b. Plant Germplasm and Genomics Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China
Manganese (Mn) is an essential constitutive element required by humans, animals, and plants. In plants, Mn is necessary for light-induced water oxidation during photosynthesis, and is required as a cofactor for various enzymes including Mn-superoxide dismutase (MnSOD) (Nickelsen and Rengstl, 2013). Under Mn-deficient conditions, plants show symptoms of decreased growth and yield, increased susceptibility to pathogens, and damage due to cold stress (Li et al., 2017). However, Mn, which is mainly taken up in the form of Mn2+ by the roots of plants, accumulates rapidly in acidic soil due to improper farming practices and environmental pollution (Guo et al., 2010; You et al., 2017). Excess Mn concentration in the above-ground tissues of plants acts as a toxic heavy metal that limits crop production and quality (Li et al., 2019). Mn toxicity may affect the growth and development of plants through various physiological processes, such as inhibition of photosynthesis, accumulation of reactive oxygen species (ROS), disruption of the activities of several key enzymes, and impairment of the uptake and transport of other mineral elements (Ducic and Polle, 2005; Lei et al., 2007; Li et al., 2010; Millaleo et al., 2010). Mn toxicity causes chlorosis and necrosis in plant leaves and death (Huang et al., 2019). Moreover, excess Mn in farmland can cause damage to animals and threaten human health, for example, by affecting the central nervous system and causing the development of Parkinson-like disorders (Lucchini et al., 2017; Nagajyoti et al., 2010). Consequently, a reliable method is required for the removal of excess Mn from environments that contain plants.
Phytoremediation is a safe and sustainable method to mitigate the harmful effects of heavy metals by reducing soil metal concentrations (Marques et al., 2009). Previous studies have identified plants with high Mn tolerance (e.g., Broussonetia papyrifera) that may serve as phytoremediators in Mn-contaminated soils (Liu et al., 2008; Pan et al., 2018; Huang et al., 2019). These Mn-tolerant plants have evolved multiple molecular, biochemical, and cellular strategies to prevent the adverse effects of excessive Mn (Shao et al., 2017). Research shows that numerous plant species cope with Mn toxicity through activation of the antioxidant system (Gonzalez et al., 1998; Fecht-Christoffers et al., 2006; Sheng et al., 2016). In addition, plants such as rice and Arabidopsis thaliana tolerate high Mn stress effectively by regulating the uptake, translocation, and distribution of Mn (Peiter et al., 2007; Shao et al., 2017; Tsunemitsu et al., 2018). For instance, Mn uptake has been shown to be alleviated through chelation of free Mn ions by organic and inorganic compounds (Chen et al., 2015, 2016). Plants can reportedly compartmentalise excess Mn into the apoplasts, vacuoles, endoplasmic reticulum, Golgi apparatus, and cell wall (Takemoto et al., 2017). In rice and A. thaliana, the sequestration of Mn into vacuoles for Mn homeostasis is mediated by the metal tolerance protein (MTP) (Yang et al., 2019).
Recent years have seen the identification of numerous genes and proteins involved in the regulation of Mn uptake and translocation and integration of specific signal pathways of Mn detoxification (Delhaize et al., 2003; Yamaji et al., 2013). In rice, excess Mn is transported from the apoplast by OsYSL6 (Sasaki et al., 2011). In Arabidopsis thaliana, mutant and overexpression studies have shown that AtMTP11 expression is positively associated with Mn tolerance (Delhaize et al., 2007). Leaf cDNA-amplified fragment length polymorphism and proteomic analyses have shown that, in plants with high tolerance to long-term Mn toxicity, phenylpropanoid pathways, photosynthesis, cell transport, and secondary metabolism are altered (Liu et al., 2019; You et al., 2014, 2017; Zhou et al., 2013, 2017). However, plants must adapt to wide fluctuations in soil Mn concentrations, which vary temporally and spatially from 20 to 3000 mg kg −1 (Shao et al., 2017). To date, few studies have examined rapid stress responses of plants to Mn toxicity, or the detoxification response of plants when Mn concentrations return to normal levels. Thus, examining the genetic mechanisms that mediate the rapid detoxification and recovery response of plants to excess Mn concentrations and their subsequent reduction may provide valuable insight into how crop cultivars adapt to complex and changeable environments. In addition, understanding the physiological and biochemical mechanisms by which plants cope with Mn toxicity may improve strategies for phytoremediation.
Arabis paniculata, which is widely distributed in mining areas of Yunnan Province, China (Tang et al., 2016), has the ability to accumulate heavy metal ions, including cadmium (Cd), lead (Pb), and zinc (Zn) (Tang et al., 2009). However, it is currently unclear whether A. paniculata can accumulate Mn, and if so, how it responds to excess Mn stress at the physiological and transcriptome levels. Previous studies have used RNA sequencing (RNA-Seq) technology to analyse the complex response of plants to environmental stress (Führs et al., 2008; Stark et al., 2019), and identify numerous genes that play roles in plant responses to Mn toxicity (Li et al., 2019).
In this study, we asked whether Arabis paniculata has high Mn tolerance and accumulation characteristics, and if so, what genes may be involved in its Mn toxicity responses. To answer these questions, we first compared Mn tolerance and accumulation in A. paniculata and Arabidopsis thaliana. We then examined how gene expression in A. paniculata responded to excess Mn and the ensuing recovery back to normal Mn concentration. Several critical responsive pathways and genes involved in Mn tolerance in A. paniculata were analyzed. The results of this study may provide a new perspective on the Mn-responsive mechanisms of hyperaccumulation.
2. Materials and methods 2.1. Plant growth and treatmentsSeeds of Arabis paniculata and Arabidopsis thaliana ecotype Columbia were sterilised with ethanol (75%, v/v) and sodium hypochlorite (5%, v/v) for 2 min each, then washed with sterile distilled water thrice. The seeds were surface sterilised and then stratified for 2 days at 4 ℃ before being sown on Murashige and Skoog (MS) medium containing 1% (w/v) sucrose. The seeds were germinated and grown at 22 ℃ with a 12-h light/12-h dark photoperiod and photosynthetic photon flux density of 120 μmol m−2·s−1 for 7 days. Fifteen seedlings each were transferred to a Petri dish with MS agar containing 0 mM (control), 1 mM or 10 mM MnCl2. After 3 days of growth, the seedlings were subjected to MS agar with normal Mn concentration (required for plants) for recovery. All experiments were conducted with five biological replicates.
To test the germination rate, sterilised seeds were transferred to a Petri dish with MS agar containing 10 mM or 20 mM MnCl2. The number of seeds that germinated was counted after 5 days at 22 ℃ with a 12-h light/12-h dark photoperiod and photosynthetic photon flux density of 120 μmol m−2·s−1.
2.2. Determination of Mn contentTo determine the Mn content of roots and shoots in mature plants, 7-day-old seedlings were transferred to a box with Hogland solution and 24-day-old plants grown hydroponically were treated with 0.1 mM MnCl2 for 72 h (Arteca and Arteca, 2000). Plants were then harvested and washed twice with ultrapure (> 18 MΩ cm−1) water, 5 mM EDTA (pH 5.0), and ultrapure water again in succession, followed by drying for 48 h at 80 ℃. All samples were digested in 1 mL 70% nitric acid at 100 ℃ for 2 h, and diluted with ultrapure water to 14 mL for metal determination. Ion contents in samples were determined using inductively coupled plasma mass spectrometry (NexION 300 ICP-MS, PerkinElmer).
2.3. RNA isolation and sequencingArabis paniculata seedlings were harvested and stored in liquid nitrogen after 1 day and 3 days of 1 mM MnCl2 treatment, and 1 day of recovery on normal growth medium. TRIzol reagent was used to extract total RNA according to the supplier's instructions (Invitrogen, Natacor, Argentina). RNA concentration was measured using a Nanodrop 2000 spectrophotometer (Agilent RNA 6000 Nano Kit; Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA integrity number above 6.5 were selected for subsequent steps. Input material for each RNA sample required 3 μg total RNA. Enrichment of poly(A) (+) mRNA was carried out using immobilized Oligo (dT). Ribosomal RNA (rRNA) removal was performed through hybridisation of rRNA using a DNA probe. Expected RNA was obtained after digestion of the DNA probe with DNaseI and a purifying step. The desired RNA in participation with the random N6 primer was used to produce double-stranded DNA (dscDNA). The ends of dscDNA were repaired with phosphate at the 5′ end and an A nucleotide at the 3′ end for adaptor ligation. Two specific primers were used to amplify the ligation product. The PCR product was denatured by heat, and the single-stranded DNA was cyclised by splint oligo and DNA ligase. Sequencing was performed on the BGISEQ platform and paired-end reads were generated.
2.4. Transcriptome assembly and gene annotationRaw data were presented in Fastq format and operated with in-house Perl scripts. In this step, clean reads were generated by removing reads containing adapter, reads containing poly-N, and low-quality reads from raw data (Q < 20) (Zhai et al., 2015). Then the Q20, Q30, GC content, and sequence duplication level of the clean data were analysed (Pei et al., 2016). All downstream analyses were derived from high-quality clean data. Transcriptome assembly was completed using the Trinity v2.0.6 de novo transcriptome assembler. Gene function was annotated based on the following seven databases: KEGG, GO, National Center for Biotechnology Information (NCBI) non-redundant protein sequences (Nr), NCBI non-redundant nucleotide sequences (Nt), SwissProt (a manually annotated and reviewed protein sequence database), Protein family (Pfam), and euKaryotic Orthologous Groups (KOG). The software and parameters for Nr, Nt, KOG, and Swiss-Prot analysis were NCBI blast 2.2.23 with default settings. The HMMSCAN 3.0 package (default) was used for Pfam prediction.
2.5. Quantification of gene expression levels and differential expression analysisGene expression levels were calculated for each sample by RNA-Seq using the Expectation Maximization software package (Dewey and Li, 2011). Clean data were mapped back onto the assembled transcriptome using Bowtie v2.0.6. The read count for each gene was generated from the mapping results described above (Dewey and Li, 2011). Read numbers mapped to each gene were counted through HTSeq v0.5.4p3. According to gene length and read counts mapped to an individual gene, reads per kilobase per million mapped reads were obtained. DESeq R package was used to analyse differential expression. Benjamini and Hochberg's approach was employed to modify the resulting p-values and limit the probability of false discovery. The standard of genes differentially expressed was obtained from an adjusted p-value counted by DESeq of less than 0.05. Enrichment of GO functions and KEGG orthology in the differentially expressed genes (DEGs) were analysed between control and treated groups. GO enrichment analysis of the DEGs was performed using the GOseq R package. The statistical enrichment of DEGs in KEGG pathways (false discovery rate [FDR] ≤ 0.05) was investigated using KEGG Orthology-Based Annotation System software.
2.6. Quantitative reverse transcription PCR analysisAfter 1 day and 3 days of 1 mM MnCl2 treatment, and 1 day of recovery on normal growth medium, samples of Arabis paniculata were harvested and frozen in liquid nitrogen immediately and stored at −80 ℃. Total RNA was isolated from samples using TRIzol reagent according to the supplier's instructions (Invitrogen). RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For each sample, 1 μg RNA was treated with DNaseI and then reverse-transcribed using the PrimeScriptTM RT Reagent Kit (Takara, Tokyo, Japan). Quantitative reverse transcription PCR (RT-qPCR) was carried out in a 10 μL reaction mix containing 0.2 μL of each primer (10 μM concentration), 1 μL cDNA sample, and 5 μL Ultra SYBR mixture reagent (with ROX). RT-qPCR was performed using 96-well plates on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the following programme: 95 ℃ for 1 min, 35 cycles of 95 ℃ for 15 s and 55 ℃ for 1 min, followed by melting curve analysis (60 ℃–95 ℃). The relative expression was calculated using the ΔΔCT method with Actin 1 expression for normalisation. Each measurement was repeated with five independent biological replicates, each of which was represented by three technical replicates. The primers used to study the expression of different genes were designed using Primer Premier 5 software; all relevant information is provided in Table S1.
3. Results and discussion 3.1. Phenotype and physiological performance of Arabis paniculata and Arabidopsis thaliana under excessive Mn treatmentTo investigate the resistance of A. paniculata and A. thaliana to excess Mn stress, we compared survival status between these two species after treatment with 1 mM and 10 mM MnCl2 (Fig. 1A). After 3 days of exposure to 1 mM MnCl2, there was no significant difference in the growth of these two species. However, after growing in 10 mM MnCl2 for 3 days most A. thaliana seedlings were markedly shrivelled or had even died. In contrast, when grown under the same conditions, 94.4% of A. paniculata seedlings survived and maintained their green colour. Moreover, the germination rate of A. paniculata seeds was significantly higher than that of A. thaliana seeds on medium supplemented with 10 mM and 20 mM MnCl2 (Fig. 1C). These results suggest that at several stages of development A. paniculata has a greater tolerance to extreme Mn toxicity than does A. thaliana.
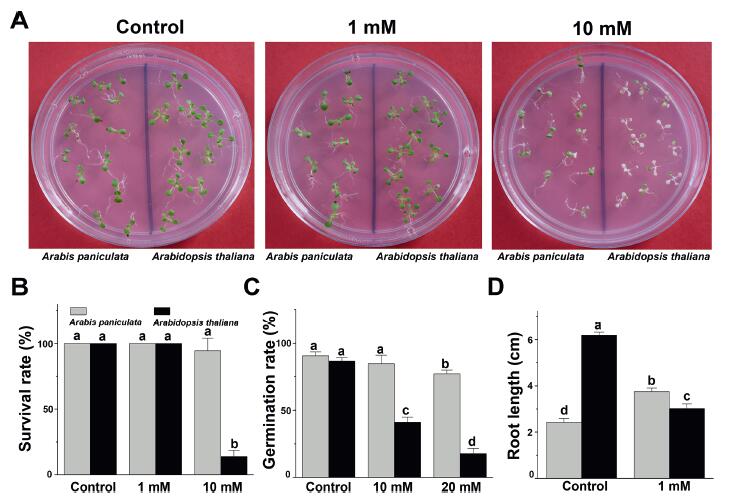
|
| Fig. 1 Arabis paniculata tolerance under Mn toxicity. (A) Phenotype of Arabis paniculata (left) and Arabidopsis thaliana (right) seedlings grown in medium containing 0 mM, 1 mM or 10 mM MnCl2 for 3 days; (B) Survival rate of Arabis paniculata and Arabidopsis thaliana seedlings grown in medium containing 0, 1, or 10 mM MnCl2 for 3 days; (C) Germination rate of Arabis paniculata and Arabidopsis thaliana seedlings grown in medium containing 0, 10, or 20 mM MnCl2 for 5 days; (D) Main root length of Arabis paniculata and Arabidopsis thaliana seedlings grown in medium containing 1 mM MnCl2 for 7 days. The letters a, b, c, and d represent significant differences according to one-way analysis of variance (ANOVA) (p < 0.05). Error bars indicate the standard error of the mean (SEM) (n = 5). |
To further examine Mn tolerance in A. paniculata, we compared the effects of moderate Mn stress on main root growth in A. paniculata and A. thaliana seedlings (Fig. 1D). Under normal conditions, the main root of A. thaliana was much longer than that of A. paniculata. However, after 7 days of 1 mM MnCl2 treatment, the root length of A. thaliana was 48.8% of the length of the control; in contrast, under the same MnCl2 treatment, the root of A. paniculata was slightly but significantly longer than that of control seedlings (p < 0.05). These findings suggest that 1 mM Mn is not stressful for A. paniculata; rather, this concentration of Mn may help absorb nutrition.
3.2. Comparison of Mn accumulation capacity between Arabis paniculata and Arabidopsis thalianaPlant heavy metal tolerance can also be determined directly by heavy metal accumulation (Zhuo et al., 2009). We determined the Mn accumulation and translocation capacity of Arabis paniculata and Arabidopsis thaliana by measuring Mn content in the stems and roots of these two species grown hydroponically and treated with 0.1 mM MnCl2 for 72 h (Fig. 2). In the roots, the Mn content of A. thaliana was 2.3 times that of A. paniculata (364.2 ± 68.8 vs. 158.0 ± 53.9 μg/g). In the shoots, the Mn content of A. paniculata was nearly twice that of A. thaliana (1480.6 ± 200.7 vs. 754.0 ± 74.2 μg/g).
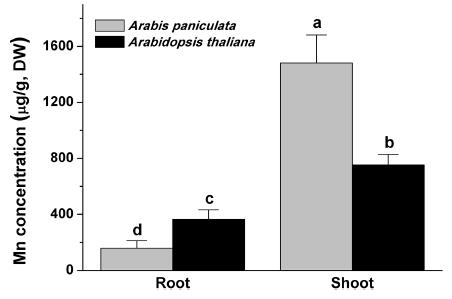
|
| Fig. 2 Mn content in the root and shoot of Arabis paniculata and Arabidopsis thaliana grown hydroponically and treated with 0.1 mM MnCl2 for 72 h. The letters a, b, c, and d represent significant differences according to one-way ANOVA (p < 0.05). Error bars indicate the SEM (n = 5). |
Generally, accumulation of Mn in the shoots was much higher than it was in the roots of the two plants. The translocation factor (TF) values of Mn to the aerial parts in Arabis paniculata and A. thaliana were 9.4 and 2.1, respectively. These results indicate that the translocation and accumulation capacities of A. paniculata are much higher than those of A. thaliana. In fact, A. paniculata has been considered a co-hyperaccumulator of multiple heavy metals, based on the fact that the TF values of Pb, Zn, and Cd in their rhizospheric soils are 1.96, 1.98, and 1.45, respectively (Tang et al., 2009). Thus, A. paniculata may be a newly discovered hyperaccumulator of Mn, with even more effective modulation of the uptake and transport of Mn than other heavy metals such as Pb, Zn, and Cd.
3.3. Transcript sequencing and assembly of Arabis paniculataTo explore the resistance mechanisms of Arabis paniculata to excess Mn, we profiled gene expression during and after recovery from Mn stress. A. paniculata seedlings showed slow growth following transfer from medium containing 1 mM MnCl2 to normal MS medium for 7 days, as shown in Fig. 1. Concurrently, the root growth of A. thaliana was significantly inhibited. Therefore, transcriptome analysis of A. paniculata was performed during exposure to and recovery from 1 mM MnCl2. The seedlings were sampled at four different points: before treatment (control), after 1 day of 1 mM MnCl 2 treatment, after 3 days of 1 mM MnCl2 treatment, and 1 day after 3 days of 1 mM MnCl2 (recovery). A range of 45.3–49.3 million clean reads were produced after removing reads covered with adapters or poly-N and low-quality reads from 51.7 to 58.8 million raw reads in total. The GC percentage was 43.1%. A total of 61, 627 unigenes with a total length of 69, 862, 281 base pairs (bp), a mean length of 1133 bp, and an N50 length of 1530 bp were assembled by the Trinity program from 12 samples. The length distribution of all the unigenes is shown in. The unigenes were annotated based on seven databases to generate comprehensive information on gene function. Of the 61, 627 unigenes, 41, 542 (67.4%), 44, 522 (72.2%), 41, 591 (67.5%), 40, 283 (65.4%), and 39, 297 (63.8%) were aligned in SwissProt, KOG, KEGG, Pfam, and GO, respectively.
3.4. Gene expression patterns during response of Mn toxicity and recovery stageTo determine the transcriptome response of A. paniculata to excessive Mn, we first analyzed the gene expression patterns during the response to Mn toxicity and recovery. Twelve gene clusters were generated by Mfuzz based on the loose clustering algorithm (Fig. 3); 80.4% of the total transcripts belonged to profiles 1–7, and profiles 9 and 12 reflected positive responses to Mn toxicity (Fig. 3A), which were divided into four general categories based on their expression patterns: initial, dose-dependent, stable, and lineage responses. In total, 15.8% of all transcripts belonging to profiles 8, 10, and 11 were downregulated by Mn toxicity.
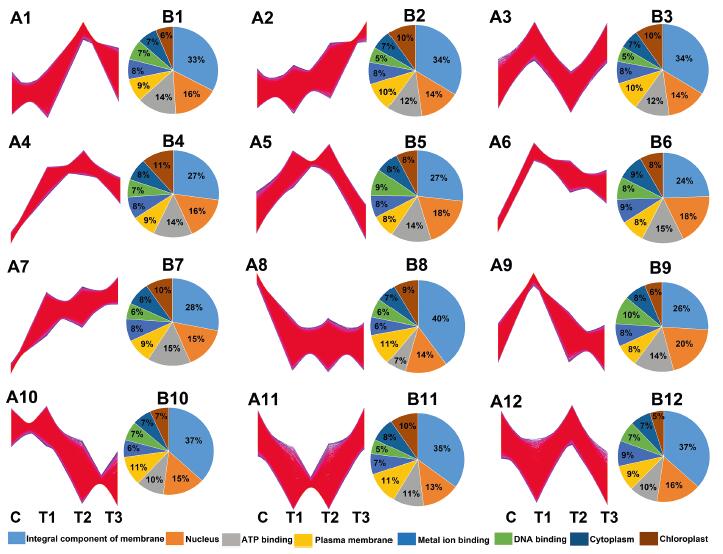
|
| Fig. 3 Analysis of Mn response genes. (A) Expression profiles of Mn response genes (A1–A12). (B) GO biological processes, cellular components, and molecular functions enriched (FDR < 0.05) among Mn response genes indicated in (A) (B1–B12). |
In all, 16, 269 (26.4% of the total) transcripts belonging to profiles 3, 6, and 9, which were initially significantly induced after 1 day of Mn stress and rapidly declined on the subsequent 3 days of Mn stress, were considered likely to be involved in the initial response. The functions of these three clusters of initial response genes were associated with the top three in terms of the number of genes involved, including integral component of membrane, nucleus, and ATP binding.
In total, 6, 258 (10.2% of the total) transcripts in profiles 1 and 12 did not change with initial Mn stress treatment, but showed significantly induced expression after 3 days of Mn stress and were predicted to be involved in dose-dependent response (Fig. 3A). Genes associated with the integral component of the membrane and responses to chitin were the most enriched in the dose-dependent response.
In total, 15, 493 (25.1% of the total) transcripts with stable responses (profiles 4 and 5) were significantly increased at initial treatment and maintained high levels during the subsequent extension of time with Mn treatment. The functions of these stable response genes were the most enriched in chloroplast and ATP binding. This gene expression pattern was consistent with the fact that the seedlings became much greener after 3 days of Mn treatment compared to the control (Fig. 1).
Lineage response (profiles 2 and 7) expression patterns of 11, 501 transcripts (18.7% of the total) increased continuously from initial treatment and subsequent prolonged Mn treatment and throughout the recovery period. In the lineage response, intracellular protein transport, chloroplast, and cytosol were the most enriched over the process as a whole. These expression patterns support the patterns of gradual growth of A. paniculata under Mn treatment.
Taken together, these results suggest that the vast majority of transcripts were affected by Mn toxicity. Furthermore, of the four general categories of responses, initial and stable response transcripts predominated during Mn toxicity. GO analysis indicated that complex functional classifications were associated with the response and recovery process of Mn toxicity, including membrane, chloroplast, protein transport, and ATP binding. Previous research has reported that dominant proteomic changes in stylo plants under Mn toxicity are associated with the GO molecular function terms of catalytic activity and binding, and the cellular component terms of cell part and membrane (Liu et al., 2019). The functional classifications inA. paniculata under Mn toxicity were roughly consistent with those in stylo plants, which suggests that this functional cataloguing of Mn-responsive genes may also reveal insights into the molecular process that underlies Mn tolerance in A. paniculata.
3.5. KEGG enrichment analysis of DEGs from excessive Mn to normal levelsTo identify biochemical pathways that regulate plant responses to Mn toxicity, KEGG pathway enrichment analyses of DEGs were performed (Fig. 4). Unigenes with significant changes (p < 0.05) in expression and expression changes greater than 1.5-fold were deemed to be regulated by Mn toxicity. We identified 22, 964 initial response DEGs (defined as genes for which transcript levels on day 1 of the Mn stress period were significantly different from their levels before Mn treatment), 23, 006 dose-dependent response DEGs (genes for which transcript levels on day 3 of the Mn stress period were significantly different from their levels before Mn treatment), and 18, 447 repair DEGs (genes for which expression during 1 day of recovery under normal growth conditions was significantly different from their levels before Mn treatment and without response genes).
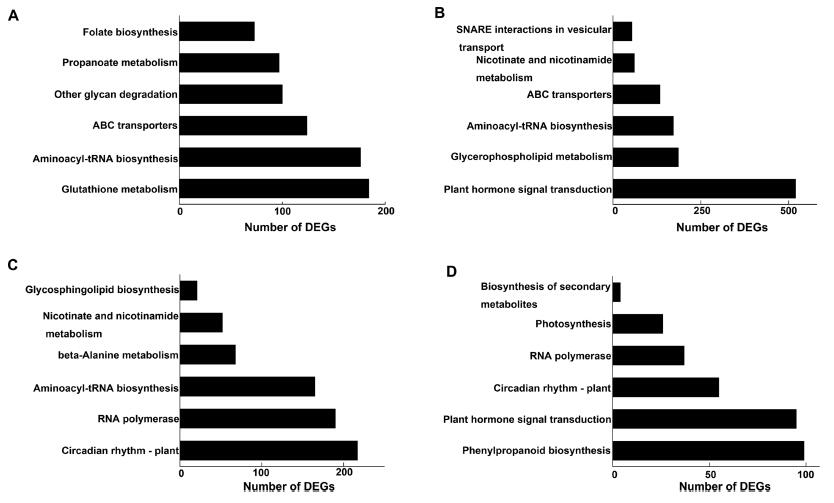
|
| Fig. 4 KEGG pathway enrichment analysis of the DEGs identified in initial response (Control vs. Treat1, A), dose-dependent response (Control vs Treat2, B), and repair process (Control vs. Treat3, C and Treat2 vs. Treat3, D). Control, before treatment; Treat1, 1 mM MnCl2 for 1 day; Treat2, 1 mM MnCl2 for 3 days; and Treat3 (recovery), 1 day after 3 days of 1 mM MnCl2. |
As shown in Fig. 4A, the dominant KEGG pathways during the initial response of A. paniculata to Mn stress include glutathione metabolism, aminoacyl-tRNA biosynthesis, and ABC transporters. Plant hormone signal transduction, glycerophospholipid metabolism, and aminoacyl-tRNA biosynthesis were the top three enriched pathways among the dose-dependent response DEGs (Fig. 4B). Circadian rhythm-plant and phenylpropanoid biosynthesis were the most enriched and had the greatest involvement in the repair process (Fig. 4C and D).
These results indicate that the response and repair process of A. paniculata under Mn stress is probably associated with glutathione metabolism, ABC transporters, glycerophospholipid metabolism, and phenylpropanoid biosynthesis. This supports previous findings that xenobiotic/antioxidative defence is involved in the response of A. paniculata to Cd stress and carbon metabolism (Zeng et al., 2011), and that phenylpropanoid biosynthesis participates in the response of stylo plants to Mn stress (Liu et al., 2019).
Although the A. paniculata transcripts that responded to Mn toxicity belong to similar KEGG pathways as the proteins that respond to Cd stress in A. paniculata and Mn-response pathways in stylo plants, these the characteristic expression patterns of these transcripts indicate that A. paniculata response to Mn toxicity is distinct.
3.6. Identification of genes that respond and recover from Mn toxicityOur GO and KEGG enrichment analyses indicate that Mn-response processes involve a complex gene regulatory network. Excess Mn in plants triggers ROS and oxidative stress (Li et al., 2019), which activates antioxidant enzymes such as glutathione reductase (GR), glutathione S-transferase (GST), non-enzymatic ascorbic acid, and glutathione (Sheng et al., 2016). In alfalfa, glutathione metabolism is known to enhance and improve Cd tolerance through activation of antioxidation and Cd chelation (Cui et al., 2020). We, therefore, profiled expression patterns of genes involved in glutathione metabolism in A. paniculata to explore their physiological functions under Mn toxicity (Fig. 5A). Glutathione metabolism had the most DEGs during the initial response stage. A large number of unigenes encoding GST (19 unigenes), glutathione peroxidase (3 unigenes), and glutathione synthase (1 unigene) were induced several-fold on day 1 of Mn treatment compared with the transcript levels of these genes before treatment, and then declined or were maintained with prolonged Mn treatment. This expression pattern was similar for profiles 6 and 9.
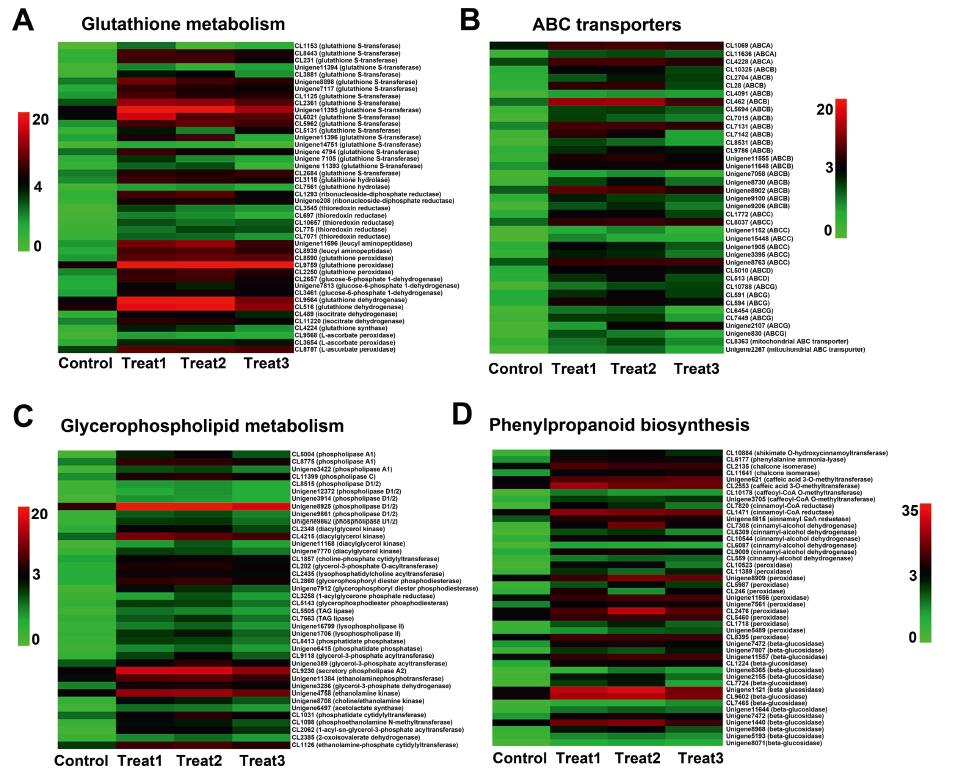
|
| Fig. 5 Overview of genes involved in glutathione metabolism (A), ABC transporters (B), glycerophospholipid metabolism (C), and phenylpropanoid biosynthesis (D). Highly expressed genes were selected and are listed (log2 (Goup1/Gourp2) > 1.5). |
In hyperaccumulators, glutathione mediates the biosynthesis of salicylic acid and jasmonate to improve Ni tolerance and Cd tolerance (Freeman et al., 2005; Tolrà et al., 2006). GSTs are a large complex family of enzymes that play vital roles in plant growth and development and tolerance to heavy metal stress (Gao et al., 2020). We therefore speculate that antioxidant enzymes and nonenzymatic glutathione are initiated as part of the early response of A. paniculata in alleviating the toxic effects of excess Mn, which might be a characteristic of A. paniculata due to its high Mn tolerance.
ABC transporters, a large group of plant proteins known to play diverse roles in growth and stress response (Andolfo et al., 2015), may be critical to plant resistance to heavy metal stress. ABC transporters are composed of two transmembrane domains and two cytosolic nucleotide-binding domains. They are classified into five subfamilies (ABC subfamily A, B, C, D, and G) according to sequence homology, domain organisation, and phylogenetic position (Lane et al., 2016). We found that ABC transporters were differentially expressed in both the initial and dose-dependent response to Mn treatment (Fig. 4). The gene expression profiles of ABC transporters were identified in order to explore their functions in Mn transport (Fig. 5B). ABC subfamily B (ABCB, 19 unigenes), ABC subfamily C (ABCC, 7 unigenes), ABC subfamily G (ABCG, 7 unigenes), and ABC subfamily A (ABCA, 3 unigenes) were up-regulated after 1 day of Mn treatment and maintained after 3 days of Mn treatment. The expression pattern was consistent with profiles 4 and 5.
Previous studies have shown that ABC transporters transport Cd in the cytoplasmic matrix or discharge Cd to the vacuole and other organelles as a type of transpiration protein (Xue et al., 2014). Expression of tobacco gene homologues for PDR8 in A. thaliana can be enhanced to detoxify Cd in tobacco plants (Fässler et al., 2011). In our study, most unigenes encoding ABC transporters of A. paniculata were upregulated during the whole response stage of Mn toxicity, consistent with the enriched functions of ATP binding and intracellular protein transport. Consequently, the translocation capacity of Mn from root to shoot is noted inA. paniculata due to the active transcript response of the ABC transporter. We speculate that ABC transporter genes might be involved in the detoxification of Mn and play essential roles in transporting excess Mn from the root to shoot in plants, which might be another feature of A. paniculata regarding its capacity for hyperaccumulation.
In plants, the barrier for Mn entry into cells and the first cellular site to sustain damage is the plasma membrane. Excessive Mn stress causes cell membrane lipid peroxidation in B. papyrifera (Huang et al., 2019). Phospholipids are the major supporting structure of the cell membrane, and phospholipid content and composition differ in their response to various stresses (Hong et al., 2016). Phospholipids can be hydrolysed into diverse products such as phosphatidic acid (PA), diacylglycerol (DAG), and lysophospholipids by phospholipase D (PLD), phospholipase C (PLC), and phospholipase A (PLA). DAG can be phosphorylated into PA by DAG kinase (DGK). A variety of stressors activate PLD and PLC in plants and specific PLD family members also respond positively to stress (Hong et al., 2008a). To discover associations between Mn toxicity and the cell membrane of A. paniculata, we examined gene expression profiles of genes involved in glycerophospholipid metabolism (Fig. 5C). After 3 days of Mn treatment, the transcript levels of PLD (6 unigenes), PLA (3 unigenes), PLC (1 unigene), and DGK (4 unigenes) increased significantly. These results indicate that dose-dependent Mn treatment can stimulate the response of the cell membrane. Activation of these enzymes during the dose-dependent response stage can improve the accumulation of PA and lysophospholipid.
The relationship between Mn toxicity and glycerophospholipid metabolism of the cell membrane might be established based on PA and lysophospholipid. PLDα1-mediated PA formation regulates desiccation sensitivity in seeds (Chen et al., 2018) and is of great importance in drought stress for the entire plant (Hong et al., 2008b). PLDγ inhibits the tolerance of A. thaliana to aluminum toxicity through membrane lipid modulation (Jian et al., 2011). In this study, A. paniculata unigenes encoding PLD, PLC, and DGK were markedly induced by Mn toxicity, which is consistent with the function of the integral component of the membrane during the Mn response stage. Abundant PA production is catalysed by PLD and DGK, which might be responsible for the notable tolerance of A. paniculata to Mn toxicity. Given that Mn toxicity produces such a severe effect in plant cells, A. paniculata might modulate the compositions of cell membrane lipids to deal with this threat.
Plants respond to metal stress by accumulating and secreting secondary metabolites (Fuhrs et al., 2012). Several important secondary metabolites are known to be synthesised under Mn toxicity, such as lignin and flavonoids in rice (Lidon et al., 2004), and callose and phenolics in cowpea (Fecht-Christoffers et al., 2006). Flavonoids are the most abundant polyphenolic secondary metabolites in plants and carry various biochemical benefits including antioxidant (Pandey and Mishra, 2012), anticancer (Brusselmans et al., 2005), and antiviral properties (Mishra et al., 2013; Cushnie and Lamb, 2005). Given that flavonoids play important roles in the response of most plants to abiotic stresses, we examined the gene expression profiles of key enzymes related to phenylpropanoid biosynthesis under excess Mn (Fig. 5D). Genes involved in phenylpropanoid biosynthesis were differentially expressed most commonly in the recovery process after Mn stress. The expression levels of the following genes were induced continuously from the response stage up to the recovery period: phenylalanine ammonia lyase (PAL, 1 unigene), chalcone isomerase (CHI, 2 unigenes), CCOMT (caffeoyl-CoA O-methyltransferase, 2 unigenes), cinnamoyl-CoA reductase (CCR, 3 unigenes), cinnamyl-alcohol dehydrogenase (CAD, 4 unigenes), and shikimate O-hydroxycinnamoyltransferase (SKOMT, 1 unigene). These expression models were similar to profile 1. Through the phenylpropanoid pathway, key flavonoid biosynthesis enzymes such as PAL and CHI are activated in Stylosanthes as an adaptive response to Mn toxicity (Liu et al., 2019). The expression of PAL and CHS is also enhanced through various stressors, including Cd, Pb, and salinity (Dai et al., 2012; Sylwia et al., 2011). In this study, the unigene expression levels of PAL, CHI, CCR, CAD, and CCO continually increased in A. paniculata during both the Mn response and recovery stages, which suggests that the production of flavonoids might be activated by these critical enzymes. Flavonoids might be responsible for repair injuries and scavenge ROS caused by Mn toxicity in A. paniculata.
3.7. Gene validation of RNA-seq with quantitative PCRFifteen candidate unigenes proposed to be involved in the resistance of A. paniculata to Mn toxicity were selected for Quantitative PCR (qPCR) analysis (Fig. 6). qPCR analysis of Mn-responsive gene expression patterns was consistent with RNA-seq. For example, the unigene expression of ABC transporters such as ABCA1/3 (CL1069 and CL4228), ABCB (CL462), ABCC (CL1772 and CL8037) and ABCG (Unigene 2107) increased strongly in response to excess Mn. The candidate unigenes involved in glycerophospholipid metabolism such as PLD1/2 (Unigene 8925), PLC (CL11399) and DGK (CL4215) were upregulated under Mn toxicity. The candidate unigenes involved in phenylpropanoid biosynthesis, such as PAL (CL6177) and chalcone isomerase (CL2135 and CL11641), were both upregulated following Mn treatment. These results indicate the reliability of the RNA-seq data.
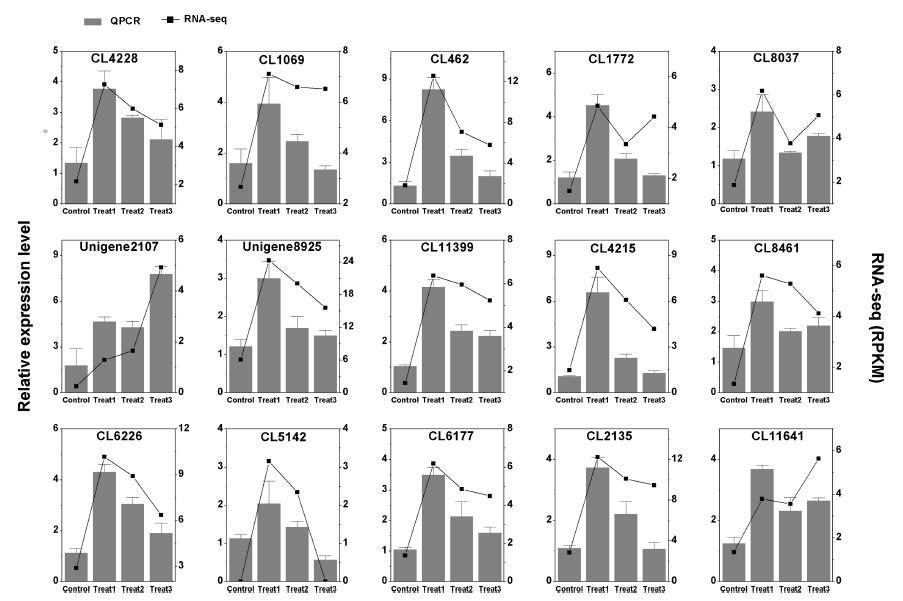
|
| Fig. 6 qPCR confirmation of 15 candidate DEGs in Arabis paniculata. |
In this study, we found that Arabis paniculata has superior tolerance to and can hyperaccumulate Mn. Our transcriptome analyses suggest that A. paniculata tolerance is likely related to early activation of glutathione metabolism to scavenge ROS, rapid adjustment of glycerophospholipid metabolism, effective transport of excess Mn from root to shoot through ABC transporter proteins, and modulation of secondary metabolite biosynthesis from the phenylpropanoid pathway for repair. These findings not only provide ideal material and genetic resources for phytoremediation in Mn-contaminated areas, but also reveal new theoretical knowledge and perspectives on mechanisms of Mn tolerance.
Author contributionsT.T. and F.Q.T. performed the experiments. W.Q.L. and T.T. designed the experiments and wrote the manuscript. The authors have read and approved the manuscript.
Declaration of Competing InterestThe authors declare that they have no conflict of interest.
AcknowledgementsThis research was supported by Natural Science Foundation of Hunan Province, China (2020JJ4293) and Scientific Research Project of the Hunan Education Department, China (18B215). The authors would like to thank Dr. Jiashi Pei for providing Mn content measurement and helpful advice. The authors would also like to thank Dr. Xudong Zhang and Dr. Yanxia Jia for providing seeds of Arabis paniculata and Arabidopsis thaliana.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.07.002.
Andolfo, G., Ruocco, M., Di Donato, A., et al., 2015. Genetic variability and evolutionary diversification of membrane ABC transporters in plants. BMC Plant Biol., 15: 51. DOI:10.1186/s12870-014-0323-2 |
R.N.Arteca, J.M.Arteca, 2000. A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol. Plantarum, 108: 188-193. DOI:10.1034/j.1399-3054.2000.108002188.x |
Brusselmans, K., Vrolix, R., Verhoeven, G., et al., 2005. Induction of cancer cell apoptosis by flavonoids is associated with their ability to inhibit fatty acid synthase activity. J. Biol. Chem., 280: 5636-5645. DOI:10.1074/jbc.M408177200 |
Chen, H., Yu, X., Zhang, X., et al., 2018. Phospholipase Dα1-mediated phosphatidic acid change is a key determinant of desiccation-induced viability loss in seeds. Plant Cell Environ., 41: 50-63. DOI:10.1111/pce.12925 |
Chen, Z., Sun, L., Liu, P., et al., 2015. Malate synthesis and secretion mediated by a manganese-enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis. Plant Physiol., 167: 176-188. |
Chen, Z., Yan, W., Sun, L., et al., 2016. Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. J. Proteomics, 143: 151-160. DOI:10.1016/j.jprot.2016.03.037 |
Cui, W., Yao, P., Pan, J., et al., 2020. Transcriptome analysis reveals insight into molecular hydrogen-induced cadmium tolerance in alfalfa: the prominent role of sulfur and (homo)glutathione metabolism. BMC Plant Biol., 20: 58. DOI:10.1186/s12870-020-2272-2 |
T.P.T.Cushnie, A.J.Lamb, 2005. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents, 26: 343-356. DOI:10.1016/j.ijantimicag.2005.09.002 |
L.P.Dai, X.J.Dong, H.H.Ma, 2012. Molecular mechanism for cadmium-induced anthocyanin accumulation in Azolla imbricata. Chemosphere, 87: 319-325. DOI:10.1016/j.chemosphere.2011.12.005 |
Delhaize, E., Gruber, B.D., Pittman, J.K., et al., 2007. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J., 51: 198-210. DOI:10.1111/j.1365-313X.2007.03138.x |
Delhaize, E., Kataoka, T., Hebb, D.M., et al., 2003. Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell, 15: 1131-1142. DOI:10.1105/tpc.009134 |
C.N.Dewey, B.Li, 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf., 12: 323. DOI:10.1186/1471-2105-12-323 |
T.Ducic, A.Polle, 2005. Transport and detoxification of manganese and copper in plants. Braz. J. Plant Physiol., 17: 103-112. DOI:10.1590/S1677-04202005000100009 |
Führs, H., Hartwig, M., Molina, L.E.B., et al., 2008. Early manganese-toxicity response in Vigna unguiculata L. - a proteomic and transcriptomic study. Proteomics, 8: 149-159. DOI:10.1002/pmic.200700478 |
Fässler, E., Plaza, S., Pairraud, A., et al., 2011. Expression of selected genes involved in cadmium detoxification in tobacco plants grown on a sulphur-amended metal-contaminated field. Environ. Exp. Bot., 70: 158-165. DOI:10.1016/j.envexpbot.2010.08.012 |
Fecht-Christoffers, M.M., Fuhrs, H., Braun, H.P., et al., 2006. The role of hydrogen peroxide-producing and hydrogen peroxide-consuming peroxidases in the leaf apoplast of cowpea in manganese tolerance. Plant Physiol., 140: 1451-1463. DOI:10.1104/pp.105.070474 |
Freeman, J.L., Garcia, D., Kim, D., et al., 2005. Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi Nickel hyperaccumulators. Plant Physiol., 137: 1082-1091. DOI:10.1104/pp.104.055293 |
Fuhrs, H., Specht, A., Erban, A., et al., 2012. Functional associations between the metabolome and manganese tolerance in Vigna unguiculata. J. Exp. Bot., 63: 329-340. DOI:10.1093/jxb/err276 |
Gao, J., Chen, B., Lin, H., et al., 2020. Identification and characterization of the glutathione S-transferase (GST) family in radish reveals a likely role in anthocyanin biosynthesis and heavy metal stress tolerance. Gene, 743: 144484. DOI:10.1016/j.gene.2020.144484 |
A.Gonzalez, K.L.Steffen, J.P.Lynch, 1998. Light and excess manganese. Implications for oxidative stress in common bean. Plant Physiol., 118: 493-504. DOI:10.1104/pp.118.2.493 |
Guo, J.H., Liu, X.J., Zhang, Y., et al., 2010. Significant acidification in major Chinese croplands. Science, 327: 1008-1010. DOI:10.1126/science.1182570 |
Hong, Y., Pan, X., Welti, R., et al., 2008a. Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell, 20: 803-816. DOI:10.1105/tpc.107.056390 |
Y.Hong, S.Zheng, X.Wang, 2008b. Dual functions of phospholipase Dα1 in plant response to drought. Mol. Plant, 1: 262-269. DOI:10.1093/mp/ssm025 |
Hong, Y., Zhao, J., Guo, L., et al., 2016. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res., 62: 55-74. DOI:10.1016/j.plipres.2016.01.002 |
Huang, H., Zhao, Y., Xu, Z., et al., 2019. Physiological responses of Broussonetia papyrifera to manganese stress, a candidate plant for phytoremediation. Ecotox. Environ. Saf., 181: 18-25. DOI:10.1016/j.ecoenv.2019.05.063 |
Jian, Z., Wang, C., Bedair, M., et al., 2011. Suppression of phospholipase Dγs confers increased aluminum resistance in Arabidopsis thaliana. PloS One, 6: e28086. DOI:10.1371/journal.pone.0028086 |
Lane, T.S., Rempe, C.S., Davitt, J., et al., 2016. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol., 16: 47. DOI:10.1186/s12896-016-0277-6 |
Y.Lei, H.Korpelainen, C.Li, 2007. Physiological and biochemical responses to high Mn concentrations in two contrasting Populus cathayana populations. Chemosphere, 68: 686-694. DOI:10.1016/j.chemosphere.2007.01.066 |
Li, J., Jia, Y., Dong, R., et al., 2019. Advances in the mechanisms of plant tolerance to manganese toxicity. Int. J. Mol. Sci., 20: 5096. DOI:10.3390/ijms20205096 |
F.C.Lidon, M.G.Barreiro, J.C.Ramalho, 2004. Manganese accumulation in rice: implications for photosynthetic functioning. J. Plant Physiol., 161: 1235-1244. DOI:10.1016/j.jplph.2004.02.003 |
Li Q., Chen L.S., Jiang H.X., et al., 2010. Effects of manganese-excess on CO2 assimilation, ribulose-1, 5-bisphosphate carboxylase/oxygenase, carbohydrates and photosynthetic electron transport of leaves, and antioxidant systems of leaves and roots in Citrus grandis seedlings. BMC Plant Biol., 10: 42. DOI:10.1186/1471-2229-10-42 |
Li, Q., Li, Y., Wu, X., et al., 2017. Metal transport protein 8 in Camellia sinensis confers superior manganese tolerance when expressed in yeast and Arabidopsis thaliana. Sci. Rep., 7: 39915. DOI:10.1038/srep39915 |
Liu, D., Zou, J., Wang, M., et al., 2008. Hexavalent chromium uptake and its effects on mineral uptake, antioxidant defence system and photosynthesis in Amaranthus viridis L. Bioresour. Technol., 99: 2628-2636. DOI:10.1016/j.biortech.2007.04.045 |
Liu, P., Huang, R., Hu, X., et al., 2019b. Physiological responses and proteomic changes reveal insights into Stylosanthes response to manganese toxicity. BMC Plant Biol., 19: 212. DOI:10.1186/s12870-019-1822-y |
Lucchini, R., Placidi, D., Cagna, G., et al., 2017. Manganese and developmental neurotoxicity. Adv. Neurobiol., 18: 13-34. |
A.P.G.C.Marques, A.O.S.S.Rangel, P.M.L.Castro, 2009. Remediation of heavy metal contaminated soils: phytoremediation as a potentially promising clean-up technology. Crit. Rev. Environ. Sci. Technol., 39: 622-654. DOI:10.1080/10643380701798272 |
Millaleo, R., Reyes-Diaz, M., Ivanov, A.G., et al., 2010. Manganese as essential and toxic element for plants: transport, accumulation and resistance mechanisms. J. Soil Sci. Plant Nutr., 10: 470-481. DOI:10.4067/S0718-95162010000200008 |
Mishra, A., Sharma, A.K., Kumar, S., et al., 2013. Bauhinia variegata leaf extracts exhibit considerable antibacterial, antioxidant, and anticancer activities. BioMed Res. Int., 2013: 1-10. |
P.C.Nagajyoti, K.D.Lee, T.V.M.Sreekanth, 2010. Heavy metals, occurrence and toxicity for plants: a review. Environ. Chem. Lett., 8: 199-216. DOI:10.1007/s10311-010-0297-8 |
J.Nickelsen, B.Rengstl, 2013. Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol., 64: 609-635. DOI:10.1146/annurev-arplant-050312-120124 |
Pan, G., Liu, W., Zhang, H., et al., 2018. Morphophysiological responses and tolerance mechanisms of Xanthium strumarium to manganese stress. Ecotox. Environ. Saf., 165: 654-661. DOI:10.1016/j.ecoenv.2018.08.107 |
A.K.Pandey, A.Mishra, 2012. Antifungal and antioxidative potential of oil and extracts derived from leaves of Indian spice plant Cinnamomum tamala. Cell. Mol. Biol., 58: 142-147. |
Peiter, E., Montanini, B., Gobert, A., et al., 2007. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. U. S. A., 104: 8532-8537. DOI:10.1073/pnas.0609507104 |
Pei, M., Niu, J., Li, C., et al., 2016. Identification and expression analysis of genes related to calyx persistence in Korla fragrant pear. BMC Genom., 17: 132. DOI:10.1186/s12864-016-2470-3 |
Sasaki, A., Yamaji, N., Xia, J., et al., 2011. OsYSL6 is involved in the detoxification of excess manganese in rice. Plant Physiol., 157: 1832-1840. DOI:10.1104/pp.111.186031 |
Shao, J.F., Yamaji, N., Shen, R.F., et al., 2017. The key to Mn homeostasis in plants: regulation of Mn transporters. Trends Plant Sci., 22: 215-224. DOI:10.1016/j.tplants.2016.12.005 |
Sheng, H., Zeng, J., Liu, Y., et al., 2016. Sulfur mediated alleviation of Mn toxicity in polish wheat relates to regulating Mn allocation and improving antioxidant system. Front. Plant Sci., 7: 1382. |
R.Stark, M.Grzelak, J.Hadfield, 2019. RNA sequencing: the teenage years. Nat. Rev. Genet., 20: 631-656. DOI:10.1038/s41576-019-0150-2 |
Takemoto, Y., Tsunemitsu, Y., Fujii-Kashino, M., et al., 2017. The tonoplast-localized transporter MTP8.2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol., 58: 1573-1582. DOI:10.1093/pcp/pcx082 |
Tang, T., Liu, P.L., Zheng, G.W., et al., 2016. Two phases of response to long-term moderate heat: variation in thermotolerance between Arabidopsis thaliana and its relative Arabis paniculata. Phytochemistry, 122: 81-90. DOI:10.1016/j.phytochem.2016.01.003 |
Tang, Y.T., Qiu, R.L., Zeng, X.W., et al., 2009. Lead, zinc, cadmium hyperaccumulation and growth stimulation in Arabis paniculata Franch. Environ. Exp. Bot., 66: 126-134. DOI:10.1016/j.envexpbot.2008.12.016 |
Tolrà, R., Pongrac, P., Poschenrieder, C., et al., 2006. Distinctive effects of cadmium on glucosinolate profiles in Cd hyperaccumulator Thlaspi praecox and non-hyperaccumulator Thlaspi arvense. Plant Soil, 288: 333-341. DOI:10.1007/s11104-006-9124-1 |
Tsunemitsu, Y., Genga, M., Okada, T., et al., 2018. A member of cation diffusion facilitator family, MTP11, is required for manganese tolerance and high fertility in rice. Planta, 248: 231-241. DOI:10.1007/s00425-018-2890-1 |
Xue, Y., Wang, Y., Yao, Q., et al., 2014. Research progress of plants resistance to heavy metal Cd in soil. Ecol. Environ. Sci., 3: 528-534. |
Yamaji, N., Sasaki, A., Xia, J.X., et al., 2013. A node-based switch for preferential distribution of manganese in rice. Nat. Commun., 4: 2442. DOI:10.1038/ncomms3442 |
Yang, S., Yi, K., Chang, M.M., et al., 2019. Sequestration of Mn into the cell wall contributes to Mn tolerance in sugarcane (Saccharum officinarum L.). Plant Soil, 436: 475-487. DOI:10.1007/s11104-019-03937-x |
You, X., Yang, L.T., Lu, Y.B., et al., 2014. Proteomic changes of Citrus roots in response to long-term manganese toxicity. Trees Struct. Funct., 28: 1383-1399. DOI:10.1007/s00468-014-1042-x |
You, X., Yang, L.T., Qi, Y.P., et al., 2017. Long-term manganese-toxicity-induced alterations of physiology and leaf protein profiles in two Citrus species differing in manganese-tolerance. J. Plant Physiol., 218: 249-257. DOI:10.1016/j.jplph.2017.08.011 |
Zeng, X.W., Qiu, R.L., Ying, R.R., et al., 2011. The differentially-expressed proteome in Zn/Cd hyperaccumulator Arabis paniculata Franch. in response to Zn and Cd.. Chemosphere, 82: 321-328. DOI:10.1016/j.chemosphere.2010.10.030 |
Zhai, C., Xu, P., Zhang, X., et al., 2015. Development of Gossypium anomalum-derived microsatellite markers and their use for genome-wide identification of recombination between the G. anomalum and G. hirsutum genomes. Theor. Appl. Genet., 128: 1531-1540. DOI:10.1007/s00122-015-2528-7 |
Zhou, C.P., Qi, Y.P., You, X., et al., 2013. Leaf cDNA-AFLP analysis of two citrus species differing in manganese tolerance in response to long-term manganese-toxicity. BMC Genom., 14: 621. DOI:10.1186/1471-2164-14-621 |
Zhou C.P., Li C.P., Liang W.W., et al., 2017. Identification of manganese-toxicity-responsive genes in roots of two citrus species differing in manganese-tolerance using cDNA-AFLP. Trees Struct. Funct., 31: 813-831. DOI:10.1007/s00468-016-1507-1 |
Zhuo, Y., Wang, Z.L., Li, B.W., et al., 2009. Promotion effects of microorganisms on phytoremediation of heavy metals-contaminated soil. Chin. J. Appl. Ecol., 20: 2025-2031. |



