b. University of Chinese Academy of Sciences, Beijing 100049, China
Roses are well-known ornamental plants that are globally beloved for their highly diverse flowers and flowering times. Flowering at a proper time is a key developmental switch that is essential for reproductive success and the life cycle (Amasino and Michaels, 2010; Baurle and Dean, 2006). Roses feature high diversity of flowering behaviors, including continuous flowering, occasionally re-blooming, and single seasonal blooming; hence, roses serve as a model for studying flowering time diversity in plants (Bendahmane et al., 2013; Dong et al., 2017). Flowering time is under complex but elegant regulation by exogenous and endogenous signaling pathways, which involve a set of gene regulatory networks (Andres and Coupland, 2012; Fornara et al., 2010). In both rose and strawberry, KSN, a homolog of Arabidopsis thaliana TFL1, has been proposed to play an essential role in regulating continuous flowering behavior (Iwata et al., 2012; Randoux et al., 2012, 2014). Duplication and functional diversification of COP1-like genes might also contribute to flowering time regulation in roses as well as other Rosaceae plants (Sun et al., 2020).
Rosa chinensis 'Old Blush' (OB) is a founder genotype in modern rose domestication and hence has been explored for many rose genetic studies (Byrne et al., 2007; Crespel et al., 2002; Hibrand Saint-Oyant et al., 2018; Li et al., 2015, 2019; Shupert et al., 2007; Spiller et al., 2011). However, Rosa wichuraiana 'Basye's Thornless' (BT) harbors important traits that differ from OB. For example, OB plants always grow erect; in contrast, before its annual flowering between April and June, BT shoots switch from prostrate growth in the vegetative stage to erect growth in the reproductive stage. This switch from prostrate to erect growth may help BT adapt to the light requirement and endogenous developmental signals upon flowering transition (Figs. 1A and S1). However, no information is available on the genetic regulatory mechanisms that underlie this switch.

|
| Fig. 1 BT expands specifically its FAR1/FRS-like genes. (A) The prostrate growth pattern of BT is shown on the left and the erect growth of R. chinensis 'Old Blush' (OB) is shown on the right (January of 2020). Note that BT showed an erect growth of its shoots upon flowering (Fig. S1). Red arrows mark flower buds of OB. (B) Specific expansion of FAR1/FRS-like genes in BT compared to other Rosaceae plants. A simplified Neighbor-Joining tree was drawn to show the phylogenetic relationships of Rosaceae plants (Fv, Fragaria vesca; Ro, Rubus occidentalis; Md, Malus domestica; Pc, Pyrus communis; Pa, Prunus armeniaca). Numbers of FAR1/FRS-like genes (FAR) and total protein coding (Total) genes are given to the right of each species. (C) Distribution of the random expansion (names in red) and syntenic FAR1/FRS-like (names in black) genes on seven BT chromosomes. Gene collinearity was defined by phylogenetic and collinearity analyses. (D) Types of FAR1/FRS-like genes on BT chromosomes. Colors mark the density of 100–50, 000 bp DNA inserts per 1 Mb of the BT genome in comparison to OB. Blue squares indicate whole-genome-duplication (WGD); orange squares, tandem duplication; purple pies, dispersed duplications; and green triangles, proximal duplications. (E) The BT expansion FAR1/FRS-like genes harbored significantly more transposon elements (TE) in regions 2000 bp up- and down-stream of coding region. P values (Wilcoxon rank sum test) show the significance levels between the BT expansion FAR1/FRS-like genes (red) and the syntenic FAR1/FRS-like genes (between BT and OB; pink), as well as the rest of the genome (whole genome; light blue). |
Light is one of the key environmental factors controlling flowering time (Liu et al., 2017, 2020; McCormac and Terry, 2002; Munnik and Nielsen, 2011; Tang et al., 2013). When plants sense a reduction in the ratio of red to far-red light, due to competition for light from the neighbor plants or other types of shade, they initiate a couple of adaptive responses, which are termed shade avoidance syndrome, including rapid shoot elongation, erect shoot, early flowering (Franklin and Whitelam, 2005). Previous studies have shown that FHY3 and FAR1, two homologous transcription factors essential for PhyA-mediated far-red light signaling, can negatively regulate Arabidopsis flowering time under both long-day and short-day conditions by promoting the expression of Early Flowering 3 and 4 (ELF3 and ELF4) (Li et al., 2011; Lin et al., 2007; Liu et al., 2019). Under fluctuating light conditions, FAR1 and FHY3 can alter their physical interaction with SPL3/4/5 transcription factors, which are key regulators in the aging pathway of flowering time control, to modulate the expression of downstream floral integrators such as AP1, FUL, LFY and miR172c, thus modifying the flowering time behaviors of A. thaliana (Xie et al., 2020).
FAR1 and FHY3 regulate a wide range of biological processes, including light signal transduction, photomorphogenesis (Liu et al., 2019; Siddiqui et al., 2016; Tang et al., 2013; Wang et al., 2016; Zhang et al., 2019), circadian clock and flowering time (Johansson and Staiger, 2015; Li et al., 2011; Liu et al., 2020; Ritter et al., 2017; Xie et al., 2020), shoot and floral development (Li et al., 2016), chloroplast and chlorophyll biosynthesis (Ouyang et al., 2011; Tang et al., 2012; Wang et al., 2016), starch synthesis (Ma et al., 2017), ABA (Tang et al., 2013) and oxidative stress responses (Ma et al., 2016) as well as immunity (Wang et al., 2016). Arabidopsis also has several FAR-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) proteins that can modulate flowering time (Bulik-Sullivan et al., 2015; Gao et al., 2013; Ma and Li, 2018; Ritter et al., 2017; Tang et al., 2013). However, the conservation and diversification of FAR1/FRS-like genes in other species, especially in woody plants, such as roses, remains to be explored in detail.
In this study, we identified FAR1/FRS-like genes in two rose genotypes, BT and OB, for which high-quality whole-genome sequences are available (Hibrand Saint-Oyant et al., 2018; Raymond et al., 2018; Zhong et al., 2020). We found that several gene duplication mechanisms, including whole-genome duplication (WGD), segmental duplication, tandem duplication and transposon element (TE) mediated duplication, especially the dispersed duplication were simultaneously involved in the expansion of FAR1/FRS-like genes in BT. Finally, we propose that the dispersed FAR1/FRS-like genes expansion and their expression pattern might associate with the regulatory of shoot-growth behavior in BT.
2. Materials and methods 2.1. Plant materials and transcriptomic profilingThe high-quality chromosome-level genome assembly for the BT genotype was constructed by ourselves (Zhong et al., 2020), and the genome sequences for haplo-OB was used for comparisons (Raymond et al., 2018). BT and OB plants were grown in the glasshouses at the Flower Research Institute of Yunnan Academy of Agricultural Sciences (Kunming, Yunnan, China). Leaf materials were collected in November (Nov) and March (Mar). Shoot apical meristem materials (SAM) with minimum leaf materials were sampled in March. RNA extraction followed by strand-specific Illumina sequencing has been described previously (Li et al., 2018).
2.2. Identification and phylogenetic analysis of FAR/FRS-like genes in rosesProtein amino acid sequences were downloaded for FAR1/FRS-like genes from TAIR10 (https://www.arabidopsis.org/), and were then used for BLAST identification of homologous sequences in BT and OB genomes with iTAK pipeline (-f 6) (Zheng et al., 2016). MAFFT was applied for sequence alignment with default parameters (Katoh et al., 2002). Phylogeny reconstructions were carried out using the Neighbor-Joining (NJ) method and 100 replicates of bootstrap simulation in RAxML 8.2.11 (stamatakis2014">Stamatakis, 2014). Chromosome distributions and annotations were plotted by TBtools (Chen et al., 2020) with house R scripts. Gene structures were plotted using Gene Structure Display Server 2.0 to show their exon/intron composition information (Hu et al., 2015) (http://gsds.cbi.pku.edu.cn//index.php).
2.3. Duplications and syntenic analysis of FAR1/RS-like genesMCSanX and related functional blocks were used to compare chromosome structural variation (Wang et al., 2012). The FAR1/FRS duplication type and collinearity were predicted with the duplicate_gene_classifier function according to protocols described in the pipeline manuals. The phylogenetically clustered gene sets were defined as syntenic only when a minimum of five genes were collinear. The remaining genes were considered non-collinear. The non-collinear genes without phylogenetic clustering signal were designated as BT expansion genes.
2.4. Exon-intron structure, conserved protein domains and motif analysesConserved protein domains were analyzed with the NCBI CD-Search Tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) using the PSSM model (maximum number of hits < 500). Conserved motifs in FAR1/FRS-like genes were identified with MEME (http://meme-suite.org/tools/meme; optimum motifs, 6-15; maximum number of motifs, 8). TBtools was used for exon-intron visualization.
2.5. Identification of insertion events between BT and OBTo find large insertion (100–50, 000 bp) events in BT compared to OB genome, each BT scaffold sequence was aligned to the OB genome with blastn. Only alignments with ≥60% identity and longer than 100 bp were kept for further analysis. Insertion density was plotted as the insertion events every 100 kb in 1 Mb window with ggplot2 in R with home scripts.
2.6. Transposon element (TE) analysisTransposon elements were analyzed with RepeatMasker (V4.1.0) (Saha et al., 2008) using plant RepBase database (https://www.girinst.org/) and Dfam 3.1 database (www.dfam.org). TEs located within 2000 bp up- and down-stream of the FAR1/FRS-like genes were counted and compared to whole genome level counts per protein-coding gene. Statistical differences between the expansion and syntenic FAR1/FRS-like genes, and the rest of the whole genome level was tested with the functionWilcoxon rank sum test in R.
2.7. Expression analysis for FAR1/FRS-like genes and their potential interactor encoding genesRNA-seq reads for shoot apical meristem (SAM), just opened young leaves in March (leaf_Mar) and November (leaf_Nov), which had been described previously (Li et al., 2018), were mapped to BT and OB genomes with HISAT2 (Kim et al., 2015) and assembled with StringTie (Pertea et al., 2015). Uniquely mapped reads (default parameters expect for -stranded = no) were used to calculate the relative expression as fragments per kilobase per million (FPKM) with ballgown package in R, and further compared for the expression pattern in different tissues and developmental stages. Normalization was carried out using the ZeroToOne method and scaled with log2. We collected proteins that interact with FAR1/FRS in A. thaliana with STRING online (https://www.expasy.org/). Protein sequences of these genes were extracted and used as seed sequences to blastp the BT and OB genomes (e-value 10-7, minimum identity 0.35, minimum length coverage 0.6). Ortholog gene pairs were then identified using OrthoFinder2 (emms2019">Emms and Kelly, 2019) in combination with the blastp, phylogeny, and synteny-based manual correction.
3. Results 3.1. BT expands significantly its FAR1/FRS-like genes than OBVia sequence similarity analysis to the 17 Arabidopsis FAR1/FRS family genes, we identified 91 and 52 FAR1/FRS-like genes in BT (Zhong et al., 2020) and OB (Raymond et al., 2018), respectively (Figs. 1B and S2; Tables S1 and S2). In BT, these genes were almost randomly dispersed on the seven chromosomes, with Chr2, 5, 6 harboring 17, 15, 16 genes, respectively. BT expanded significantly the FAR1/FRS-like genes, especially on Chromosomes 4, 5, 6, in comparison to OB genotype as well as other Rosaceae plants (Yates' chi-square test, P < 0.001; Fig. 1B; Table S2). Gene-collinearity and phylogenetic reconstruction analysis using the OB genotype as a reference indicated that 31 of the FAR1/FRS-like genes in BT are the result of gene expansion events (Fig. 1C).
3.2. Random duplication dominates BT expansion of RwFAR1FRS-like genesNext, we examined the potential mechanisms underlying the FAR1/FRS-likes expansion in BT. We classified these genes into four types of duplication events: whole-genome-duplication (WGD; two events), tandem duplication (nine events), proximal duplication (six events) and dispersed duplication (69 events). For the apparently randomly-duplicated gene sets, the gene structure and organization, including untranslated regions (UTRs), exons and introns, differed significantly from each other (Fig. S3). This pattern indicates that these were real tandemly duplicated gene sets not by products of erroneous annotation or assembly. The detection of identical numbers of WGD and tandem duplication events for OB suggests that random dispersion played a dominant role (29 among 31) in the expansion of BT RwFAR1/FRS-like genes (Fig. 1D; Table S3). However, random insertion of 100–50, 000 bp DNA fragments did not correlate with the expansion.
3.3. Dispersed transposon insertion accompanies the expansion of RwFAR1/FRS-likesBecause FAR1/FRS-like genes are derived from transposases, we next asked whether expansion of RwFAR1/FRS-like genes in BT is correlated with transposon element distribution (Hudson et al., 1999; Lin et al., 2007; Lin and Wang, 2004). We counted the TE insertion events within two kilobases up- and down-stream of BT expansion (non-syntenic) and syntenic RwFAR1/FRS-like genes. TE distance in expansion RwFAR1/FRS-like genes was not significantly different than in other genes throughout the genome (Fig. S4). Interestingly, the number of TEs was significantly higher in the expansion RwFAR1/FRS-like genes than in both the syntenic RwFAR1/FRS-like genes and in other genes in the genome (Fig. 1E; Table S4). This increased number of TEs indicates that the expansion of RwFAR1/FRS-like genes may have been accompanied by transposon activity.
3.4. Expanded RwFAR1/FRSs feature diversified protein structures and motifsWe examined the characteristics of proteins encoded by the genes of the RwFAR1/FRS-like expansion. The gene structure of syntenic and expansion RwFAR1/FRS-like genes (e.g., exon-intron numbers and organization) were very similar (Figs. S5 and S6). Motif analysis revealed that proteins encoded by expansion RwFAR1/FRS-like genes have between two to eight motif types, similar to proteins of syntenic RwFAR1/FRS-like genes (Fig. S7). Proteins encoded by expansion RwFAR1/FRS-like genes contain FAR1, FHY3 and DDE_Tnp_ISL3 domains individually or in combination. These patterns of protein motifs do not differ from those of proteins encoded by syntenic RwFAR1/FRS-like genes (Fig. S6), which corroborates the hypothesis that expansion RwFAR1/FRS-like genes were dispersed via random transposon insertion.
3.5. Lineage-specific expansion of RwFAR1/FRS-like genes mainly occurs in clade I and IITo understand whether the RwFAR1/FRS-like gene expansion had any further features, we performed a phylogenetic analysis with protein sequence alignment. This analysis grouped the FAR1/FRSs into five clades (I–V) with all Arabidopsis proteins clustered in clades III-V and both FAR1 and FHY3 sitting within clade IV (Fig. 2A). Both BT and OB had similar numbers of FAR1/FRSs to Arabidopsis within clades III-V. However, BT (54) doubled its FAR1/FRSs in clades I and II in comparison to OB (25; Yates' chi-square test, P < 0.001) with most of them randomly dispersed on the seven BT chromosomes (Fig. 2A). Twenty-three among the 31 (74.2%) expanded RwFAR1/FRS-like genes were grouped in clades I and II (Fig. 2A), indicating that BT featured a lineage-specific expansion of RwFAR1/FRS-like genes.
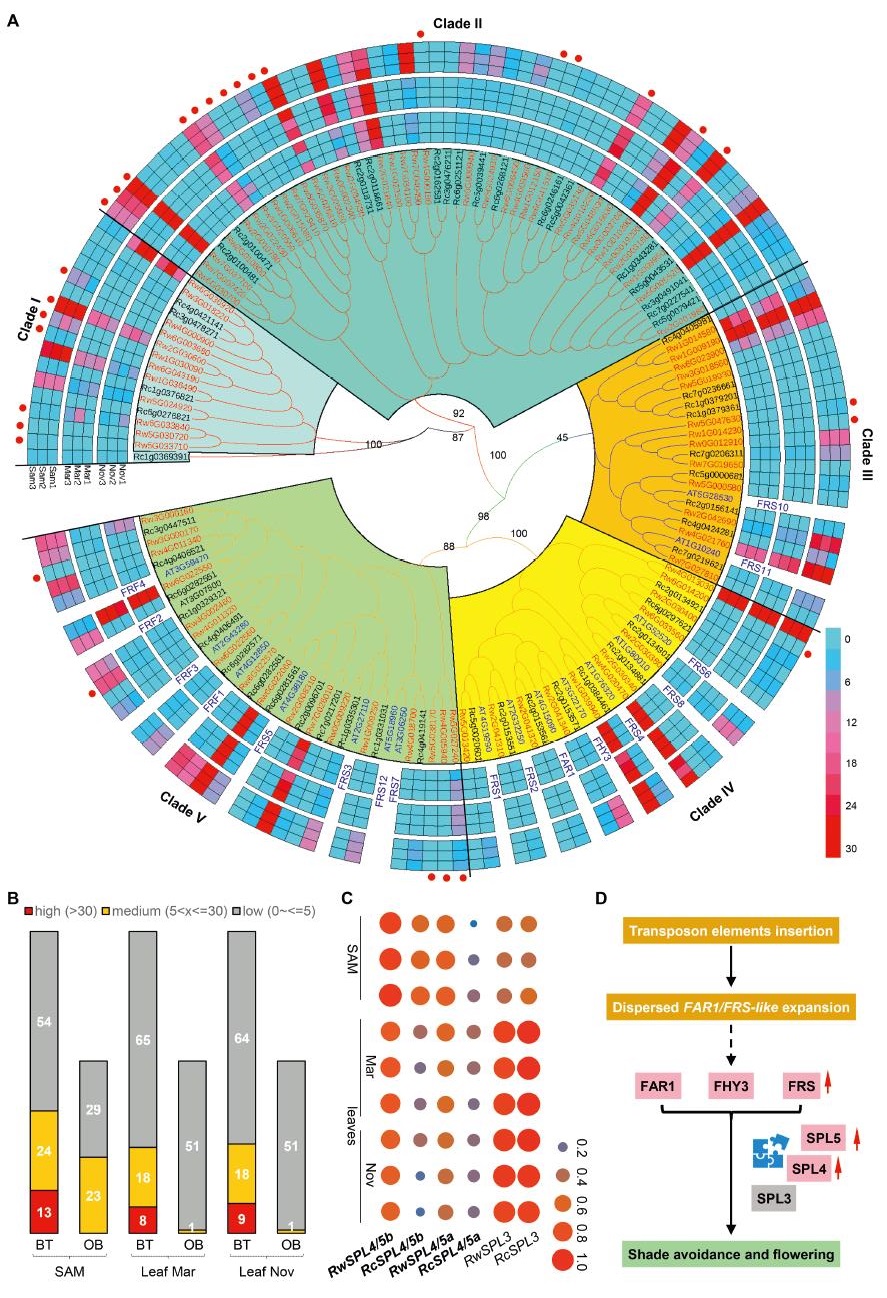
|
| Fig. 2 Lineage-specific expansion and expression diversification of FAR1/FRS-like genes between Rosa wichuraiana 'Basye's Thornless' (BT) and R. chinensis 'Old Blush' (OB). (A) Lineage-specific expansion and expression diversification of FAR1/FRS-like genes. A Neighbor-Joining tree is shown with numbers on branches indicating the bootstrap supports. Five clades were identified (Clade I to V). Except for three genes (all in clade II on Chr2), all members of clades I and II were of dispersed/random origin (Fig. 1D). Roses featured two specific clades (I and II) of FAR/FRS-like genes, while sharing three (clades III, IV and V) of FAR/FRS-like genes with Arabidopsis. Circles from outside to inside indicate the relative expression levels in FPKM values in shoot apical meristem (SAM) (1–3), March leaves (leaf Mar) (1–3), and November leaves (leaf Nov) (1–3). Genes marked with red dots indicate genes that are the result of gene expansion in BT. Note that most of the OB genes showed relatively lower expression than their corresponding phylogenetic orthologs of BT. (B) Summary of the expression levels for FAR1/FRS-like genes in BT and OB. Mean FPKM values of three biological replicates per tissue were arbitrarily classified into high (> 30), medium (5 < x ≤ 30), and low (x < 5) levels. Numbers in bars indicate the number of genes per level for each genotype. (C) Expression of SPL4/5-like genes, not SPL3-like genes, differed between BT and OB. Circle size and color indicate the relative expression levels in FPKM values scaled with log2 in shoot apical meristem (SAM) and leaves (Mar and Nov; for each tissue, three biological replicates were included). (D) Hypothesized model that shows how RwFAR1/FRS-like gene expansion in roses has led to flowering time regulation and shade avoidance through the interaction of SPL-like proteins. Names in pink blocks show genes that are differentially in BT and OB, while gray blocks indicate no significant variation in gene expression. Red upward arrows indicate that gene expression is relatively higher in BT than in OB. The blue icon indicates potential dynamic protein interactions. |
In BT, several genes showed a relatively high expression (FPKM values more than 30) in SAM (13), March leaves (8) and November leaves (9). No OB genes were expressed at relatively high levels. Several OB genes were expressed at medium levels in SAM (23), March leaves (1), and November leaves (1); in contrast, there were a greater number of BT genes expressed at medium levels in SAM (24), March leaves (18), and November (18) (Fig. 2B; Table S5). Three BT genes (Rw6G036920 in clade I, Rw1G014580 in clade III, and Rw3G000170 in clade V) were only expressed in leaves (Fig. 2B; Table S5). Notably, BT paralogs were always expressed at relatively higher levels than OB genes in those expressed FAR1/FRS-like genes. Additionally, the BT expansion genes showed a variety of expression patterns, including expression restricted to either SAM or leaves, expression in both tissues, or no expression (Fig. 2A). Only two genes (Rw2G034530 in clade II and Rw3G000160 in clade V) showed differential expression between March and November leaves, indicating that these genes might have specialized function. Taken together, these expression patterns indicate that FAR1/FRS-like gene expression differs significantly in BT and OB.
3.7. Expression of FAR1/FRS-like protein interaction partners differs between BT and OBArabidopsis FAR1 and FHY3 have several known interaction partners and their interactions play essential roles in light-signaling processes (Fig. S8). However, among these potential interacting partners, only homologous genes encoding SPL4/5-like proteins showed differential expression between BT and OB in different tissues (Fig. 2C; Fig. S9; Table S6). Because the molecular interaction between FAR1/FHY3 and SPL4/5 plays an important role in flowering time regulation in Arabidopsis (Xie et al., 2020), we hypothesize that the coordinated variation in expression between RwFAR1/FRS-like genes and RwSPL4/5-like genes might play a role in mediating the switch from prostrate-to-erect growth in shoots, and therefore flowering time in BT plants (Fig. 2D).
4. DiscussionIn this study, transposon element (TE)-mediated duplication was detected, a pattern has been reported in many other plants (Cannon et al., 2004; Dodsworth et al., 2016; Flagel and Wendel, 2009; Li et al., 2017; Panchy et al., 2016; Qiao et al., 2019). We found that FAR1/FRS-like genes diversified in their gene structure, motifs, and protein domain compositions after TE-associated dispersed expansion. More interestingly, we observed strong variation in gene expression between rose genotypes for this important family of transcription factors. Previous studies have shown that various FAR1/FRS-like genes have different DNA-binding activities and functional features involved in regulating plant development and environmental adaptation (Li et al., 2016; Ma and Li, 2018; O'Malley et al., 2016; Ritter et al., 2017); thus, we speculate that variation in gene expression may account for shoot growth differences and flowering time changes between rose genotypes. Confirmation of this hypothesis requires further functional characterization. FAR1/FRS-like genes play important roles in adaptation to varying light and other environmental conditions in A. thaliana, but the evolutionary pattern of this important family of transcriptional regulators has been poorly investigated especially in woody plants. Our study thus represents one of the first examples linking FAR1/FRS-like gene evolution with mechanisms in the generation and maintenance of plant diversity (Grierson et al., 2011; Pennisi 2005).
In conclusion, the use of high-quality chromosome-level genome sequences (Hibrand Saint-Oyant et al., 2018; Raymond et al., 2018; Zhong et al., 2020) has increased our understanding of the evolutionary patterns of key transcription factors, the FAR1/FRS-like family. Our findings provide an important starting point for dissecting the molecular genetic regulation of many morphological novelties in non-model plants, especially woody plants (Dong et al., 2017). Combining data described in this research with further genetic analyses (e.g., QTL and GWAS technologies) may also offer important information for marker-assisted breeding of non-model crops with interesting traits.
Author contributionsJ-Y H and M-C Z conceptualized the project; X-D J and W–H C collected the samples, extracted the genomic DNA and total RNA. M-C Z analyzed and visualized the data. J-Y H and M-C Z drafted the manuscript with contributions from all authors. All authors have read and approved the final manuscript.
Declaration of competing interestThe authors declare no competing interest.
AcknowledgementsThis work was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences to J-Y H (XDB31000000); the CAS Pioneer Hundred Talents Program to J-Y H (292015312D11035) and Yunnan Recruitment Program of Experts in Science to J-Y H. We acknowledge the support from Dan Wang and Yi-Bo Sun. We appreciate Shubin Li, Shulan Chen, Hongyuan Yu and Yuanlin Lv for the help with rose cultivation. This work is partially facilitated by the Germplasm Bank of Wild Species of China and Kunming Botanical Garden, Kunming Institute of Botany, Chinese Academy of Sciences.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.11.002.
R.M.Amasino, S.D.Michaels, 2010. The timing of flowering. Plant Physiol., 154: 516-520. DOI:10.1104/pp.110.161653 |
F.Andres, G.Coupland, 2012. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet., 13: 627-639. DOI:10.1038/nrg3291 |
I.Baurle, C.Dean, 2006. The timing of developmental transitions in plants. Cell, 125: 655-664. DOI:10.1016/j.cell.2006.05.005 |
Bendahmane, M., Dubois, A., Raymond, O., et al, 2013. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot., 64: 847-857. DOI:10.1093/jxb/ers387 |
Bulik-Sullivan, B.K., Loh, P.R., Finucane, H.K., et al, 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet., 47: 291-295. DOI:10.1038/ng.3211 |
Byrne, D.H., Anderson, N., Pemberton, H.B., 2007. The use of Rosa wichurana in the development of landscape roses adapted to hot humid climates. in H. B. Pemberton (ed. ), Proceedings of the Ivth International Symposium on Rose Research and Cultivation (Int Soc Horticultural Science: Leuven vol. 1).
|
Cannon, S.B., Mitra, A., Baumgarten, A., et al, 2004. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol., 4: 10. DOI:10.1186/1471-2229-4-10 |
Chen, C., Chen, H., Zhang, Y., et al, 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant, 13: 1194-1202. DOI:10.1016/j.molp.2020.06.009 |
Crespel, L., Chirollet, M., Durel, C., et al, 2002. Mapping of qualitative and quantitative phenotypic traits in Rosa using AFLP markers. Theoret. Theoret. Appl. Genetics., 105: 1207-1214. DOI:10.1007/s00122-002-1102-2 |
S.Dodsworth, M.W.Chase, A.R.Leitch, 2016. Is post-polyploidization diploidization the key to the evolutionary success of angiosperms?. Bot. J. Linn. Soc., 180: 1-5. DOI:10.1111/boj.12357 |
Dong, X., Jiang, X., Kuang, G., et al, 2017. Genetic control of flowering time in woody plants: roses as an emerging model. Plant Divers., 39: 104-110. DOI:10.1016/j.pld.2017.01.004 |
D.M.Emms, S.Kelly, 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol., 20: 238. DOI:10.1186/s13059-019-1832-y |
L.E.Flagel, J.F.Wendel, 2009. Gene duplication and evolutionary novelty in plants. New Phytol., 183: 557-564. DOI:10.1111/j.1469-8137.2009.02923.x |
Fornara, F., de Montaigu, A., Coupland, G., 2010. SnapShot: control of flowering in Arabidopsis. Cell 141, 550, 550e1-e2.
|
K.A.Franklin, G.C.Whitelam, 2005. Phytochromes and shade-avoidance responses in plants. Ann. Bot., 96: 169-175. DOI:10.1093/aob/mci165 |
Gao, Y., Liu, H., An, C., et al, 2013. Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J., 75: 795-807. DOI:10.1111/tpj.12240 |
Grierson, C. S., Bames, S. R., Chase, M. W., et al, 2011. One hundred important questions facing plant science research. New Phytol., 192: 6-12. DOI:10.1111/j.1469-8137.2011.03859.x |
Hibrand Saint-Oyant, L., Ruttink, T., Hamama, L., et al, 2018. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants., 4: 473-484. DOI:10.1038/s41477-018-0166-1 |
Hu, B., Jin, J., Guo, A.Y., et al, 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics, 31: 1296-1297. DOI:10.1093/bioinformatics/btu817 |
Hudson, M., Ringli, C., Boylan, M.T., et al, 1999. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev., 13: 2017-2027. DOI:10.1101/gad.13.15.2017 |
Iwata, H., Gaston, A., Remay, A., et al, 2012. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J., 69: 116-125. DOI:10.1111/j.1365-313X.2011.04776.x |
M.Johansson, D.Staiger, 2015. Time to flower: interplay between photoperiod and the circadian clock. J. Exp. Bot., 66: 719-730. DOI:10.1093/jxb/eru441 |
Katoh, K., Misawa, K., Kuma, K., et al, 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res., 30: 3059-3066. DOI:10.1093/nar/gkf436 |
D.Kim, B.Landmead, S.L.Salzberg, 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12: 357-360. DOI:10.1038/nmeth.3317 |
Li, D., Fu, X., Guo, L., et al, 2016. FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A., 113: 9375-9380. DOI:10.1073/pnas.1602960113 |
Li, G., Siddiqui, H., Teng, Y., et al, 2011. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol., 13: 616-622. DOI:10.1038/ncb2219 |
Li, J.M., Qin, M.F., Qiao, X., et al, 2017. A new insight into the evolution and functional divergence of sweet transporters in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. Plant Cell Physiol., 58: 839-850. DOI:10.1093/pcp/pcx025 |
Li, S., Yang, G., Yang, S., et al, 2019. The development of a high-density genetic map significantly improves the quality of reference genome assemblies for rose. Sci. Rep., 9: 5985. DOI:10.1038/s41598-019-42428-y |
Li, S., Zhong, M., Dong, X., et al, 2018. Comparative transcriptomics identifies patterns of selection in roses. BMC Plant Biol., 18: 371. DOI:10.1186/s12870-018-1585-x |
Li, S., Zhou, N., Zhou, Q., et al, 2015. Inheritance of perpetual blooming in Rosa chinensis 'Old Blush'. Hortic. Hortic. Plant J., 1: 108-112. |
Lin, R., Ding, L., Casola, C., et al, 2007. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science, 318: 1302-1305. DOI:10.1126/science.1146281 |
R.Lin, H.Wang, 2004. Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol.. Plant Physiol., 136: 4010-4022. DOI:10.1104/pp.104.052191 |
Liu, Y., Ma, M., Li, G., et al, 2020. Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell, 32: 1464-1478. DOI:10.1105/tpc.19.00981 |
Liu, Y., Wei, H., Ma, M., et al, 2019. Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell, 31: 2089-2106. DOI:10.1105/tpc.18.00991 |
Liu, Y., Xie, Y., Wang, H., et al, 2017. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell, 29: 2269-2284. DOI:10.1105/tpc.17.00268 |
L.Ma, G.Li, 2018. FAR1-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) family proteins in Arabidopsis growth and development. Front. Plant Sci., 9: 692. DOI:10.3389/fpls.2018.00692 |
Ma, L., Tian, T., Lin, R., et al, 2016. Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant, 9: 541-557. DOI:10.1016/j.molp.2015.12.013 |
Ma, L., Xue, N., Fu, X., et al, 2017. Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol., 213: 1682-1696. DOI:10.1111/nph.14300 |
A.C.McCormac, M.J.Terry, 2002. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J., 32: 549-559. DOI:10.1046/j.1365-313X.2002.01443.x |
T.Munnik, E.Nielsen, 2011. Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol., 14: 489-497. DOI:10.1016/j.pbi.2011.06.007 |
R.C.O'Malley, S.S.C.Huang, L.Song, 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 165: 1280-1292. DOI:10.1016/j.cell.2016.04.038 |
Ouyang, X., Li, J., Li, G., et al, 2011. Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell, 23: 2514-2535. DOI:10.1105/tpc.111.085126 |
N.Panchy, M.Lehti-Shiu, S.H.Shiu, 2016. Evolution of gene duplication in plants. Plant Physiol., 171: 2294-2316. DOI:10.1104/pp.16.00523 |
ElizabethPennisi, 2005. What determins species diversity?. Science, 309: 90. DOI:10.1126/science.309.5731.90 |
Pertea, M., Pertea, G.M., Antonescu, C.M., et al, 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol., 33: 290-295. DOI:10.1038/nbt.3122 |
Qiao, X., Li, Q.H., Yin, H., et al, 2019. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol., 20: 38. DOI:10.1186/s13059-019-1650-2 |
Randoux, M., Daviere, J.M., Jeauffre, J., et al, 2014. RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol., 202: 161-173. DOI:10.1111/nph.12625 |
Randoux, M., Jeauffre, J., Thouroude, T., et al, 2012. Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J. Exp. Bot., 63: 6543-6554. DOI:10.1093/jxb/ers310 |
Raymond, O., Gouzy, J., Just, J., et al, 2018. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet., 50: 772-777. DOI:10.1038/s41588-018-0110-3 |
Ritter, A., Inigo, S., Fernández-Calvo, P., et al, 2017. The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat. Commun., 8: 15235. DOI:10.1038/ncomms15235 |
S.Saha, S.Bridges, Z.V.Magbanua, D.G.Peterson, 2008. Empirical comparision of ab initio repeat finding programs. Nucleic Acids Res., 36: 2284-2294. DOI:10.1093/nar/gkn064 |
Shupert, D.A., Byme, D.H., Pemberton, H.B., 2007. Inheritance of flower traits, leaflet number and prickles in roses. in H. B. Pemberton (ed. ), Proceedings of the Ivth International Symposium on Rose Research and Cultivation (Int Soc Horticultural Science: Leuven vol. 1).
|
Siddiqui, H., Khan, S., Rhodes, B.M., et al, 2016. FHY3 and FAR1 act downstream of light stable phytochromes. Front. Plant Sci., 7: 175. |
Spiller, M., Linde, M., Hibrand-Saint Oyant, L., et al, 2011. Towards a unified genetic map for diploid roses. Theoret. Appl. Genetics., 122: 489-500. DOI:10.1007/s00122-010-1463-x |
A.Stamatakis, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Sun, Y.B., Zhang, X.J., Zhong, M.C., et al, 2020. Genome-wide identification of WD40 genes reveals a functional diversification of COP1-like genes in Rosaceae. Plant Mol. Biol., 104: 81-95. DOI:10.1007/s11103-020-01026-7 |
Tang, W., Ji, Q., Huang, Y., et al, 2013. FAR-RED ELONGATED HYPOCOTYl3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol., 163: 857-866. DOI:10.1104/pp.113.224386 |
Tang, W., Wang, W., Chen, D., et al, 2012. Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell, 24: 1984-2000. DOI:10.1105/tpc.112.097022 |
Wang, W., Tang, W., Ma, T., et al, 2016. A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol., 58: 91-103. DOI:10.1111/jipb.12369 |
Wang, Y.P., Tang, H.B., DeBarry, J.D., et al, 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res., 40: e49. DOI:10.1093/nar/gkr1293 |
Xie, Y., Zhou, Q., Zhao, Y., et al, 2020. FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering. Mol. Plant, 13: 483-498. DOI:10.1016/j.molp.2020.01.013 |
Zhang, R., Yang, C., Jiang, Y., et al, 2019. A PIF7-CONSTANS-Centered molecular regulatory network underlying shade-accelerated flowering. Mol. Plant, 12: 1587-1597. DOI:10.1016/j.molp.2019.09.007 |
Zheng, Y., Jiao, C., Sun, H.H., et al, 2016. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant, 9: 1667-1670. DOI:10.1016/j.molp.2016.09.014 |
Zhong, M. -C., Jiang, X. -D., Yang, G. -Q. et al., 2020. Genomic hitchhiking with moisture adaptation patterns stem prickleless in rose. bioRxiv. https://doi.org/10.1101/2020.07.16.207795
|



