b. Department of Botany, National Museum of Natural History, MRC 166, Smithsonian Institution, Washington, DC, 20013-7012, USA;
c. Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, 510650, China
The family Solanaceae consists of approximately 21 tribes, 100 genera and 2500 species (Hunziker, 2001; Olmstead and Bohs, 2007), including many of the world's most important and economically significant crop species (e.g., potatoes, tomatoes, eggplants, and chili peppers). In addition to their agricultural importance, many species in the family are of high ornamental value (e.g., petunias, jimsonweeds, and nightshade) or have significant medicinal potential (e.g., species in the Atropina lineage; Olmstead et al., 2008). The highest species diversity of Solanaceae is found in the western hemisphere, particularly in South America (Olmstead, 2013; Dupin et al., 2017). Molecular phylogenetic analyses of Solanaceae have yielded 19 main clades, three of which are found outside the New World (Anthocercideae in Australia, and Hyoscyameae and Mandragora L. in Eurasia), yet relationships within some of the main clades still need to be further explored (Olmstead et al., 2008; Olmstead, 2013; Särkinen et al., 2013).
The tribe Hyoscyameae sensu lato is one of the most important medicinal groups in Solanaceae. The tribe, which is mainly restricted to Eurasia and has diversity centers in the Mediterranean-Turanian region and the Qinghai-Tibet Plateau (QTP) region (Tu et al., 2010), includes nearly 40 species in seven genera (the berry fruit-bearing Atropa L. and the capsule fruit-bearing Hyoscyameae sensu stricto, including Anisodus Link ex Spreng., Atropanthe Pascher, Hyoscyamus L., Physochlaina G. Don, Przewalskia Maxim., and Scopolia Jacq.) (Lu and Zhang, 1986; D'Arcy and Zhang, 1992). Of these genera, Physochlaina and Scopolia are distributed in Europe and Asia. A few species of Hyoscyamus (~20 spp.), the largest genus in the tribe, extend to Africa. Some genera, such as Anisodus, Atropanthe and Przewalskia, are endemic to the QTP and adjacent areas. Phytochemical studies have suggested that genera in Hyoscyameae are closely related (Tetenyi, 1987; Gemeinholzer and Wink, 2001) and molecular phylogenetic studies have indicated that Hyoscyameae is monophyletic (Olmstead et al., 2008; Tu et al., 2010; Särkinen et al., 2013; Sanchez-Puerta and Abbona, 2014). However, inter-generic phylogenetic relationships within Hyoscyameae are still obscure. Specifically, researchers have yet to consistently position the monotypic genus Atropanthe, which is endemic to central and southwest China, within Hyoscyameae (Yuan et al., 2006; Olmstead et al., 2008; Tu et al., 2010). Furthermore, the relationship between Anisodus and Hyoscyamus remains uncertain (Fig. 1). The reconstruction of these unresolved relationships within Hyoscyameae may be aided by plastome data, which have provided new insights into the evolutionary history of other plant lineages (e.g., Guo et al., 2019; Mu et al., 2020).
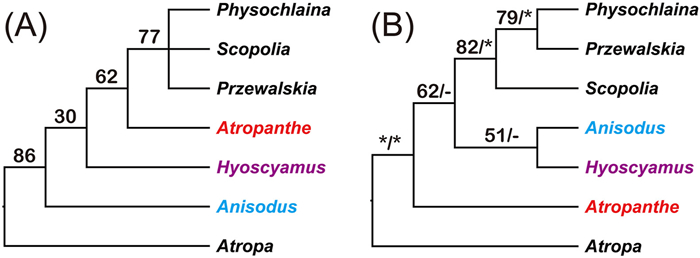
|
| Fig. 1 Two phylogenetic trees of the tribe Hyoscyameae adapted from (A) Olmstead et al. (2008) and (B) Tu et al. (2010). Support values with maximum parsimony analysis (BSMP) are shown in (A), and both BSMP and posterior probabilities of Bayesian analysis (PPBI) are presented in (B). Stars denote BSMP = 100 and PPBI = 1.00, and hyphens denote BSMP < 50 or PPBI < 0.95. |
Divergence times within Hyoscyameae are also obscure. Olmstead (2013) suggested that Solanaceae diversified at a time that coincided with the isolation of South America from the Late Cretaceous through the Cenozoic. A relatively young stem age of Solanaceae, 48.64 million years ago (Ma), has been estimated based on fossil calibration, and the crown ages of several clades are estimated to be young, including Hyoscyameae (~12 Ma) and Physalinae (~9 Ma) (Dupin et al., 2017). However, a well-preserved fossil of the lantern fruit (Physalis infinemundi Wilf) from the Eocene Patagonia (52.2 Ma) has recently been reported, and represents the earliest unequivocal fossil of Solanaceae to date (Wilf et al., 2017). Considering the relatively derived phylogenetic position of the Physalinae clade within Solanaceae, and that fossils only offer minimum estimates of certain clades, the origin of Solanaceae is likely to be older than the Eocene (Särkinen et al., 2018).
Plant diversification in Hyoscyameae sensu stricto is thought to be linked to the Himalayan uplift (Lu and Zhang, 1986). Cytological studies suggest an early evolution occurred before the orogeny of the Himalaya (Tu et al., 2005), while all genera in the tribe Hyoscyameae are estimated to have diversified during the middle to late Miocene (Tu et al., 2010). Elucidating biogeographical histories of plant groups requires well-resolved phylogenies and clear paleontological evidence. In this regard, combining the recent discovery of thePhysalis infinemundi fossil with a robust phylogeny represents a promising approach to determining whether the uplift of the QTP and the diversification of Hyoscyameae are correlated.
In this study, we reconstructed the phylogeny of the tribe Hyoscyameae by using whole chloroplast genome data of all seven genera in the tribe. We then took advantage of a recent discovery of the earliest fossil P. infinemundi from the Gondwanan Patagonia to re-estimate the divergence time of each genus in tribe Hyoscyameae. Finally, we used these divergence times to assess whether the diversification of the Hyoscyameae is correlated with the orogeny of the QTP and adjacent areas. The explicit estimations of both phylogeny and divergence times obtained here are not only important for further understanding the biogeographic history of Solanaceae, but also for providing empirical data to better understand the mechanisms of speciation and plant diversification on the QTP and adjacent areas.
2. Materials and methods 2.1. Taxon sampling and data retrievalNine species representing all genera of the tribe Hyoscyameae were sampled. For the genus Physochlaina, we sequenced the whole chloroplast genome of Physochlaina physaloides (L.) G. Don (Tong et al., 2019), which is distributed in the most eastward range of the genus, aiming to make a comparison with Physochlaina orientalis G. Don which is distributed in the most westward range. Whole chloroplast genomes of an additional 20 species representing the main clades of Solanaceae were selected based on Särkinen et al. (2013). Related sequences were downloaded from NCBI (Table S1).
2.2. Phylogenetic reconstructionThe substitution saturation test was performed for the data matrix in DAMBE7 (Xia, 2018). Phylogenetic analyses were performed using maximum likelihood (ML) analyses implemented in IQ-TREE v.1.6.12 (Nguyen et al., 2015), and Bayesian inference (BI) in MrBayes v.3.2.6 (Ronquist et al., 2012). Plastid sequences were aligned using MAFFT (Katoh et al., 2019) and refined using Gblocks v.0.91b (Castresana, 2000) to exclude all gaps. The bootstrap support values (BSML) of the ML analyses were estimated from 1000 replicates using UFBoot2 (Hoang et al., 2018). For the BI analyses, the best model was provided by jModeltest v.2.1.10 (Darriba et al., 2012). Then the Markov chain Monte Carlo (MCMC) algorithm was applied to two datasets, with three hot chains and one cold chain for 4 × 107 generations in a parallel mode. Trees were sampled every 1000 generations beginning with a random tree. The run was accomplished and the average standard deviation of split frequencies was less than 0.01 in all cases. Bayesian posterior probabilities (PPBI) were calculated as the 50% majority-rule consensus of all sampled trees, with the first 25% trees discarded as burn-in.
2.3. Divergence time estimationSeveral confirmed fossils of Solanaceae are documented (Särkinen et al., 2013, 2018; Wilf et al., 2017). Among them, the earliest fossil with well-preserved fruit morphology, P. infinemundi (Wilf et al., 2017), was employed for node constraint in this study. Because Physalis is paraphyletic (Olmstead et al., 2008; Särkinen et al., 2013), and the inflated calyx occurs in multiple solanoid lineages, the fossil P. infinemundi was used to constrain the Solanoideae node. An XML format file was generated using the BEAUti in the BEAST v.2.6.0 (www.beast2.org) package and then performed on the CIPRES website (Miller et al., 2010). A lognormal relaxed molecular clock was selected with a general time-reversible substitution model with gamma site heterogeneity, four rate categories, and a birth-death process tree prior. An offset of 52.2 Ma (mean = 1, Stdev = 1.25) was set for the crown age of Solanoideae clade with the 95% highest posterior density (HPD) intervals (52.5–73.4 Ma). The MCMC length was 2 × 109 with sampling every 4000 generations for analyses. BEAST log files were analyzed with Tracer v.1.7 (Rambaut et al., 2018) for convergence with the first 10% removed as burn-in. When an effective sample size for parameters in the log file was more than 200, the maximum clade credibility (MCC) tree with median heights was generated by TreeAnnotator in the BEAST package with the initial 20% of trees discarded as burn-in. The final tree was edited in FigTree v.1.4.4 (Rambaut et al., 2018) to view related parameters.
3. Results 3.1. Phylogenetic relationships within HyoscyameaeA data matrix with a length of 144, 352 bp was obtained, and the substitution saturation test performed on DAMBE7 supports the downstream analysis (Iss < Iss. c). The best-fit model (GTR + I + G) was selected by AICc with jModeltest. The tribe Hyoscyameae was supported to be monophyletic with full support (BSML = 100, PPBI = 1.00, Fig. 2). Within the tribe Hyoscyameae, the first clade to diverge was the berry genus Atropa. Anisodus and Hyoscyamus formed a clade with strong support (BSML = 84, PPBI = 1.00). The monotypic genus Atropanthe was sister to the clade of (Scopolia (Physochlaina, Przewalskia)) with strong support (BSML = 99, PPBI = 1.00). Another monotypic genus endemic to the QTP, Przewalskia, grouped with the second largest genus in the tribe, Physochlaina, with strong support (BSML = 83, PPBI = 1.00).
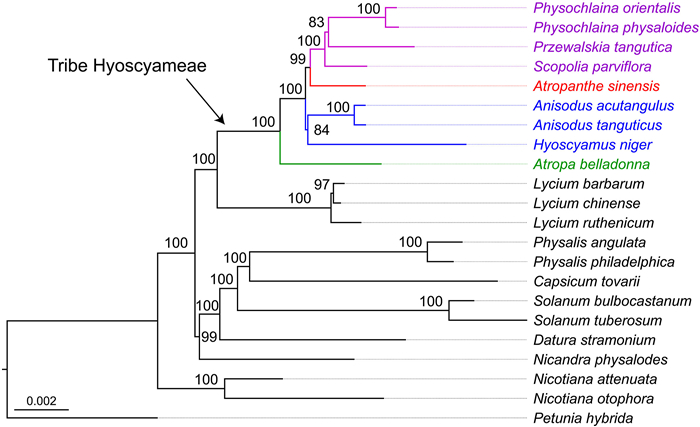
|
| Fig. 2 Phylogeny of the tribe Hyoscyameae inferred from maximum likelihood analyses based on whole chloroplast genome data of Solanaceae. Support values generated from the analysis of maximum likelihood are indicated on branches, posterior probabilities of the Bayesian inference are 1.00 for all nodes. |
The stem age of the tribe Hyoscyameae was dated to be 47.11 Ma (95% HPD: 36.75–57.86 Ma), and its crown age was estimated to be 30.28 Ma (95% HPD: 21.19–40.35 Ma) (Fig. 3). The crown age of the clade consisting of the six capsule-type genera (Hyoscyameae sensu stricto) was inferred as 22.52 Ma (95% HPD: 15.19–30.53 Ma). The split time between Anisodus and Hyoscyamus was dated as 21.17 Ma (95% HPD: 13.96–29.11 Ma). The crown ages of Atropanthe and Scopolia were dated as 20.67 Ma (95% HPD: 13.48–28.35 Ma) and 16.24 Ma (95% HPD: 10–23.14 Ma), respectively.Physochlaina and Przewalskia diverged at 14.82 Ma (95% HPD: 8.5–21.22 Ma).
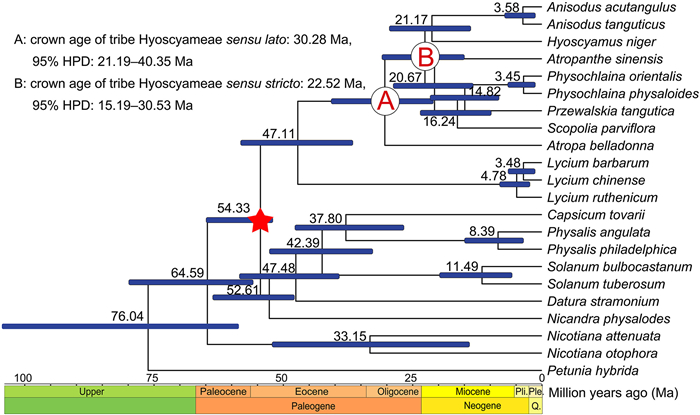
|
| Fig. 3 BEAST chronogram of Solanaceae inferred from whole chloroplast genome data. Blue bars represent the 95% highest posterior density intervals of node ages. The fossil Physalis infinemundi Wilf was used for calibration and is marked by a red star in the tree. Pli.: Pliocene, Ple.: Pleistocene, Q.: Quaternary. |
A robust phylogeny of the tribe Hyoscyameae was constructed in this study. The delimitation of the tribe Hyoscyameae has been debated (Yuan et al., 2006). Atropa with its berry fruits had been recognized as a member of the tribe Solaneae (D'Arcy, 1991), whereas the six remaining genera of the tribe bearing capsule fruits have been treated to constitute the tribe Hyoscyameae sensu stricto (Lu and Zhang, 1986; D'Arcy and Zhang, 1992). In this study, Atropa was resolved as sister to Hyoscyameae sensu stricto with strong support (Fig. 2). This is also supported by morphological characteristics, such as the thin inner side of the testa cell (vs. thick in Hyoscyameae sensu stricto) (Yang, 2002). Previously, the phylogenetic relationship between Anisodus and Hyoscyamus was uncertain (Olmstead et al., 2008), or resolved as a sister clade with weak support (Tu et al., 2010; Sanchez-Puerta and Abbona, 2014). In the current study, Anisodus and Hyoscyamus were resolved as sister genera with strong support (BSML = 84, PPBI = 1.00). The testa morphology in these two genera is brain-striate (cerebroid), while it is net-like in Atropanthe, Scopolia, Physochlaina, and Przewalskia (Yang, 2002).
Atropanthe is a monotypic genus endemic to central and southwest China. It grouped with the (Scopolia (Physochlaina, Przewalskia)) clade with weak support (BSMP = 62) (Olmstead et al., 2008), yet it was strongly supported as the first clade to diverge within Hyoscyameae sensu stricto (Tu et al., 2010). Based on a data matrix consisting of ten plastid markers (11, 610 bp) representing the most variable regions in the chloroplast genome, Sanchez-Puerta and Abbona (2014) constructed the backbone of Hyoscyameae, but the phylogenetic positions of Atropanthe and Anisodus were unresolved. By using a larger dataset consisting of whole plastid sequences, we demonstrated that Atropanthe is robustly resolved as sister to the clade (Scopolia (Physochlaina, Przewalskia)) (BSML = 99, PPBI = 1.00; Fig. 2). The phylogeny and its support values of Hyoscyameae at the genus level are well-resolved in this study, which demonstrates the great potential of whole chloroplast genome data in phylogenomic study of certain plant lineages.
4.2. Divergence time estimations of the tribe HyoscyameaeBased on whole chloroplast genome data and the newly found lantern fossil from the Eocene Patagonia, we estimated the stem and crown ages of the tribe Hyoscyameae to be 47.11 Ma (95% HPD: 36.75–57.86 Ma) and 30.28 Ma (95% HPD: 21.19–40.35 Ma), respectively (Fig. 3). Previous studies have indicated that the tribe Hyoscyameae is closely related to New World lineages (Tu et al., 2010; Dupin et al., 2017). During the Paleocene and early Eocene, Trans-Atlantic dispersal between the New World and Eurasia was possible, such as with the extinct genus Lagokarpos McMurran & Manchester (Tang et al., 2019). Thus, our divergence time estimates suggest that ancestral Hyoscyameae might have landed in Europe from the New World and dispersed eastward to Asia; during this period, their fruit type transformed from berries (as in Atropa) to capsules (as in Hyoscyameae sensu stricto).
Our dating of the crown ages of Hyoscyameae clade except for Atropa to the early Miocene (22.52 Ma, 95% HPD: 15.19–30.53 Ma, Fig. 3) supports previous hypotheses that the diversification of Hyoscyameae sensu stricto is closely linked to the orogeny of the QTP (~40–35 Ma, Favre et al., 2015; Lu and Zhang, 1986; Wen et al., 2014). Well-preserved fossils of animals and plants discovered from the QTP indicate that this region underwent drastic uplift (from 2300 m to ca. 3000 m) during the early to middle Miocene (Deng et al., 2019; Su et al., 2019). Our inferred divergence time of ancestral Hyoscyameaesensu stricto clade (22.52 Ma) is consistent with this important geologic event.
Because of the rapid uplift of the QTP and the transformation of its climatic ecosystem from typical tropical or subtropical to a plateau-type biotic assemblage at the Paleogene/Neogene boundary, diverse and complicated local habitat and climate conditions occurred, which might have triggered and facilitated speciation and diversification of the ancestral Hyoscyameae. Members of Hyoscyameae diverged and evolved both intrinsically (ploidy, chromosome base number, and chromosome structure) and extrinsically (fruit type, inflorescence, and other morphology). Plants distributed in the alpine and arctic regions with harsh and cold environments may easily turn to polyploidy (Brochmann et al., 2004; Madlung, 2013; Van de Peer et al., 2017). Cytological studies have demonstrated a large proportion of polyploidy and complicated variation in chromosome number and structure in most members of Hyoscyameae (Tu et al., 2005, and references therein). Besides berries and capsules, several members in the Atropina lineage have atypical Solanaceae fruit types, such as mericarps in Nolana L. ex L.f. and dry indehiscent fruit in Sclerophylax Miers, and they occupy a continuous coastal desert habitat in Peru and Chile (Olmstead et al., 2008; Dillon et al., 2009). The diverse fruit morphology in the Atropina lineage implies a great genetic potential for fruit variation and evolution in Solanaceae, and repeated evolution of fruit morphology has been detected, such as the gain and loss of balloon-like fruit calyx in the tribe Physalideae (Deanna et al., 2019). High morphological and genetic diversity of fruit in the Atropina lineage may have contributed to the diversification and evolutionary success of ancestral Hyoscyameae when encountering diverse habitat and climate condition.
4.3. Rapid evolutionary radiation of PhysochlainaMany plant lineages exhibit the out-of-QTP pattern, including Gentiana (Tourn.) L. (Favre et al., 2016), Hippophae L. (Jia et al., 2012), Incarvillea Juss (Rana et al., 2021), Lilieae (Huang et al., 2018), Rhodiola L. (Zhang et al., 2014), and Saxifraga Tourn. ex L. (Ebersbach et al., 2017). Physochlaina and Przewalskia, two of the youngest genera in Hyoscyameae, show great variation in species numbers and distribution ranges. Physochlaina consists of 12 species distributed from the Far East to Central Asia, whereas Przewalskia is a monotypic genus endemic to high altitudes on the QTP. Thus, although both lineages originated on the QTP, Przewalskia is restricted to this region, while Physochlaina has diversified and expanded its range out of the QTP. Possible factors that may have facilitated the rapid diversification of Physochlaina include karyotype, ploidy level, and related fertility ability (densely-flowered vs. 1–3 flowers). Physochlaina is a hexaploid with a basic chromosome number of x = 7; in contrast, Przewalskia is a tetraploid with a basic chromosome number of x = 11 (Tu et al., 2005). Furthermore, Physochlaina has the type 2B karyotype, whereas Przewalskia has the type 2A karyotype. The type 2A karyotype is considered relatively primitive compared to type 2B (Stebbins, 1971), and empirical studies indicate that hexaploidy is more competitive than tetraploidy (Ramsey, 2011).
The two Physochlaina species sampled in this study represent a broad geographic range covering eastern Asia (P. physaloides, Tong et al., 2019) and central Asia (P. orientalis, Gandini et al., 2019). These two species are also the most derived sister species in the genus (Tu et al., 2010; Särkinen et al., 2013; Sanchez-Puerta and Abbona, 2014). The divergence time between Physochlaina and Przewalskia was inferred to be 16.24 Ma (95% HPD: 10–23.14 Ma), and the two Physochlaina species diverged at 3.45 Ma (95% HPD: 1.48–6.21 Ma, Fig. 3). Considering the young lineage age and vast distribution range from northeast to Central Asia, Physochlaina is an exemplar in Hyoscyameae demonstrating the rapid evolutionary radiation pattern on the QTP and adjacent areas. Integrated effects from the fast uplift of the QTP since the late Paleogene may have contributed to habitat differentiation (e.g., slopes, rock fissures, and understory) among species within Physochlaina. It is of great interest to further investigate the phylogeny, divergence times, and biogeography of the tribe Hyoscyameae at the species level, particularly exploring the disjunct distributional pattern in the Mediterranean, eastern Asia, and Africa. Further findings should advance our understanding of the mechanisms of species diversification on the QTP and adjacent areas, and the dynamics of the evolutionary diversifications of Hyoscyameae and Solanaceae.
5. ConclusionsBased on whole chloroplast genome data, we obtained a well-resolved phylogeny of the tribe Hyoscyameae in Solanaceae. Atropa was resolved as sister to Hyoscyameae sensu stricto, the phylogenetic position of Atropanthe was strongly resolved as sister to the clade (Scopolia, Physochlaina, Przewalskia), and together they are sister to the strongly supported Anisodus–Hyoscyamus clade. Fossil-calibrated divergence times within Hyoscyameae were re-estimated based on the recently discovered Eocene Patagonia Physalis fossil. The stem age of the tribe was dated to the Eocene, and Hyoscyameae sensu stricto diversified during the early Miocene. These findings suggest a strong correlation between the lineage diversification and the rapid uplift of the QTP during the early Miocene. Future work needs to emphasize the species-level evolutionary diversification of the Hyoscyameae, particularly exploring the evolution of the species disjunctively distributed in Africa, Asia, and Europe.
Author's contributionsXYM and TYT conceived the work, FWL, LT, and YXZ prepared the datasets and carried out the analyses, FWL and XYM wrote the manuscript, XYM and JW revised the manuscript.
Declaration of competing interestThe authors declare no conflict of interest.
AcknowledgmentsWe thank Dr. Xiao-Feng Chi and Dr. Ofelia Vargas-Ponce for contributing the whole chloroplast genome sequences of Anisodus tanguticus (Maxim.) Pascher and Physalis philadelphica Lam. before the release in NCBI. This work was supported by the Beijing Natural Science Foundation (Grant No. 5192012), National Natural Science Foundation of China (grant number 32070235) and the China Scholarship Council (Grant No. 201906515009).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.17660/ActaHortic.2007.745.11.
Brochmann C., Brysting A.K., Alsos I.G., et al, 2004. Polyploidy in arctic plants. Biol. J. Linn. Soc, 82: 521-536. DOI:10.1111/j.1095-8312.2004.00337.x |
Castresana J., 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol, 17: 540-552. DOI:10.1093/oxfordjournals.molbev.a026334 |
D'Arcy W.G., 1991. The Solanaceae since 1976 with a review of its biogeography. In: Hawkes, J.G., Lester R.N., Nee M., Estrada-Ramos, N. (Eds. ), Solanaceae Ⅲ: Taxonomy, Chemistry, Evolution.. London: Royal Botanic Garden, Kew.
|
D'Arcy W.G., Zhang Z.Y., 1992. Notes on the Solanaceae of China and neighboring areas. Novon, 2: 124-128. DOI:10.2307/3391672 |
Darriba D., Taboada G.L., Doallo R., et al, 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 9: 722. DOI:10.1038/nmeth.2109 |
Deanna R., Larter M.D., Barboza G.E., et al, 2019. Repeated evolution of a morphological novelty: a phylogenetic analysis of the inflated fruiting calyx in the Physalideae tribe (Solanaceae). Am. J. Bot, 106: 270-279. DOI:10.1002/ajb2.1242 |
Deng T., Wang X.M., Wu F.X., et al, 2019. Review: implications of vertebrate fossils for paleo-elevations of the Tibetan Plateau. Global Planet. Change, 174: 58-69. DOI:10.1016/j.gloplacha.2019.01.005 |
Dillon M.O., Tu T.Y., Xie L., et al, 2009. Biogeographic diversification in Nolana(Solanaceae), a ubiquitous member of the atacama and Peruvian deserts along the western coast of South America. J. Syst. Evol, 47: 457-476. DOI:10.1111/j.1759-6831.2009.00040.x |
Dupin J., Matzke N.J., Särkinen T., et al, 2017. Bayesian estimation of the global biogeographical history of the Solanaceae. J. Biogeogr, 44: 887-899. DOI:10.1111/jbi.12898 |
Ebersbach J., Muellner-Riehl A.N., Michalak I., et al, 2017. In and out of the Qinghai-Tibet Plateau: divergence time estimation and historical biogeography of the large arctic-alpine genus Saxifraga L. J. Biogeogr, 44: 900-910. DOI:10.1111/jbi.12899 |
Favre A., Paeckert M., Pauls S.U., et al, 2015. The role of the uplift of the QinghaiTibetan Plateau for the evolution of Tibetan biotas. Biol. Rev, 90: 236-253. DOI:10.1111/brv.12107 |
Favre A., Michalak I., Chen C., et al, 2016. Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J. Biogeogr, 43: 1967-1978. DOI:10.1111/jbi.12840 |
Gandini C.L., Garcia L.E., Abbona C.C., et al, 2019. The complete organelle genomes of Physochlaina orientalis: insights into short sequence repeats across seed plant mitochondrial genomes. Mol. Phylogenet. Evol, 137: 274-284. DOI:10.1016/j.ympev.2019.05.012 |
Gemeinholzer B., Wink M., 2001. Solanaceae: occurrence of secondary compounds versus molecular phylogeny. In: van den Berg R.G., Barendse G.W.M., van der Weerden G.M., Mariani, C. (Eds. ), Solanaceae V: Advances in Taxonomy and Utilization.. Nijmegen: Nijmegen University Press: 165-177.
|
Guo C., Guo Z.H., Li D.Z., 2019. Phylogenomic analyses reveal intractable evolutionary history of a temperate bamboo genus (Poaceae: bambusoideae). Plant Divers, 41: 213-219. DOI:10.1016/j.pld.2019.05.003 |
Hoang D.T., Chernomor O., von Haeseler A., et al, 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol, 35: 518-522. DOI:10.1093/molbev/msx281 |
Huang J., Yang L.Q., Yu Y., et al, 2018. Molecular phylogenetics and historical biogeography of the tribe Lilieae (Liliaceae): bi-directional dispersal between biodiversity hotspots in Eurasia. Ann. Bot, 122: 1245-1262. DOI:10.1093/aob/mcy138 |
Hunziker A., 2001. Genera Solanacearum: the Genera of Solanaceae Illustrated, Arranged According to a New System.. ARG Gantner Verlag KG, Ruggell.. |
Jia D.R., Abbott R.J., Liu T.L., et al, 2012. Out of the Qinghai-Tibet Plateau: evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophae rhamnoides (Elaeagnaceae). New Phytol, 194: 1123-1133. DOI:10.1111/j.1469-8137.2012.04115.x |
Katoh K., Rozewicki J., Yamada K.D., 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf, 20: 1160-1166. DOI:10.1093/bib/bbx108 |
Lu A.M., Zhang Z.Y., 1986. Studies of the subtribe hyoscyaminae in China. In: D'Arcy, W.G. (Ed. ), Solanaceae: Biology and Systematics. New York: Columbia University Press: 56-78.
|
Madlung A., 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity, 110: 99-104. DOI:10.1038/hdy.2012.79 |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE) (New Orleans, LA).
|
Mu X.Y., Tong L., Sun M., et al, 2020. Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol. Phylogenet. Evol, 147: 106802. DOI:10.1016/j.ympev.2020.106802 |
Nguyen L., Schmidt H.A., von Haeseler A., et al, 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol, 32: 268-274. DOI:10.1093/molbev/msu300 |
Olmstead, R.G., Bohs, L., 2007. A summary of molecular systematic research in Solanaceae: 1982-2006. In: Spooner, D.M., Bohs, L., Govannoni, J., Olmstead, R.G., Shibata, D. (Eds. ), Ⅵ International Solanaceae Conference: Genomics Meets Biodiversity, vol. 745. ISHS Acta Horticulturae, Leuven, pp. 255-268.https://doi.org/10.17660/ActaHortic.2007.745.11.
|
Olmstead R.G., Bohs L., Migid H.A., et al, 2008. A molecular phylogeny of the Solanaceae. Taxon, 57: 1159-1181. DOI:10.1002/tax.574010 |
Olmstead R.G., 2013. Phylogeny and biogeography in Solanaceae, Verbenaceae and Bignoniaceae: a comparison of continental and intercontinental diversification patterns. Bot. J. Linn. Soc, 171: 80-102. DOI:10.1111/j.1095-8339.2012.01306.x |
Rambaut A., Drummond A.J., Xie D., 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1. 7. Sys. Biol, 67: 901-904. DOI:10.1093/sysbio/syy032 |
Ramsey J., 2011. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. U.S. A, 108: 7096-7101. DOI:10.1073/pnas.1016631108 |
Rana S.K., Luo D., Rana H.K., et al, 2021. Geoclimatic factors influence the population genetic connectivity of Incarvillea arguta (Bignoniaceae) in the Himalaya Hengduan Mountains biodiversity hotspot. J. Syst. Evol, 59: 151-168. DOI:10.1111/jse.12521 |
Ronquist F., Teslenko M., van der Mark P., et al, 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys029 |
Sanchez-Puerta M.V., Abbona C.C., 2014. The chloroplast genome of Hyoscyamus niger and a phylogenetic study of the tribe Hyoscyameae (Solanaceae). PLoS One, 9: e98353. DOI:10.1371/journal.pone.0098353 |
Särkinen T., Bohs L., Olmstead R.G., et al, 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol. Biol, 13: 214. DOI:10.1186/1471-2148-13-214 |
Särkinen T., Kottner S., Stuppy W., et al, 2018. A new commelinid monocot seed fossil from the early Eocene previously identified as Solanaceae. Am. J. Bot, 105: 95-107. DOI:10.1002/ajb2.1009 |
Stebbins G.L., 1971. Chromosome Evolution in Higher Plants.. London: Edward Arnold Ltd.
|
Su T., Farnsworth A., Spicer R.A., et al, 2019. No high Tibetan plateau until the neogene. Sci. Adv, 5: v2189. DOI:10.1126/sciadv.aav2189 |
Tang H., Liu J., Wu F.X., et al, 2019. Extinct genus Lagokarpos reveals a biogeographic connection between Tibet and other regions in the Northern Hemisphere during the Paleogene. J. Systemat. Evol, 57: 670-677. DOI:10.1111/jse.12505 |
Tetenyi P., 1987. A chemotaxonomic classification of the Solanaceae. Ann. Mo. Bot. Gard, 74: 600-608. DOI:10.2307/2399328 |
Tong L., Zhu Y.X., Lei F.W., et al, 2019. The complete chloroplast genome of Physochlaina physaloides (Solanaceae), an important medicinal plant. Mitochond. DNA B. Res, 4: 3427-3428. DOI:10.1080/23802359.2019.1674730 |
Tu T.Y., Sun H., Gu Z.J., et al, 2005. Cytological studies on the Sino-Himalayan endemic Anisodus and four related genera from the tribe Hyoscyameae (Solanaceae) and their systematic and evolutionary implications. Bot. J. Linn. Soc, 147: 457-468. DOI:10.1111/j.1095-8339.2005.00384.x |
Tu T.Y., Volis S., Dillon M.O., et al, 2010. Dispersals of Hyoscyameae and mandragoreae (Solanaceae) from the new world to Eurasia in the early Miocene and their biogeographic diversification within Eurasia. Mol. Phylogenet. Evol, 57: 1226-1237. DOI:10.1016/j.ympev.2010.09.007 |
Van de Peer Y., Mizrachi E., Marchal K., 2017. The evolutionary significance of polyploidy. Nat. Rev. Genet, 18: 411-424. DOI:10.1038/nrg.2017.26 |
Wen J., Zhang J.Q., Nie Z.L., et al, 2014. Evolutionary diversifications of plants on the Qinghai-Tibetan plateau. Front. Genet, 5: 4. DOI:10.3389/fgene.2014.00004 |
Wilf P., Carvalho M., Gandolfo M., et al, 2017. Eocene lantern fruits from Gondwanan Patagonia and the early origins of Solanaceae. Science, 355: 71-75. DOI:10.1126/science.aag2737 |
Xia X.H., 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol, 35: 1550-1552. DOI:10.1093/molbev/msy073 |
Yang D.Z., 2002. Tribe Hyoscyameae of the Solanaceae. Structure, Differentiation and Phylogenetic Relationship. Institute of Botany, the Chinese Academy of Sciences, Beijing.. PhD thesis.. |
Yuan Y.W., Zhang Z.Y., Chen Z.D., et al, 2006. Tracking ancient polyploids: a retroposon insertion reveals an extinct diploid ancestor in the polyploid origin of belladonna. Mol. Biol. Evol, 23: 2263-2267. DOI:10.1093/molbev/msl099 |
Zhang J.Q., Meng S.Y., Allen G.A., et al, 2014. Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol.Phylogenet. Evol, 77: 147-158. DOI:10.1016/j.ympev.2014.04.013 |



