried leaves for each species. We constructed Illumina paired-end libraries with an insert size of 500 base pairs (bp) and sequenced these libraries through the HiSeq X Ten System. For each sample, we generated four gigabytes (Gb) of 2 × 150 bp short read data. We removed those reads with a Phred quality score < 7 and 5% ambiguous nucleotides. These clear reads were used to assemble the de novo plastome of targeted species by using NOVOplasty v.3.7.2 (Dierckxsens et al., 2017): mapping the whole-genome sequencing reads to the reference plastome, extracting the mapped organelle reads and connecting them together. To examine whether different reference plastomes affect the final assembly quality, we used plastomes of two distantly related species, Fagopyrum luojishanense J.R. Shao (Wang et al., 2017) and Rheum palmatum L. (Fan et al., 2016) (GenBank accession numbers are NC037706 and KR816224 respectively.) as references to assemble plastomes of P. tibeticum and A. bracteata. For C. ebinuricum, we only used the plastome of Rumex acetosa L. as the reference. The BWA v.0.7.12 (Li and Durbin, 2009) and SAMtools v.1.3.1 (Li et al., 2009) were used to build an index and map all plastome sequences to the reference plastome via the mem algorithm and convert and sort the output files, respectively. Geneious v.8.1.4 (Kearse et al., 2012) was also used to compare and adjust the assembled plastome sequences manually. We annotated plastomes with Plann v.1.0 (Huang and Cronk, 2015) and checked the quality with Sequin v.15.10 (Clark et al., 2016). We drew the plastome gene map through OGDRAW (Lohse et al., 2007). We used MAFFT v.7.221 (Katoh and Standley, 2013) and MEGA7 (Kumar et al., 2016) to examine whether using different plastome references to assemble plastomes of the same species alters sequence variations.
The whole genome sequence data of C. ebinuricum, A. bracteata and P. tibeticum reported in this paper have been deposited in the Genome Warehouse in National Genomics Data Center (Zhang et al., 2020), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number GWHAOPN01000000, GWHAOPP01000000 and GWHAOPO01000000, respectively. They are publicly accessible at https://bigd.big.ac.cn/gwh.
2.2. Comparative analyses of plastomes in the family PolygonaceaeWe chose plastomes of Atraphaxis bracteata and Parapteropyrum tibeticum (assembled based on the reference plastome of Fagopyrum luojishanense) and downloaded nine plastomes of the representative genera and species in the Polygonaceae from the National Center for Biotechnology Information (NCBI) for comparative analyses (Table 1). These downloaded species comprised Fagopyrum tataricum (L.) Gaertn (Liu et al., 2016), F. luojishanense (Wang et al., 2017), Fagopyrum dibotrys (D. Don) H. Hara (Wang et al., 2017), Fagopyrum esculentum Moench (Logacheva et al., 2008), Muehlenbeckia australis (G. Forst.) Meisn (Schuster et al., 2018), Oxyria sinensis Hemsl (Luo et al., 2017), Rumex japonicus Houtt (Gurusamy et al., 2020), R. acetosa L. (Gui et al., 2018) and R. palmatum L. (Fan et al., 2016). A total of 12 plastomes were used for final analyses.
| Species | Genome Size (bp) | GC (%) | LSC (bp) | SSC (bp) | IR (bp) | Gene | CDS | tRNA | rRNA | Pseudogenes | Data sources | GenBank or NGDC accession number |
| Parapteropyrum tibeticum | 159, 968 | 37.72 | 84, 855 | 13, 497 | 30, 808 | 130 | 83 | 37 | 8 | 2 | This study | GWHAOPO01000000 |
| Calligonum ebinuricum | 164, 270 | 37.44 | 89, 028 | 13, 512 | 30, 865 | 130 | 80 | 37 | 8 | 5 | This study | GWHAOPN01000000 |
| Atraphaxis bracteata | 164, 264 | 37.43 | 88, 854 | 13, 620 | 30, 895 | 130 | 82 | 37 | 8 | 3 | This study | GWHAOPP01000000 |
| Fagopyrum tataricum | 159, 272 | 37.88 | 84, 397 | 13, 241 | 30, 817 | 131 | 86 | 37 | 8 | – | NCBI | KX085498 |
| Fagopyrum luojishanense | 159, 265 | 37.84 | 84, 431 | 13, 094 | 30, 870 | 130 | 85 | 37 | 8 | – | NCBI | NC037706 |
| Fagopyrum dibotrys | 159, 320 | 37.93 | 84, 422 | 13, 264 | 30, 817 | 129 | 84 | 37 | 8 | – | NCBI | KY275181 |
| Fagopyrum esculentum | 159, 599 | 37.98 | 84, 888 | 13, 343 | 30, 684 | 130 | 83 | 37 | 8 | 2 | NCBI | EU254477 |
| Muehlenbeckia australis | 163, 484 | 37.44 | 88, 166 | 13, 486 | 30, 916 | 131 | 83 | 37 | 8 | 3 | NCBI | MG604297 |
| Oxyria sinensis | 160, 404 | 37.54 | 85, 501 | 13, 133 | 30, 885 | 131 | 83 | 37 | 8 | 3 | NCBI | NC032031 |
| Rumex japonicus | 159, 292 | 37.50 | 85, 028 | 13, 006 | 30, 629 | 129 | 84 | 37 | 8 | – | NCBI | MN720269 |
| Rumex acetosa | 160, 269 | 37.20 | 86, 135 | 13, 128 | 30, 503 | 129 | 83 | 36 | 8 | 2 | NCBI | NC042390 |
| Rheum palmatum | 161, 541 | 37.32 | 86, 518 | 13, 111 | 30, 956 | 131 | 84 | 37 | 8 | 2 | NCBI | KR816224 |
| Note: NGDC, Genome Warehouse in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences. | ||||||||||||
We compared plastome size, GC content, the length of four areas in plastomes, and the number of genes (CDS) and pseudogenes using Python Script. We then compared the boundaries of the Large single copy region (LSC), Small single copy region (SSC) and Inverted repeats (IR, contains IRA and IRB) regions in these species. We used mVISTA software (Frazer et al., 2004) for the genome-wide comparison analysis with Fagopyrum luojishanense as reference.
2.3. SSR search and comparisonWe used Krait software (Du et al., 2018) to search for SSRs in plastomes. We set the parameter for minimum repeat numbers to 10, 5, 4, 3, 3, 3 for six different repeats: mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide and hexanucleotide respectively (Zhao et al., 2019). We also set up reverse SSRs that can be combined with their complementary SSRs (i.e., GTA is a reverse motif of ATG).
2.4. Phylogenetic analysesWe downloaded the plastome sequence of Limonium aureum (L.) Hill (Liedo et al., 1998) from NCBI as outgroup. We used the total plastome sequences as the first data set and extracted all gene sequences as the second data set. The third data set comprised only LSC and SSC sequences. We used jModelTest v.2.1.4 (Posada, 2008) to search for the optimal model for Bayesian analyses, and found that GTR+I+R was the best model for all three data sets. Bayesian analyses were conducted in MrBayes v.3.12 (Huelsenbeck et al., 2001). We set up four Monte Carlo Markov Chain (MCMC), ran 1 million generations and saved a tree every 100 generations. We finally set up a 20% burn fraction and synthesized the consistent trees with the remaining samples. We also used RAxML v.8.2.9 (Stamatakis, 2014) to build trees based on each of three data sets, respectively, using the most suitable GTRGAMMA model and setting 1000 bootstrap tests.
3. Result 3.1. Plastome assembly based on different referencesAfter reference-guided assembly, we obtained three plastome sequences without gaps. The assembly information of the three newly sequenced plastomes is listed in Table 1. These plastomes all have the typical quadripartite structure with lengths ranging from 159, 968 bp to 164, 270 bp (Fig. 1). We used plastomes of F. luojishanense and R. palmatum as two different references to assemble plastomes of A. bracteata and P. tibeticum. F. luojishanense and R. palmatum are distantly related, which allowed us to identify any negative effects caused by reference selection. We found that the coding genes of A. bracteata and P. tibeticum plastomes assembled using two different references are largely the same. Also, the whole plastome length showed no difference between these two annotated plastomes when assembled using different references. Small differences between plastomes were mainly found in the IR and SSC regions, which may derive from the different boundary determinations between these two regions. In addition, some CDS genes may have been annotated as pseudogenes or vice versa (Table 2). For example, when we used F. luojishanense as a reference plastome, the IR and SSC regions of the assembled P. tibeticum plastome were 30, 808 bp and 13, 497 bp, respectively, and the number of the CDS genes and pseudogenes were 83 and 2. When we used R. palmatum as the reference plastome, the IR regions of the assembled P. tibeticum plastome was slightly shorter (28, 579 bp), the SSC region was longer (17, 955 bp), there were two fewer CDS genes (81) and two more pseudogenes (4). When we aligned these two P. tibeticum plastomes, they showed no nucleotide variation or loss. Therefore, the effects of using different reference plastomes appears to be limited to plastome annotation. Similarly, when we used different reference plastomes to assemble two annotated A. bracteata plastomes, the IR and SSC regions varied slightly. The two assembled A. bracteata plastomes showed no alignment differences.
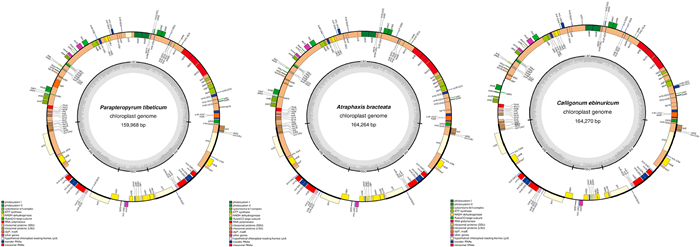
|
| Fig. 1 Gene maps of three plastomes in Parapteropyrum tibeticum (assembled using Fagopyrum luojishanense as reference plastome), Atraphaxis bracteata (assembled using Fagopyrum luojishanense as reference plastome) and Calligonum ebinuricum (assembled using Rumex acetosa as reference plastome). Genes outside the circle are transcribed clockwise; genes inside are transcribed counterclockwise. Different colors represent different functional groups. The inner circle shows GC content in dashed grey area. LSC, large single copy; IRB, inverted repeat B; SSC small single copy; and IRA, inverted repeat A. |
| Species | Genome Size (bp) | GC (%) | LSC (bp) | SSC (bp) | IR (bp) | Gene | CDS | tRNA | rRNA | Pseudogenes | Reference plastomes in tde GenBank |
| Parapteropyrum tibeticum | 159, 968 | 37.72 | 84, 855 | 13, 497 | 30, 808 | 130 | 83 | 37 | 8 | 2 | NC037706 |
| 159, 968 | 37.72 | 84, 855 | 17, 955 | 28, 579 | 130 | 81 | 37 | 8 | 4 | KR816224 | |
| Atraphaxis bracteata | 164, 264 | 37.43 | 88, 854 | 33, 828 | 20, 791 | 130 | 81 | 37 | 8 | 4 | KR816224 |
| 164, 264 | 37.43 | 88, 854 | 13, 620 | 30, 895 | 130 | 82 | 37 | 8 | 3 | NC037706 |
In contrast to plastomes that were assembled using R. palmatum as a reference plastome, the two plastomes assembled using F. luojishanense as a reference plastome had no alignment differences and fewer pseudogenes. More importantly, although all plastomes from Polygonaceae have IR regions > 30, 000 bp (Fan et al., 2016; Wang et al., 2017; Gui et al., 2018), this regions became shorter in the assembled plastomes that used R. palmatum as a reference. Because the IR region is generally conserved within a family, we choose to use plastomes assembled with F. luojishanense as a reference for further analyses.
3.2. Basic characteristics of plastomes of PolygonaceaeThe plastome lengths of 12 species of Polygonaceae range from 159, 265 bp to 164, 270 bp. The F. luojishanense plastome is the shortest, whereas the C. ebinuricum plastome, which is 5005 bp longer, is the longest. Polygonaceae plastome GC content ranges from 37.20% to 37.98%, with the smallest GC content in R. acetosa and the largest in F. esculentum (Table 1). All Polygonaceae plastomes contain four regions (i.e., LSC, SSC and two IR regions), with four boundaries (i.e., LSC/IRB, IRB/SSC, SSC/IRA, IRA/LSC) (Fig. 2). The LSC regions range from 84, 422 bp to 89, 028 bp with the smallest forF. dibotrys and the largest for C. ebinuricum with a difference of 4606 bp. The lengths of the SSC regions range from 13, 006 bp to 13, 620 bp, with the smallest in R. japonicus and the largest in A. bracteata, with a difference of 614 bp. The length of IR regions ranges from 30, 503 bp to 30, 956 bp, with the smallest in R. acetosa and the largest in R. palmatum, with a difference of 453 bp. The total number of the annotated genes in the 12 Polygonaceae plastomes examined ranges from 129 to 131, and the number of coding CDS genes from 80 to 86. All Polygonaceae plastomes have eight rRNA genes, while having either 36 or 37 tRNA genes.
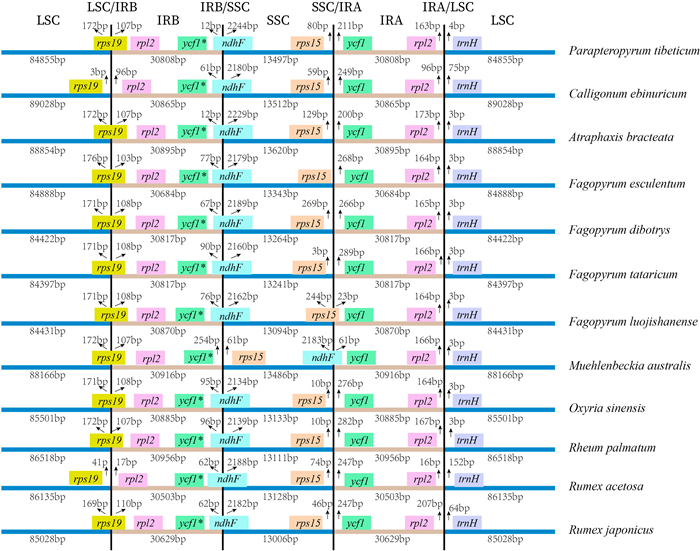
|
| Fig. 2 Comparison of the boundaries of LSC, SSC, IR regions in plastomes of 12 Polygonaceae species. |
Plastome sequences of all 12 Polygonaceae species are highly similar (Fig. 3). Most variations were confined to intergenic spacer sequences, indicating that the available plastomes across Polygonaceae are relatively well-conserved. The LSC/IRB boundary in Polygonaceae species is located between the rps19 gene and rpl2 genes. The length of rps19 gene in the IRB region ranges from 103 bp to 110 bp. The boundaries range from 3 bp to 41 bp from rps19 gene or 96 bp to 17 bp from rpl2 gene, respectively. At the IRB/SSC boundary, the length of ndhF gene in the IRB region ranges from 12 bp to 96 bp. The only exception is in the M. australis plastome, in which the SSC sequence is completely inverted, placing the IRB/SSC boundary adjacent to the rps15 gene in the SSC region. The SSC/IRA boundary of 10 Polygonaceae species is located between the rps15 gene and ycf1 gene, ranging from 0 bp to 269 bp after rps15 gene or from 23 bp to 289 bp before the ycf1 gene. For M. australis, which has an inverted SSC region, the SSC/IRA boundary occurs within ndhF gene, which enters the IRA region. The SSC/IRA boundary of F. luojishanense is in the rps15 gene. All 12 Polygonaceae species have IRA/LSC boundaries between the rpl2 and trnH genes, ranging from 16 bp to 207 bp after the rpl2 gene and from 3 bp to 152 bp before the trnH gene.
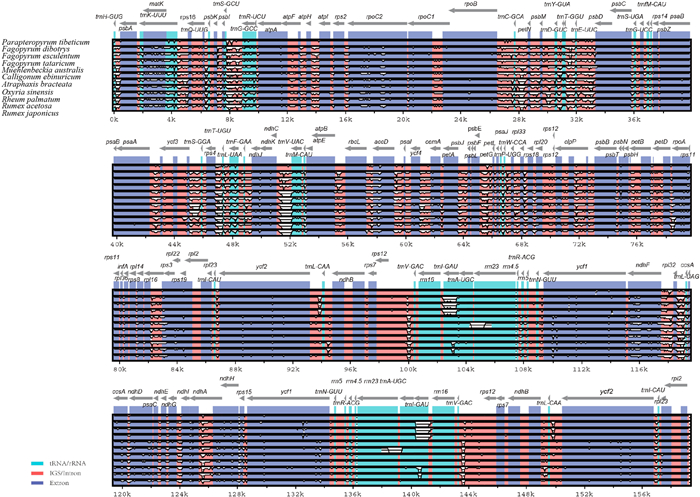
|
| Fig. 3 Plastome alignments of 12 Polygonaceae species using mVISTA. The y-axis represents the range from 50% to 100%. |
The number of SSRs found in 12 plastomes varies greatly from 48 to 77 (Table 3; Fig. 4). The smallest number was found for F. dibotrys and C. ebinuricum while the largest for R. palmatum. The number of the mononucleotide SSRs was the largest in 12 species, ranging from 27 (R. acetosa) to 47 (M. australis). The number of the dinucleotide varies from 9 (C. ebinuricum) to 18 (R. japonicus). Trinucleotide SSR number ranges from 2 (P. tibeticum) to 12 (R. japonicus). The number of the tetranucleotide SSRs varies from 4 (five species) to 9 (R. palmatum). The pentanucleotide SSRs are absent in both F. luojishanense and F. esculentum, whereas there are 3 in R. acetosa. The only plastome that has a hexanucleotide SSR is that of O. sinensis, which has one.
| Species | Mono- | Di- | Tri- | Tetra- | Penta- | Hexa- | Total SSRs | Total lengtd (bp) | Relative abundance (loci/Mb) | Relative density (bp/Mb) | Sequence covered by SSRs (%) |
| Parapteropyrum tibeticum | 30 | 13 | 2 | 4 | 1 | 0 | 50 | 551 | 312.56 | 3, 444.44 | 0.35 |
| Fagopyrum dibotrys | 28 | 11 | 3 | 4 | 2 | 0 | 48 | 552 | 301.28 | 3, 464.73 | 0.35 |
| Fagopyrum esculentum | 33 | 16 | 4 | 4 | 0 | 0 | 57 | 618 | 357.15 | 3, 872.28 | 0.39 |
| Fagopyrum luojishanense | 38 | 15 | 4 | 4 | 0 | 0 | 61 | 653 | 383.01 | 4, 100.08 | 0.42 |
| Fagopyrum tataricum | 30 | 11 | 3 | 4 | 1 | 0 | 49 | 552 | 307.65 | 3, 465.77 | 0.35 |
| Calligonum ebinuricum | 28 | 9 | 5 | 5 | 1 | 0 | 48 | 541 | 292.20 | 3, 293.36 | 0.33 |
| Atraphaxis bracteata | 32 | 11 | 4 | 7 | 1 | 0 | 55 | 638 | 334.83 | 3, 884.02 | 0.39 |
| Muehlenbeckia australis | 47 | 14 | 6 | 5 | 1 | 0 | 73 | 806 | 446.53 | 4, 930.15 | 0.50 |
| Oxyria sinensis | 32 | 11 | 7 | 6 | 2 | 1 | 59 | 666 | 367.82 | 4, 152.02 | 0.42 |
| Rumex japonicus | 35 | 18 | 12 | 7 | 1 | 0 | 73 | 815 | 458.28 | 5, 116.39 | 0.52 |
| Rumex acetosa | 27 | 17 | 10 | 5 | 3 | 0 | 62 | 699 | 386.85 | 4, 361.42 | 0.44 |
| Rheum palmatum | 40 | 17 | 10 | 9 | 1 | 0 | 77 | 898 | 476.66 | 5, 558.96 | 0.56 |
| Note: Mono-, mononucleotide; Di, dinucleotide; Tri-, trinucleotide; Tetra-, tetranucleotide; Penta-, pentanucleotide; and Hexa-, hexanucleotide. | |||||||||||
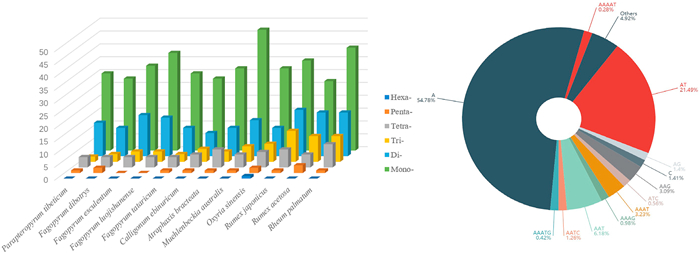
|
| Fig. 4 Distribution of different SSRs types in plastomes across 12 Polygonaceae species (left) and the 12 SSR repeat units that occur most frequently (right). |
Polygonaceae plastomes had 13 classes of high frequency SSR (Fig. 4). Poly(A) and (C) mononucleotide repeats account for 56.19% of the total. Dinucleotides (22.89% of repeats) were the next most abundant repeat with frequent occurrence of repeat motifs AG and AT. Trinucleotides (9.83% of repeats) consisted of repeat motifs AAG, AAT and ATC. Tetranucleotides (5.47% of repeats) included three repeat motifs: AATC, AAAG and AAAT, while pentanucleotides (0.7% of repeats) consisted of two repeat motifs: AAATG and AAAAT. These SSRs, which were identified in single plastomes, should be examined for polymorphisms at the population level.
3.4. Phylogenetic analysesFor phylogenetic analyses of the family Polygonaceae, we first created two plastome data sets: one consisted of whole plastome sequences and the second consisted of plastome gene sequences. We created a third data set by extracting the sequences of LSC and SSC regions. When we assembled the plastomes of P. tibeticum and A. bracteata with different references, the IR regions of both species varied. Therefore, in our phylogenetic analyses both annotated plastomes of each of these two species were used.
Our first phylogenetic analyses used plastome sequences of P. tibeticum and A. bracteata that had been assembled and annotated with F. luojishanense as a reference. The ML and BI trees based on all three data sets had the same topology, with three major clades, each clade and subclade highly supported (bootstrap values and Bayes posterior probabilities) (Fig. 5) Four Fagopyrum species and P. tibeticum clustered into one large clade, with P. tibeticum and F. luojishanense forming a subclade. A. bracteata was sister to C. ebinuricum and together these two species clustered into another large clade with M. australis. A third clade consisted of one subclade comprising two Rumex species and another comprising O. sinensis and R. palmatum. When we used plastomes of P. tibeticum and A. bracteata that had been assembled and annotated with R. palmatum as a reference, tree topology and phylogenetic relationships were unchanged.
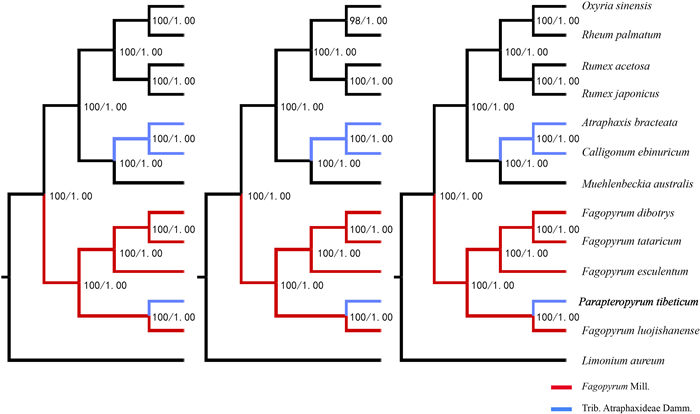
|
| Fig. 5 Phylogenetic trees of 12 Polygonaceae species reconstructed by ML/BI analyses based on the total plastome sequence data sets (left), gene data sets (middle) and LSC-SSC sequence data sets (right). Numbers before slashes represent ML bootstrap values and numbers after slashes are Bayes posterior probabilities. |
Over the past several years, numerous species have been used as references to assemble plastomes of targeted species (Dierckxsens et al., 2017; Guo et al., 2017). Phylogenetic relationships of orders and families have been established based on these plastome sequences (Yang et al., 2018, 2019). In fact, plastome data has been widely used to construct genus-level phylogenies within one family (Parks et al., 2009; Zhang et al., 2018b, Zhang et al., 2018a). However, the quality of plastome data must be examined to assure that assembled and annotated plastomes of the targeted species are not affected by randomly selected reference plastomes.
Our examination of two species using two different references indicated that different reference plastomes might slightly affect plastome annotation of targeted species in two ways. First, when we used different reference plastomes, the sequence at the boundary between IR and SSC regions changed. Second, using different reference plastomes led to the annotation of coding genes as pseudogenes and vice versa. However, we found that two aligned plastomes show no nucleotide variation or loss. Therefore, the reference plastome mainly affects the plastome annotation of the targeted species. For phylogenetic reconstruction, it is better to use whole plastomes, as small annotation variation may distort true phylogenetic relationships when only using coding genes, SSC, or IR regions. In addition, it should be noted that we only used NOVOplasty and Plann v.1.0 for our plastome assembly and annotation of two species. It remains unknown whether other methods may affect such results when different plastomes are selected as the reference and whether such effects vary in different taxonomical groups.
4.2. General plastome characteristics of the family PolygonaceaePlastomes of 12 species from eight genera in the Polygonaceae have generally conserved quadripartite structure, similar length differences, number of genes and GC content. The small change in the plastome length derives mainly from the LSC length variation. Future studies should widely assemble plastomes for various genera of a family, which will be highly useful for phylogenetic constructions. The contraction and expansion of the IR region is common in the plastome evolution (Hansen et al., 2007). However, we found that the IR region of the plastomes in the Polygonaceae is relatively stable, although with small contractions and expansions. The most interesting finding in the plastomes of the Polygonaceae involves the highly varied number of SSRs in the sampled species. For example, the number of SSRs inCalligonum, Parapteropyrum and Atraphaxis (48–55) is smaller than that in other genera (59–77) except Fagopyrum. These SSRs are highly useful in designing primers to examine phylogenetic relationships between closely related species and intraspecific populations. Further studies should focus on the selection of those SSRs with genetic polymorphisms at the population level. The plastome SSRs for the three species first reported here may prove relevant for further population genetic studies, which is critical for the conservation of these endangered species.
4.3. Phylogenetic relationship of the woody Parapteropyrum tibeticumAlthough morphological evidence suggests that the monotypic Parapteropyrum is closely related to the wood genera of the Atraphaxideae (Ronse De Craene and Akeroyd, 2008; Bao and Li, 1993; Hong, 1995; Li et al., 1998), both chromosomal and molecular studies have shown that this species nests within the genus Fagopyrum as an endangered and woody buckwheat (Tian et al., 2009, 2011; Sanchez and Kron, 2009; Tavakkoli et al., 2010; Sun et al., 2014). Our phylogenetic analyses based on plastomes provide robust statistical support for this inference. Therefore, this species should be transferred and placed under Fagopyrum (Tian et al., 2011). After excluding Parapteropyrum, we found that two woody genera of the Atraphaxideae, Calligonum and Atraphaxis, were sister to each other, but together sister to the woody Muehlenbeckia of another tribe. Therefore, these findings suggest that woodiness might have originated multiple times in the Polygonaceae and the tribe Atraphaxideae requires further circumscription based on further evidence.
Tian et al. (2011) proposed that insular woodiness may explain the origin of the woody Parapteropyrum (Fagopyrum) tibeticum from the herbal Fagopyrum. Although this phenomenon is generally used to explain the origin of woody plants on islands, it may also explain the origin of woody plants in the Qinghai-Tibet Plateau. In addition, Parapteropyrum (Fagopyrum) tibeticum is a hexaploid (Tian et al., 2009). Parapteropyrum (Fagopyrum) tibeticum is estimated to have diverged from its sister herbal ancestor within Fagopyrum from 6.35 million to 14.8 million years ago, during the period when the Qinghai-Tibet Plateau experienced extensive uplifts and habitat changes (Harrison et al., 1992; Li et al., 1995; Shi et al., 1998; Spicer et al., 2003). The woody habit of Parapteropyrum (Fagopyrum) tibeticum might have evolved in response to these dramatic habitat changes during polyploidization (Tian et al., 2009, 2011). Further studies are needed to examine how polyploidization and arid habitat selection together shaped such a special trait.
This woody perennial buckwheat is highly relevant as a newly domesticated crop. The cultivation of perennial woody crops saves labor-force for annual planting and harvesting and reduces environmental damage. In addition, woody crops such as Parapteropyrum (Fagopyrum) tibeticum can be developed into horticultural plants for arid regions where it naturally occurs. Artificial planting of this species will restore vegetation coverage in arid regions and provide basic fruits for fruit-eating birds and other animals, which will accelerate the establishment of an aridity tolerance and ecological balance.
Author contributionsBibo Yang performed data analyses and wrote manuscript. Liangda Li performed the experiment and helped with manuscript preparation. Jianquan Liu helped revise the manuscript and conception of the study with constructive discussions. Lushui Zhang contributed to the conception of the study, collected the samples, revised the tables, figures and manuscript.
Declaration of competing interestThe authors declare no conflict of interest.
AcknowledgementsThis work was funded by the National Natural Science Foundation of China (grant No. 31590821).
Bao B.J., Li A.R., 1993. A study of the genus Atraphaxis in China and the system of Atraphaxideae (Polygonaceae). Acta Phytotaxon. Sin, 31: 127-139. |
Carbonell-Caballero J., Alonso R., Iba#241;ez V., et al, 2015. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Mol. Biol. Evol, 32: 2015-2035. DOI:10.1093/molbev/msv082 |
Clark K., Karsch-Mizrachi I., Lipman D.J., et al, 2016. GenBank. Nucleic Acids Res, 44: D67-D72. DOI:10.1093/nar/gkv1276 |
Dierckxsens N., Mardulyn P., Smits G., 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res, 45: e18. DOI:10.1093/nar/gkw1060 |
Doyle J.J., Doyle J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19: 11-15. |
Du L.M., Zhang C., Liu Q., et al, 2018. Krait: an ultrafast tool for genome-wide survey of microsatellites and primer design.. Bioinformatics, 3: 681-683. |
Fan K., Sun X.J., Huang M., et al, 2016. The complete chloroplast genome sequence of the medicinal plant Rheum palmatum L. (Polygonaceae).. Mitochondrial DNA, 27: 2935-2936. DOI:10.3109/19401736.2015.1060448 |
Frazer K.A., Pachter L., Poliakov A., et al, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res, 32: W273-W279. DOI:10.1093/nar/gkh458 |
Gui L.J., Jiang S.F., Wang H.P., et al, 2018. Characterization of the complete chloroplast genome of sorrel (Rumex acetosa). Mitochondrial DNA B Res, 3: 902-904. DOI:10.1080/23802359.2018.1501297 |
Guo X.Y., Liu J.Q., Hao G.Q., et al, 2017. Plastome phylogeny and early diversification of Brassicaceae. BMC Genom, 18: 176. DOI:10.1186/s12864-017-3555-3 |
Gurusamy R., Cho S.J., Park S.J., 2020. The complete plastome sequence of Rumex japonicus Houtt. : a medicinal plant. Mitochondrial DNA B Res, 5: 439-440. DOI:10.1080/23802359.2019.1704194 |
Hansen D.R., Dastidar S.G., Cai Z.Q., et al, 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol. Phylogenet. Evol, 45: 547-563. DOI:10.1016/j.ympev.2007.06.004 |
Harrison T.M., Copeland P., Kidd W.S.F., et al, 1992. Raising tibet.. Science, 255: 1663-1670. DOI:10.1126/science.255.5052.1663 |
Hong S.P., 1995. Pollen morphology of Parapteropyrum and some putatively related genera (Polygonaceae-Atraphaxideae).. Grana, 37: 153-159. |
Huang D.I., Cronk Q.C., 2015. Plann: a command-line application for annotating plastome sequences. Appl. Plant Sci, 38: 1500026. |
Huelsenbeck J.P., Ronquist F., Nielsen R., et al, 2001. Bayesian inference of phylogeny and its impact on evolutionary biology.. Science, 294: 2310-2314. DOI:10.1126/science.1065889 |
Jansen R.K., Ruhlman T.A., 2012. Plastid genomes of seed plants. In: Bock R., Knoop, V. (Eds.), Genomics of Chloroplasts and Mitochondria. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes).. Springer, Dordrecht, pp: 103-126. |
Katoh K., Standley D.M., 2013. MAFFT multiple sequence alignment software version 7:improvements in performance and usability. Mol. Biol. Evol, 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse M., Moir R., Wilson A., et al, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7:molecular evolutionary genetics analysis version 7. 0 for bigger datasets.. Mol. Biol. Evol, 33: msw054. |
Li H., Durbin R., 2009. Fast and accurate short read alignment with BurrowsWheeler transform.. Bioinformatics, 25: 1754-1760. DOI:10.1093/bioinformatics/btp324 |
Li H., Handsaker B., Wysoker A., et al, 2009. The sequence alignment/map format and SAMtools.. Bioinformatics, 25: 2078-2079. DOI:10.1093/bioinformatics/btp352 |
Li A.R., 1981. Parapteropyrum A. J. Li-Unum genus novum Polygonacearum sinicum. Acta Phytotaxon. Sin, 19: 330-332. |
Li A.R., 1998. Flora Republicae Popularis Sinicae, vol. 25. Polygonaceae.. Science Press, Beijing, pp: 120-143: 120-143, 1.
|
Li J.J., Shi Y.F., Li B.Y., 1995. Uplift of the Qinghai-Xizang (Tibet) Plateau and Global Change. Lanzhou University Press, Lanzhou, pp: 1-207. |
Liedo M.D., Crespo M.B., Cameron K.M., et al, 1998. Systematics of Plumbaginaceae based upon cladistic analysis of rbcL sequence data. Syst. Bot, 23: 21-29. DOI:10.2307/2419571 |
Liu M.Y., Zheng T.R., Ma Z.T., et al, 2016. The complete chloroplast genome sequence of Tartary buckwheat cultivar miqiao 1 (Fagopyrum tataricum Gaertn. ). Mitochondrial DNA B Res, 1: 577-578. DOI:10.1080/23802359.2016.1197056 |
Logacheva M.D., Samigullin T.H., Dhingra A., et al, 2008. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale- A wild ancestor of cultivated buckwheat.. BMC Plant Biol, 8: 59. DOI:10.1186/1471-2229-8-59 |
Lohse M., Drechsel O., Bock R., 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet, 52: 267-274. DOI:10.1007/s00294-007-0161-y |
Luo X., Wang T.J., Hu H., et al, 2017. Characterization of the complete chloroplast genome of Oxyria sinensis. Conserv. Genet. Res, 9: 1-4. DOI:10.1007/s12686-016-0602-3 |
Parks M., Cronn R., Liston A., 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol, 7: 84. DOI:10.1186/1741-7007-7-84 |
Posada D., 2008. jModeltest: phylogenetic model averaging. Mol. Biol. Evol, 25: 1253-1256. DOI:10.1093/molbev/msn083 |
Raubeson L.A., Jansen R.K., 2005. Chloroplast genomes of plants. In: Henry, R.J.(Ed.), Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants.. CABI Publishing, Wallingford, pp: 45-68. |
Ronse De Craene L.P., Akeroyd J.R., 2008. Generic limits in Polygonum and related genera (Polygonaceae) on the basis of floral characters. Bot. J. Linn. Soc, 98: 321-371. |
Sanchez A., Schuster T.M., Kron K.A., 2009. A large-scale phylogeny of Polygonaceae based on molecular data. Int. J. Plant Sci, 170: 1044-1055. DOI:10.1086/605121 |
Schuster T.M., Gibbs M.D., Bayly M.J., 2018. Annotated plastome of the temperate woody vine Muehlenbeckia australis (G. Forst.) Meisn. (Polygonaceae). Mitochondrial DNA B Res, 3: 399-400. |
Shi Y.F., Li J., Li B., et al, 1998. Uplift and Environmental Changes of QinghaiTibetan Plateau in the Late Cenozoic Period. Guangzhou: Guangdong Science and Technology Press: 434-435.
|
Spicer R.A., Harris N.B.W., Widdowson M., et al, 2003. Constant elevation of southern Tibet over the past 15 million years.. Nature, 421: 622-624. DOI:10.1038/nature01356 |
Stamatakis A., 2014. RAxML version 8:a tool for phylogenetic analysis and postanalysis of large phylogenies.. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Sun W., An C., Zheng X.L., et al, 2014. Phylogenetic analysis of Polygonum L. s. lat. and related genera (Polygonaceae) inferred from nrDNA internal transcribed spacer (ITS) sequences. Plant Sci. J, 32: 228-235. |
Tavakkoli S., Osaloo S.K., Maassoumi A.A., 2010. The phylogeny of Calligonum and Pteropyrum (Polygonaceae) based on nuclear ribosomal DNA ITS and chloroplast trnL-F sequences. Iran. J. Biotechnol, 8: 7-15. |
Tian X.M., Liu R.R., Tian B., et al, 2009. Karyological studies of Parapteropyrum and Atraphaxis (Polygonaceae).. Caryologia, 62: 261-266. |
Tian X.M., Luo J., Wang A.L., et al, 2011. On the origin of the woody buckwheat Fagopyrum tibeticum (=Parapteropyrum tibeticum) in the Qinghai-Tibetan Plateau. Mol. Phylogenet. Evol, 61: 515-520. DOI:10.1016/j.ympev.2011.07.001 |
Wang C.L., Ding M.Q., Zou C.Y., et al, 2017. Comparative analysis of four buckwheat species based on morphology and complete chloroplast genome sequences. Sci. Rep, 7: 6514. DOI:10.1038/s41598-017-06638-6 |
Yang X.Y., Qian X.Y., Wang Z.F., 2018. The complete chloroplast genome of Mimosa pudica and the phylogenetic analysis of mimosoid species. Mitochondrial DNA B Res, 3: 1265-1266. DOI:10.1080/23802359.2018.1532831 |
Yang X.Y., Wang Z.F., Luo W.C., et al, 2019. Plastomes of Betulaceae and phylogenetic implications. J. Syst. Evol, 57: 508-518. DOI:10.1111/jse.12479 |
Zhang L., Xi Z.X., Wang M.C., et al, 2018a. Plastome phylogeny and lineage diversification of Salicaceae with focus on poplars and willows. Ecol. Evol, 8: 1-8. DOI:10.1002/ece3.3360 |
Zhang Z., Ma L., Abbasi A., et al, 2020. Database resources of the national genomics data center in 2020. Nucleic Acids Res, 48: D24-D33. DOI:10.1093/nar/gkz1210 |
Zhang L.S., Yang X.Y., Mao X.X., et al, 2018b. The complete chloroplast genome of Antiaris toxicaria, a medicinal and extremely toxic species. Mitochondrial DNA Part B Res, 3: 1100-1101. DOI:10.1080/23802359.2018.1516121 |
Zhao Y.M., Yang Z.Y., Zhao Y.P., et al, 2019. Chloroplast genome structural characteristics and phylogenetic relationships of Oleaceae. Chin. Bull. Bot, 54: 441-454. |



