b. Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla 666303, Yunnan, China;
c. Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China
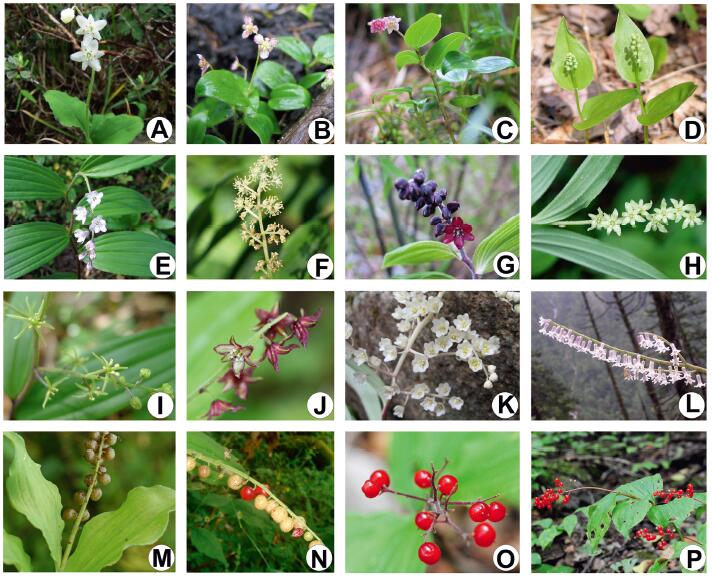
Maianthemum F.H. Wigg. is a medium-sized genus from tribe Polygonateae, which originally belonged to Liliaceae and was finally placed in the recently defined subfamily Nolinoideae of Asparagaceae after a series of taxonomic changes (Chen and Tamura, 2000; APG IV, 2016). This genus is usually rhizomatous understory herbs with more than 35 species widely distributed from eastern Asia and Europe to North and Central Americas (Therman, 1956; LaFrankie, 1986a; Meng et al., 2005), and morphologically featured with underground rhizomes, simple aerial stems, terminal paniculate to racemose inflorescences, and red berries at maturity (Fig. 1). Traditionally, two genera were recognized for species of Maianthemum based on their flower morphology, one was Maianthemum sensu stricto characterized by four tepals, four stamens, two carpels and the other was Smilacina Desf. with six tepals, six stamens, and three carpels. The merge of the two taxa and the monophyly of the expanded Maianthemum are strongly supported by morphological evidence (LaFrankie, 1986a, b) and molecular phylogenetic data (Kim and Lee, 2007; Meng et al., 2008; Kim et al., 2017).
|
| Fig. 1 Morphological diversity in Maianthemum. A–L: flowers, M–P: fruits. A, M. lichiangense; B, M. gongshanense; C, M. fusciduliflorum; D, M. canadense; E, M. szechuanicum; F, M. racemosum; G, M. purpureum; H, M. atropurpureum; I, M. tatsienense; J, M. atropurpureum; K, M. oleraceum; L, M. henryi; M, M. atropurpureum; N, M. henryi; O–P, M. japonicum. |
Kim and Lee (2007) supported the monophyly of the broadly circumscribed Maianthemum based on partial trnK sequences, but their study had limited sampling and a single chloroplast marker with low resolution of relationships within the genus. Meng et al. (2008) conducted a first comprehensive study on the genus with a sampling of 22 species and six closely related outgroup taxa using eight chloroplast and nuclear markers (trnL-F, rps16, rpl16, psbA-trnH, rbcL, ndhF, trnK, and ITS). The phylogenetic results strongly supported the monophyly of Maianthemum with most species from the New World as the first diverged lineages. On the other hand, the majority of taxa from the eastern Himalayan region in SW China form a well-supported clade. The other species from central to NE China and Japan were resolved as several independent groups located between the New World and the SW China (Meng et al., 2008). Recently, Kim et al. (2017) also sampled many species from the genus for phylogenetic analyses, but their study was focused on the dimerous Maianthemum sensu stricto group.
The eastern Himalayas to Hengduan Mountains of SW China are biologically among the world's richest and most diverse regions with a high level of endemism and have been recognized as the biologically richest temperate region in the world (Wu, 1988; Boufford and Van Dyck, 1999; Myers et al., 2000; Sun et al., 2017). This area is the most diversified region for Maianthemum within eastern Asia, which harbors about 13 species with a high level of morphological variation among species and even within a species (Hara, 1987; Li, 1990; Meng et al., 2008). Although most species from SW China are well supported as a natural group, species relationship within this clade remains totally unclear. Furthermore, many species with wide distribution, such as Maianthemum henryi (Baker) LaFrankie and M. atropurpureum (Franch.) LaFrankie, are found to be non-monophyly (Meng et al., 2008). Because evolutionary radiations and species diversifications have not been well analyzed for plants from SW China, it is necessary to include Maianthemum as a case to study the divergence pattern within this biodiversity hotspot.
In this study, we use a large number of species of Maianthemum across its entire distribution range to generate a well-resolved molecular phylogeny based on a combined set of nuclear ribosomal (nrDNA) and chloroplast (cpDNA) sequences. Although our previous study supported the monophyly of the broadly circumscribed Maianthemum, the species sampling, especially from SW China, was limited. With a broader level of taxon sampling within Maianthemum, including multiple individuals, and an extensive character sampling from eight DNA regions, our paper intends to reconstruct phylogenetic species relationships and biogeographic history within Maianthemum and test whether the species, especially those from SW China, are reciprocally grouped according to their taxonomic species.
2. Materials and methods 2.1. Taxon samplingA total of 125 accessions were sampled, representing 91 taxa (29 species) of Maianthemum and 34 outgroup taxa sampled widely from the subfamily of Nolinoideae (Appendix A). Our sampling covers the biogeographic distribution of the genus worldwide, including 24 species from eastern Asia and Europe, three from North America, and two from Central America. Particularly, all 13 species from SW China were included with up to 16 individuals sampled for the widespread distributed species (Table 1). The outgroup taxa were selected to include representatives from other genera in tribe Polygonateae (i.e., Heteropolygonatum M.N. Tamura & Ogisu, Disporopsis Hance, and Polygonatum Mill.; 13 species) and the other members in Nolinoideae of Asparagaceae (21 species) based on previous broader analyses (Rudall et al., 2000; Kim et al., 2010; Chen et al., 2013).
| Taxa | Individual number | Monophyly | Clade |
| M.gigas | 1 | – | New World |
| M.paniculatum | 2 | yes | New World |
| M.racemosum | 5 | yes | New World |
| M.stellatum | 3 | yes | New World |
| M.canadense | 2 | yes | North Temperate |
| M.dahuricum | 4 | yes | North Temperate |
| M.formosanum | 2 | yes | North Temperate |
| M.japonicum | 5 | no | North Temperate |
| M.nanchuanense | 1 | – | North Temperate |
| M.robustum | 1 | – | North Temperate |
| M.stenolobum | 1 | – | North Temperate |
| M.tatsienense | 7 | no | North Temperate |
| M.trifolium | 2 | yes | North Temperate |
| M.yesoense | 1 | – | North Temperate |
| M.bicolor | 1 | – | North Temperate |
| M.bifolium | 4 | yes | North Temperate |
| M.dilatatum | 4 | no | North Temperate |
| M.atropurpureum | 5 | no | SW China |
| M.forrestii | 1 | – | SW China |
| M.fusciduliflorum | 1 | – | SW China |
| M.fuscum | 1 | – | SW China |
| M.gongshanense | 1 | – | SW China |
| M.henryi | 13 | no | SW China |
| M.lichiangense | 1 | – | SW China |
| M.oleraceum | 1 | – | SW China |
| M.purpureum | 16 | no | SW China |
| M. sp. nov. | 1 | – | SW China |
| M.szechuanicum | 1 | – | SW China |
| M.tubiferum | 3 | no | SW China |
Total DNAs were extracted from about 15 mg silica-gel dried leaf tissue using the modified CTAB method (Doyle and Doyle, 1987) or the DNeasy Plant Mini Kits (QIAGEN, Mississauga, Ontario, Canada) following the manufacturer's protocol. Eight molecular regions (trnL-F, rps16, rpl16, psbA-trnH, rbcL, trnK, trnC-petN and nrITS) were sequenced to reconstruct phylogenetic relationships within Maianthemum and related taxa in Nolinoideae. For primers and sequence amplification, we followed protocols described in Meng et al. (2008), except for trnC-petN sequences amplified according to Lee and Wen (2004).
The PCR products were purified using the polyethylene glycol precipitation procedure and cycle sequencing reaction was carried out using the following profile: 35 cycles of 97 ℃ for 15 s, 50 ℃ for 5 s, and 60 ℃ for 4 min. The products of cycle-sequencing were cleaned using the Sephadex columns (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) and dried at 60 ℃ in a vacuum evaporator. The sequences were generated on an ABI prism 3730 capillary sequencer (Applied Biosystems, Foster City, California, USA).
To provide a more complete taxa coverage, sequences available from the GenBank were combined to our dataset with most derived from our previous studies (Meng et al., 2008, 2014). Information on the voucher specimens and the corresponding GenBank accession numbers are provided in. Sequence alignment was performed in MAFFT 6 using the default alignment parameters (Katoh and Toh, 2008) followed by manual adjustment in PhyDE 1.0 (Müller et al., 2010).
2.3. Phylogenetic analysisPhylogenetic analyses were performed using both maximum likelihood (ML) and Bayesian inference (BI) methods. We performed a combined analysis of all chloroplast and nrITS sequences because of the low variation of all markers for Maianthemum and the non-significance of incongruence test. The ML trees were inferred using RAxML v.8.2.12 (Stamatakis, 2006) with data partitioned into gene-regions allowing for independent parameter estimates on each partition and branch length estimates were optimized across all gene regions. The ML analyses employed a GTRCAT model for each partition followed by a final parameter optimization step using the GTR substitution distribution and Γ distribution of among-site rate variation. Bootstrap supports (BS) were estimated using a rapid bootstrapping algorithm and 1000 replicates in RAxML.
Bayesian inference was conducted using MrBayes v.3.2.1 (Ronquist and Huelsenbeck, 2003) with nucleotide substitution models selected based on the Akaike Information Criterion (AIC) determined by MrModelTest v.2.3 (Nylander, 2004). Independent model tests were performed on each marker and model parameters (statefreq, revmat and shape) were unlinked between partitions. The Bayesian Markov chain Monte Carlo (MCMC) analyses were run for 10, 000, 000 generations with 4 incrementally heated chains starting from random trees and sampling one out of every 1000 generations. The burn-in and convergence diagnostics were graphically assessed using AWTY (Nylander et al., 2008) and the remaining trees were assumed to be representative of the posterior probability (PP) distribution.
2.4. Dating the times of divergenceWe also estimated the tree topology and node ages of Maianthemum using a Bayesian relaxed clock model as implemented in BEAST v.1.8.5 (Drummond and Rambaut, 2007). Since our interests were focused on the ages of major clades and lineages shown intercontinental disjunction, the dataset used for time estimation was reduced from the phylogenetic data above with only one accession included for each species. The dating dataset was partitioned using BEAUti 1.8.5 to generate input files for BEAST. Rate variation among sites was modeled using a gamma distribution with four rate categories in the GTR model with a Yule speciation tree prior and an uncorrelated lognormal distributed relaxed clock model (Drummond et al., 2006). Two independent MCMC runs were performed for 50 million generations each and sampling every 5000 generations. Tracer 1.6 (Rambau et al., 2014) was used to check for convergence between the runs with results considered reliable when the effective sampling size for all parameters exceeded 200 (Rambau et al., 2014). After the discarding of ca. 10% burn-in, the rest sampled posterior trees were summarized to generate a maximum clade credibility tree using the program TreeAnnotator 1.8.5 (Drummond and Rambaut, 2007) with a PP limit of 0.5 and mean node heights. The means and 95% higher posterior densities (HPD) of age estimates were obtained from the combined outputs using Tracer.
It is better to estimate the ages of molecular phylogenetic trees with calibration of multiple fossils (Forest, 2009), but there are very few reliable fossils reported from the monocots Asparagaceae (Friis et al., 2011). Thus, we used two secondary-calibration points in our analyses, following the approach described in recent studies on Tecophilaeaceae and Asparagaceae subfamily Scilloideae (Buerki et al., 2012, 2013) and Polygonatum (Wang et al., 2016). The stem and crown ages of Asparagaceae were constrained using a normal distribution with a mean of 58.3 and 56.4 million years ago (Ma), respectively, both with a standard deviation of 6.0.
3. ResultsThe final dataset comprised an aligned matrix of 916 base pairs (bp) with 54 bp variables and 30 bp potentially parsimony-informative for rps16, 1447 bp with 130 variable and 66 potentially parsimony-informative for rbcL, 729 bp with 47 variable and 27 potentially parsimony-informative for psbA-trnH, 1117 bp with 98 variable and 40 potentially parsimony-informative for rpl16, 994 bp with 101 variable and 59 potentially parsimony-informative for petN-trnC, 1839 bp with 363 variable and 169 potentially parsimony-informative for trnK, 957 bp with 64 variable and 39 potentially parsimony-informative for trnL-F, and 745 bp with 291 variable and 211 potentially parsimony-informative for nrITS. The total number of characters included in the combined dataset is 8744 bp with 1137 variable and 641 parsimony-informative. Due to the low variations of both the chloroplast and nrITS sequences and no hard conflicts (BS > 50) found among them, all the sequences were directly combined following Meng et al. (2008) and Kim et al. (2017).
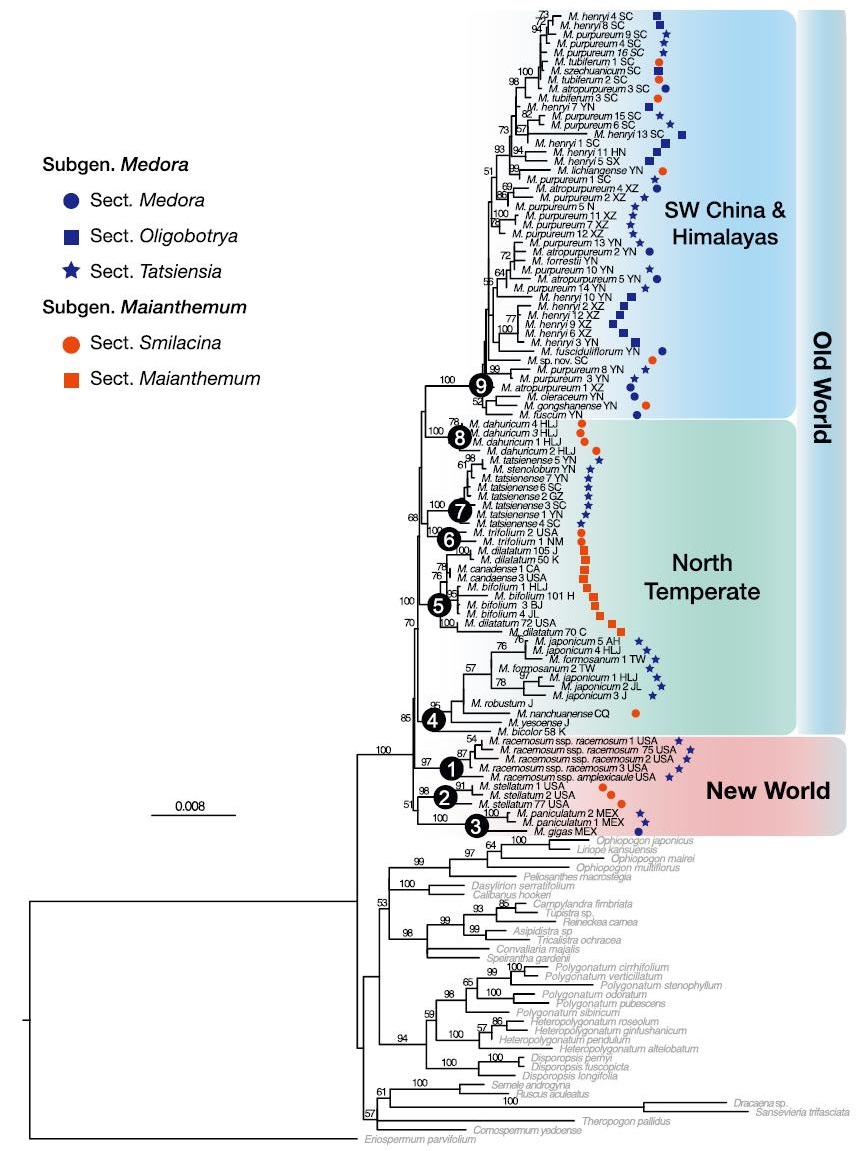
Both the ML best tree (Fig. 2) and the BI consensus phylogram (Fig. S1) suggested the monophyly of Maianthemum with recognition of nine strongly supported clades, but phylogenetic relationships among these nine clades are largely unresolved in both ML and BI analyses (clades 1–9, Figs. 2 & S1). Clades 1–3 consist of taxa all from the New World including the two common species of Maianthemum racemosum (L.) Link (BS = 100, PP = 1, clade 1) and Maianthemum stellatum (L.) Link (BS = 100, PP = 1, clade 2) from North America and the others from tropical America (BS = 100, PP = 1, clade 3). Members in clades 4–8 are sampled from north temperate areas and all the clades are robustly supported with BS ranging from 85 to 100 and PP = 1 (Figs. 2 & S1). The traditional Maianthemum sensu stricto is well supported (BS = 100, PP = 1, clade 5 in Figs. 2 & S1). The clade 9 is a well resolved monophyletic group mostly restricted and diversified in the eastern Himalayas to the Hengduan Mountains of SW China (Fig. 3). Phylogenetic relationships within this group remain unclear except for a well-supported subclade with species mostly from north Hengduan Mountains (Fig. 3). The widely sampled species from this region do not form a monophyletic group at the specific level, such as Maianthemum henryi and Maianthemum purpureum (Wall.) LaFrankie (Fig. 3).
|
| Fig. 2 The Maximum likelihood tree of Maianthemum based on eight combined chloroplast and nuclear sequences (trnL-F, rps16, rpl16, psbA-trnH, rbcL, trnK, trnC-petN and ITS). The bootstrap values in 1000 replicates are shown by circles with different colors on each node. |

|
| Fig. 3 Phylogenetic relationships for the SW China clade (extracted from clade 9 of Fig. 2) with taxa localities shown on a map. The black arrow indicates a possible migration route from the eastern Himalayas to the Hengduan Mountains. |
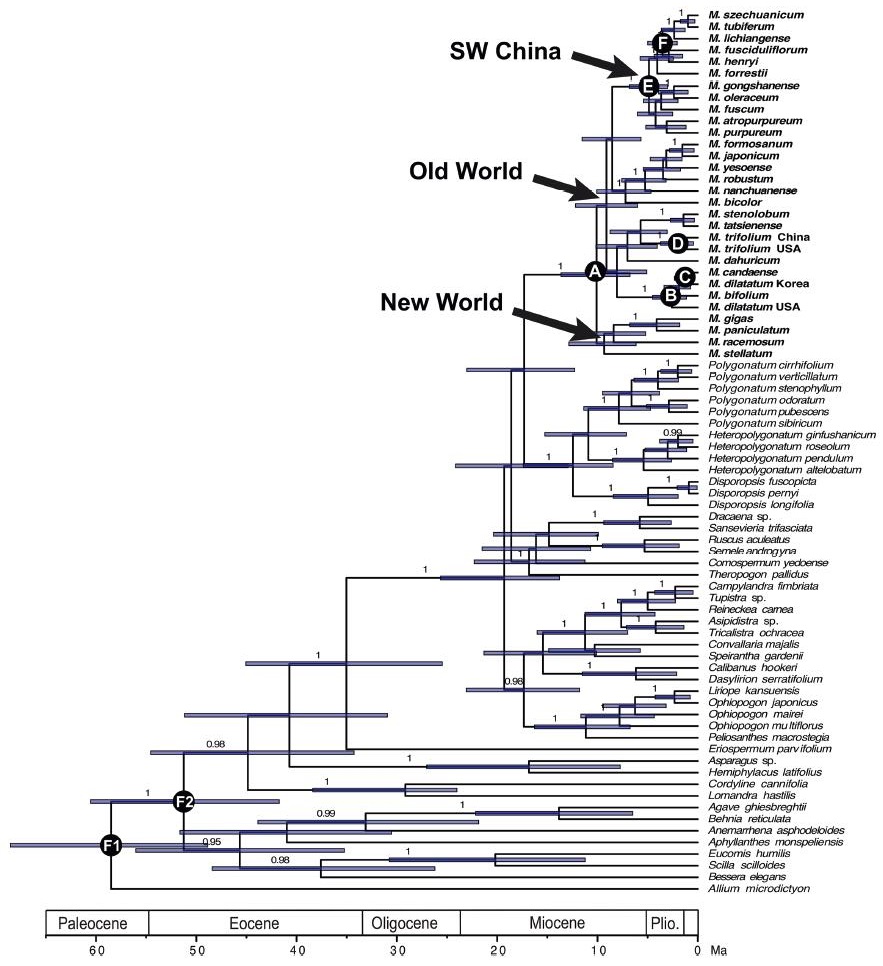
The Bayesian dating analyses based on two calibration points suggested an origin of the Maianthemum crown lineage at 10.09 Ma with a 95% HPD of 6.77–13.64 Ma (node A, Fig. 4). The disjunction between Eurasian and North American species was estimated at 2.79 (1.46–4.38; Fig. 4) or 3.31 (1.65–5.18) Ma. The disjunct age between the New World Maianthemum dilatatum (Alph. Wood) A. Nelson & J.F. Macbr. and the rest Maianthemum sensu stricto was estimated to be 2.59 (95% HPD: 1.13–4.51) Ma (node B in Fig. 4) and the Old World M. dilatatum diverged from the New World M. canadense Desf. at 1.23 (95% HPD: 0.37–2.34) Ma (node C in Fig. 4). The infraspecific disjunction within Maianthemum trifolium (L.) Sloboda is also recent with an estimation of 1.94 (95% HPD: 0.44–3.71) Ma (node D in Fig. 4). The crown age of the SW clade was estimated as 4.87 (3.03–6.83) Ma and the eastern Hengduan Mountains clade was inferred to be 3.52 (2.06–4.99) Ma.
|
| Fig. 4 Maximum clade credibility tree of Maianthemum and the closely related taxa derived from a BEAST analysis. Posterior estimates of divergence times were inferred using two fixes as normal age constraints (F1: 58.3 Ma; F2: 56.4 Ma). Nodes are posterior mean ages with node bars representing 95% highest posterior density intervals. A-F indicate nodes with interests and discussion in the text. |
Infrageneric classifications of Maianthemum were proposed by many authors on the base of morphology and geographic distribution. Hara (1987) divided the traditional genus of Smilacina into four sections based on the combination of several significant flower features but not included the New World species of Smilacina and Maianthemum sensu stricto. Li's (1990) classification was different from Hara (1987), recognizing two subgenera (Medora and Maianthemum) and five sections (Medora, Oligobotrya, Tatsiensia, Smilacina, and Maianthemum), which includes all taxa from Smilacina and Maianthemum sensu stricto. Representatives from all subgenera and sections versus Li (1990) were sampled in our analyses, and the phylogenetic results indicate that both subgenera Medora and Maianthemum and all sections are not monophyletic, except for the section Maianthemum (Fig. 2). Similar results are also found from Kim and Lee (2007), Meng et al. (2008), and Kim et al. (2017), which exhibited low resolution to the infrageneric classification and suggested non-monophyly of most sections recognized by Li (1990). Therefore, our phylogenetic analyses further confirmed its inconsistent with the morphology-based classifications and thus traditional morphological characters (e.g., rhizome morphology, corolla shape, and flower color) are not phylogenetically informative.
Although the deep relationship of the genus is still unclear, our results suggested that all samples of Maianthemum were recognized into nine clades, but phylogenetic relationships among these lineages are unresolved (Fig. 2). These main lineages may have arisen from a rapid initial radiation in the late Miocene (Fig. 4), as evidenced by short branch lengths subtending longer branches that lead to the crown clades of 1–9, respectively (Figs. 2 & S1). This pattern suggests short periods of rapid diversification after the divergence from a common ancestor followed by long intervals of relatively constant evolution as suggested by Kim et al. (2017). Our results further indicated that these nine clades are recognizable into two clusters according to their biogeographic distribution, i.e., the Old and New World (Fig. 2). The Old World group can be further distinguishable into a monophyletic SW China lineage and a paraphyletic north temperate complex (Fig. 2).
The New World group is composed of three lineages as suggested by the ML and Bayesian trees (clades 1–3 in Figs. 2 & S1). The two North American species of Maianthemum racemosum (clade 1) and M. stellatum (clade 2) are supported to be monophyly, respectively. M. racemosum is a species native to North America widespread found from every US state except Hawaii, and from every Canadian province and territory except Nunavut, as well as from Mexico (LaFrankie, 1986a). All samples of M. racemosum ssp. racemosum from eastern North America grouped into a clade sister to the western North American subspecies M. racemosa ssp. amplexicaule (Fig. 2). M. stellatum is the other species widely distributed in North America. It is a woodland herbaceous perennial plant, different from its close relative M. racemosum with smaller, more open inflorescences, flowers with stamens shorter rather than longer than the petals, and somewhat narrower and more curved leaves (LaFrankie, 1986a). Both of them show the characteristic zigzag of the stem between the alternate leaves. The mountains of tropical America is another important center of species diversification for the genus with more than ten species are reported from this region (LaFrankie, 1986a). Only two species were sampled from Central America and they were grouped as the third lineage (clade 3, Fig. 2). Kim et al. (2017) sampled four species and also found this group is monophyletic.
The diversification and relationship of the New World Maianthemum are still unclear. The Central American taxa are suggested to be close to the Maianthemum stellatum in the ML and Bayesian trees (Fig. 2), other than to the M. racemosum in the BEAST tree (Fig. 4). The unresolved relationship among these three lineages, perhaps is resulted from the present limited sampling in Central America, and/or the possible long isolation of the Central American taxa in the early evolutionary history of the genus. More molecular data and samples from Central America are needed to explore their phylogenetic relationships, which are also important to understand the early evolutionary history of the genus.
As a transition between the New World and SW China groups, the north temperate taxa is a complex group composing of five lineages (clades 4–8 in Fig. 2), which are mostly occurred from northeastern Asia with a few found from Europe and North America. The north temperate group might present fast radiation with short branch lengths among lineages, and the relationships of these five clades remain uncertain (Fig. 2).
Clade 4 represents species ranging from central and NE China to Japan, corresponding to clade C in Meng et al. (2008), which sampled only two species. Six species were included in this study with five individuals of Maianthemum japonicum (A. Gray) LaFrankie from NE China and Japan. Species in this clade share the synapomorphic characters of 4–9 ovate to elliptic leaves and white, linear petals. M. japonicum is an erect, herbaceous perennial plant with stems 30–60 cm tall, and producing a clump of unbranched stems from a creeping rhizome. This species is widely found in northeastern Asia from the Russian Far East and NE China to Korea and Japan. It is not monophyletic because Maianthemum formosanum (Hayata) LaFrankie from Taiwan is nested within it. The taxonomic status of this species needs further evidence and examination.
Maianthemum sensu stricto (clade 5) has been traditionally recognized as a separate genus, characterized by dimerous flowers with linear petal and small herbs with only 2–4 cordiform leaves (Meng et al., 2008). This clade contains only three species disjunct between North America and Eurasia. Our analyses support the monophyly of this clade and the paraphyletic Maianthemum dilatatum into two well-defined groups from western North and northeast Asia, the same as in Kim et al. (2017) which conducted a specific phylogenetic analysis of this clade with extensive samplings. M. canadense is sister to M. bifolium (L.) F. W. Schmidt in Kim et al. (2017). However, M. canadense is closer to eastern Asian M. dilatatum than M. bifolium in this study (Fig. 2). The relationships within Maianthemum sensu stricto need to be tested further with more evidences.
Maianthemum trifolium is a species disjunctly distributed in northeastern North America and northeastern Asia. M. trifolium is similar to Maianthemum sensu stricto as sharing common vegetative form, growth habit, speckled fruits when immature, and inflorescence architecture (LaFrankie, 1986a). Maianthemum sensu stricto and M. trifolium have only two or three leaves on the floral stem and occupy a similar geographic range (eastern Asia and North America). Despite these similarities, Maianthemum trifolium has six (not four) tepals and the species has been segregated from Maianthemum sensu stricto (LaFrankie, 1986b). Molecular phylogeny also supports their close relationship but with weak supports (Kim et al., 2017). Our study also suggested the monophyly of Maianthemum trifolium (clade 6), but did not support its sister relationship to Maianthemum sensu stricto.
The two species of Maianthemum tatsienense (Franch.) LaFrankie and M. stenolobum (Franch.) S.C. Chen & Kawano grouped together (clade 7) are morphologically distinguishable in having green flowers with narrow petals, and shining leaves and stems, perhaps representing synapomorphies of this clade (Fig. 1I). Since M. stenolobum is nested within M. tatsienense, M. tatsienense seems to be paraphyletic and its taxonomy needs further attention with more samples of M. stenolobum.
Clade 8 represents by only Maianthemum dahuricum (Fisch. & C.A. Mey.) LaFrankie, which is an unusual species with leaves abaxially densely pubescent and flowers in clusters of 2–4 and restricted in northeastern Asia. The position of the Old World M. dahuricum remained unresolved (Meng et al., 2008) or is close to the SW China clade (Kim et al., 2017). Similar to Kim et al. (2017), we found it is close to the SW China clade though they are remotely separated. The isolated status of the species should be further examined with more data.
4.2. Divergence pattern within SW China cladeThe Hengduan Mountains to the eastern Himalayas in SW China is a diversification center for the genus with more than thirteen species (Li, 1990; Meng et al., 2008). All species from this region have been comprehensively sampled with more than ten individuals for some widespread species (e.g., Maianthemum henryi and M. purpureum, ). Similar to previous studies (Meng et al., 2008; Kim et al., 2017), our results showed that all of them were strongly supported as a monophyletic clade with exception of M. tatsienense and M. stenolobum which is separated as an independent lineage (clade 7 in Fig. 2). Morphologically, the SW China clade is characterized by having large inflorescences and flowers with conspicuous petals with color ranging from dark purple, light purple, to white, e.g., Fig. 1 (E, G, H, J-L).
As indicated by previous studies (Meng et al., 2008; Kim et al., 2017), phylogenetic relationships within this clade are not consistent with morphology, as shown individuals from the same species are grouped into difference lineages and different species from the same area are grouped together (Fig. 3). For example, Maianthemum henryi is easily distinguishable with long-tubed flowers (Fig. 1L), but molecular data did not support the monophyly of this species with thirteen accessions separated into at least six lineages (Fig. 3; Table 1). M. atropurpureum and M. purpureum are another two species widely found in SW China. They are morphologically similar with flowers white to purplish red, but M. atropurpureum usually with perianth connate proximally forming a tube and M. purpureum's free. The two species are also non-monophyly respectively. The three collections of M. tubiferum (Batalin) LaFrankie is not grouped together either. Similar results were found by Meng et al. (2008) and Kim et al. (2017). SW China, including the Hengduan Mountains and the eastern Himalayas, is one of the biologically richest temperate regions in the world (Wu, 1988; Sun, 2002). Heterogeneity and complexity of high montane habitats in SW China may explain the high level of morphological divergence and rich biodiversity of its floras (Xiang et al., 2004; Nie et al., 2005).
Phylogenetic relationships within this clade are extremely low and species relationship within this group is not well resolved (Fig. 3). This lineage may have arisen from rapid initial radiation at 4.87 (3.03–6.83) Ma (node E in Fig. 4) in the eastern Himalayas following with a second one at 3.52 (2.06–4.99) Ma (node F in Fig. 4) in the Hengduan Mountains, as also evidenced by short branch lengths subtending longer branches that lead to the crown groups (Fig. 3). More and more evidence suggest that the Qinghai-Tibetan Plateau (QTP) is a very recent construct and its modern high elevation only came into being since the middle Miocene (Harrison et al., 1992; Shi et al., 1998; Spicer et al., 2003, 2020). The diversification of Maianthemum largely coincides with the topographically complex landscape of the QTP with deep valleys and high mountains formed since the Miocene, and is also similar to the scenarios estimated for the major divergence of many other plant groups from this region (Wang et al., 2009; Liu et al., 2014; Favre et al., 2015; Ebersbach et al., 2017).
Both phylogenetic and biogeographic dating results indicated that the fast radiation of SW China clade was occurred in the eastern Himalayas to the south Hengduan Mountains (south Xizang and northwest Yunnan), followed with subsequent radiation in the eastern Hengduan Mountains (from west Sichuan to part of northwest Yunnan) in the Pliocene (Fig. 4). Thus, our data suggest a west-to-east dispersal or migration route from eastern Himalayas to the Hengduan Mountains (Fig. 3), which is a common mechanism shaping the geographical distribution of plants as well as their speciation and diversification in this area (Wen et al., 2014; Ebersbach et al., 2017). For example, Solms-laubachia Muschler, which has originated in central Asia, and subsequently migrated eastward into the Hengduan Mountains via eastern Himalayas (Yue et al., 2009). The dispersal direction of Incarvillea Jussieu within the Sino-Himalaya was from central Asia to the eastern Himalayas, then via the Hengduan area to eastern Asia (Chen et al., 2005; Rana et al., 2021). Cyananthus Wallich ex Bentham provides another example of a west-to-east dispersal route from the eastern Himalayas to the Hengduan Mountains (Zhou et al., 2013). Interestingly, an opposite route is also common as found in Syncalathium Lipschitz sensu stricto which was dispersed from the northern edge of the Hengduan Mountains (e.g., Gansu and Qinghai) to the central Hengduan area, and more recently into the eastern Himalayas (Zhang et al., 2011).
4.3. Intercontinental disjunction between North America and eastern AsiaIntercontinental disjunctions of Maianthemum in the Northern Hemisphere appear to have occurred multiple times during the late Miocene to the Pliocene (Fig. 4). The crown genus was split into two lineages, representing an early disjunction between the New World group and all the others mostly from the Old World estimated at 10.09 Ma in the late Miocene (node A in Fig. 4). This result is similar to that estimated in Meng et al. (2008), which estimated the divergence time of crown Maianthemum to be no earlier than the late Miocene (8.30 Ma) based on the rbcL gene sequence data sampled from the Asparagales and other monocots (Meng et al., 2008). Three recent disjunct lineages from the Pliocene were detected by our phylogenetic analyses, including two disjunct lineages occurred withinMaianthemum sensu stricto (nodes B & C in Fig. 4) and one found from Maianthemum trifolium (node D in Fig. 4). The disjunctions between the New World M. dilatatum and the rest at 2.59 (1.13–4.51) Ma and the other between M. canadense and M. dilatatum (Asia) at 1.23 (0.37–2.34) Ma are recent in the Pliocene, similar to the divergence time estimated in Meng et al. (2008). Kim et al. (2017) also estimated similar but a little older ages in the Pliocene (5.0 Ma and 3.5 Ma) for the intercontinental disjunctions within this clade. There are very few examples of intercontinental disjunction within a species (Nie et al., 2006). M. trifolium is disjunct distributed between northeastern Asia and northern North America. The disjunction within M. trifolium is first reported with a recent age of 1.94 (0.44–3.71) Ma.
The disjunction between eastern Asian and North American taxa are likely the result of a combination of vicariance and long-distance dispersal events. The early disjunction within Maianthemum should be resulted by a migration between the Old and the New World in the late Miocene via the Beringia, which is common pattern found in many temperate plants disjunct in the Northern Hemisphere (Wen, 1999; Donoghue and Smith, 2004; Milne, 2006; Wen et al., 2010). It seems that dispersal via long distance possibly across the Beringia is the most likely explanation for the recent disjunction in Maianthemum since the Beringia was no longer available for direct exchanges of most north temperate plants after about 3.5 Ma (Hopkins, 1967; Tiffney, 1985). The facilitation of long-distance dispersal in the genus is also supported by its effective long-distance dispersals by birds (Meng et al., 2008; Kim et al., 2017). As shown in Fig. 1(M–P), young fruits are fleshy berries with purple dots that become red at maturity, which are dispersed by birds and/or migrating animals (Piper, 1989; Conran and Tamura, 1998). Long-distance dispersal may be one of the key adaptations that have allowed Maianthemum species to succeed in high latitudes during the widely fluctuating climates of the Pliocene and Pleistocene (Kim et al., 2017). Several recent studies of other taxa in Asparagaceae based on molecular data have supported dispersal as the dominant factor responsible for transoceanic or inland distributions (Lu and Morden, 2014; Wang et al., 2016; Wang and Yang 2018).
Author contributionsYM, Z-LN and Y-PY designed the research and collected the materials. RM, Z-LN and YM performed the experiments and data analysis. All authors have contributed to interpretation of the results and writing the manuscript.
Declaration of competing interestThe authors declare there is no conflict of interest regarding this manuscript.All the authors are agreed to submit this manuscript.
AcknowledgementsThis study was supported by grants from Natural Sciences Foundation of China (31760055), Natural Sciences Foundation of Hunan Province (2019JJ40232 and 2019JJ40233), Comprehensive Scientific Investigation of Biodiversity from the Wuling Mountains (2014FY110100), the Hunan Provincial Innovation Foundation for Postgraduate (CX2018B724), and the John D.and Catherine T.MacArthur Foundation.We thank Drs.Jun Wen, David E.Boufford, Akiko Soejima, Jin Murata, Esteban Martinez, Ting-Shuang Yi, Chien-Ti Chao, Tao Deng, Yang Niu, Guang-Wan Hu for samples collection and Dr.Guang-Yan Wang for lab assistance.The authors also thank two anonymous reviewers for their helpful comments and suggestions.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.02.001.
APG IV, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 18: 1-20. |
Boufford, D.E., Van Dyck, P.P., 1999. South-Central China, In: Mittermeier, R.A., Myers, N., Mittermeier, C.G. (Eds. ), Hotspots: Earth's Biologically Richest and Most Endangered Terrestrial Ecoregions. CEMEX, Mexico City, pp. 338-351.
|
Buerki, S., Jose, S., Yadav, S.R., et al, 2012. Contrasting biogeographic and diversification patterns in two Mediterranean-type ecosystems. PLoS One, 7: e39377. DOI:10.1371/journal.pone.0039377 |
S.Buerki, J.C.Manning, F.Forest, 2013. Spatio-temporal history of the disjunct family Tecophilaeaceae: a tale involving the colonization of three Mediterranean-type ecosystems. Ann. Bot., 111: 361-373. DOI:10.1093/aob/mcs286 |
Chen, S., Guan, K., Zhou, Z., et al, 2005. Molecular phylogeny of Incarvillea (Bignoniaceae) based on ITS and trnL-F sequences. Am. J. Bot., 92: 625-633. DOI:10.3732/ajb.92.4.625 |
Chen, S.C., Kim, D.K., Chase, M.W., et al, 2013. Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLoS One, 8: e59472. DOI:10.1371/journal.pone.0059472 |
Chen, X. -Q., Tamura, M.N., 2000. Polygonatum, In: Wu, Z. -Y., Raven, P.H. (Eds. ), Flora of China. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, pp. 223-232.
|
Conran, J.G., Tamura, M.N., 1998. Convallariaceae, In: Kubitzki, K. (Ed. ), The families and genera of vascular plants, Vol. 3. Springer-Verlag, Berlin.
|
M.J.Donoghue, S.A.Smith, 2004. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos. Trans. R. Soc. London Biol., 359: 1633-1644. DOI:10.1098/rstb.2004.1538 |
J.J.Doyle, J.L.Doyle, 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull., 19: 11-15. |
Drummond, A.J., Ho, S.Y., Phillips, M.J., et al, 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol., 4: e88. DOI:10.1371/journal.pbio.0040088 |
A.J.Drummond, A.Rambaut, 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol., 7: 214. DOI:10.1186/1471-2148-7-214 |
Ebersbach, J., Muellner-Riehl, A.N., Michalak, I., et al, 2017. In and out of the Qinghai-Tibet Plateau: divergence time estimation and historical biogeography of the large arctic-alpine genus Saxifraga L. J. Biogeogr., 44: 900-910. DOI:10.1111/jbi.12899 |
Favre, A., Packert, M., Pauls, S.U., et al, 2015. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev., 90: 236-253. |
F.Forest, 2009. Calibrating the Tree of Life: fossils, molecules and evolutionary timescales. Ann. Bot., 104: 789-794. DOI:10.1093/aob/mcp192 |
Friis, E.M., Crane, P.R., Pedersen, K.R., 2011. Early Flowers and Angiosperm Evolution. Cambridge University Press, Cambridge.
|
H.Hara, 1987. Notes towards a revision of the Asiatic species of the genus Smilacina. J. Fac. Sci. Univ. Tokyo Sect. III Bot., 14: 137-159. |
Harrison, T.M., Copeland, P., Kidd, W.S.F., et al, 1992. Raising tibet. Science, 255: 1663-1670. DOI:10.1126/science.255.5052.1663 |
Hopkins, D.M., 1967. The Bering Land Bridge. Stanford University Press, Stanford, California.
|
K.Katoh, H.Toh, 2008. Recent developments in the MAFFT multiple sequence alignment program. Briefings Bioinf., 9: 286-298. DOI:10.1093/bib/bbn013 |
C.Kim, K.M.Cameron, J.-H.Kim, 2017. Molecular systematics and historical biogeography of Maianthemum s.s. Am. J. Bot., 104: 939-952. DOI:10.3732/ajb.1600454 |
Kim, J.-H., Kim, D.-K., Forest, F., et al, 2010. Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Ann. Bot., 106: 775-790. DOI:10.1093/aob/mcq167 |
S.C.Kim, N.S.Lee, 2007. Generic delimitation and biogeography of Maianthemum and Smilacina (Ruscaceae sensu lato): preliminary results based on partial 3′ matK gene and trnK 3′ intron sequences of cpDNA. Plant Syst. Evol., 265: 1-12. DOI:10.1007/s00606-007-0517-2 |
J.V.LaFrankie, 1986. Morphology and taxonomy of the New World species of Maianthemum (Liliaceae). J. Arnold Arbor., 67: 371-439. DOI:10.5962/bhl.part.27393 |
J.V.LaFrankie, 1986. Transfer of the species of Smilacina to Maianthemum (Liliaceae). Taxon, 35: 584-589. DOI:10.2307/1221922 |
C.Lee, J.Wen, 2004. Phylogeny of Panax using chloroplast trnC-trnD intergenic region and the utility of trnC-trnD in interspecific studies of plants. Mol. Phylogenet. Evol., 31: 894-903. DOI:10.1016/j.ympev.2003.10.009 |
H.Li, 1990. Infrageneric system of the genus Maianthemum. Act. Bot. Yunnan, S3: 1-12. |
Liu, J.-Q., Duan, Y.-W., Hao, G., et al, 2014. Evolutionary history and underlying adaptation of alpine plants on the Qinghai–Tibet Plateau. J. Syst. Evol., 52: 241-249. DOI:10.1111/jse.12094 |
P.-L.Lu, C.W.Morden, 2014. Phylogenetic relationships among Dracaenoid genera (Asparagaceae: Nolinoideae) inferred from chloroplast DNA loci. Syst. Bot., 39: 90-104. DOI:10.1600/036364414X678035 |
Meng, Y., Nie, Z.-L., Deng, T., et al, 2014. Phylogenetics and evolution of phyllotaxy in the Solomon's seal genus Polygonatum (Asparagaceae: Polygonateae). Bot. J. Linn. Soc., 176: 435-451. DOI:10.1111/boj.12218 |
Meng, Y., Nie, Z.-L., Yang, Y.-P., et al, 2005. Karyomorphology of Maianthemum sensu lato (Polygonatae, Ruscaceae). J. Plant Res., 118: 155-162. DOI:10.1007/s10265-005-0205-7 |
Meng, Y., Wen, J., Nie, Z.-L., et al, 2008. Phylogeny and biogeographic diversification of Maianthemum (Ruscaceae: Polygonatae). Mol. Phylogenet. Evol., 49: 424-434. DOI:10.1016/j.ympev.2008.07.017 |
R.I.Milne, 2006. Northern hemisphere plant disjunctions: a window on tertiary land bridges and climate change?. Ann. Bot., 98: 465-472. DOI:10.1093/aob/mcl148 |
Muller, K., Muller, J., Quandt, D., 2010. PhyDE: Phylogenetic Data Editor, version 1.0. http://beast.bio.ed.ac.uk/tracer (2010). Accessed 23 Nov 2010.
|
Myers, N., Mittermeier, R.A., Mittermeier, C.G., et al, 2000. Biodiversity hotspots for conservation priorities. Nature, 403: 853-858. DOI:10.1038/35002501 |
Nie, Z.-L., Sun, H., Beardsley, P.M., et al, 2006. Evolution of biogeographic disjunction between eastern Asia and eastern North America in Phryma (Phrymaceae). Am. J. Bot., 93: 1343-1356. DOI:10.3732/ajb.93.9.1343 |
Nie, Z.-L., Wen, J., Gu, Z.-J., et al, 2005. Polyploidy in the flora of the Hengduan Mountains hotspot, southwestern China. Ann. Mo. Bot. Gard., 92: 275-306. |
Nylander, J.A.A., 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
|
Nylander, J.A.A., Wilgenbusch, J.C., Warren, D.L., et al, 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics, 24: 581-583. DOI:10.1093/bioinformatics/btm388 |
J.K.Piper, 1989. Light, flowering, and fruiting within patches of Smilacina racemosa and Smilacina stellata (Liliaceae). Bull. Torrey Bot. Club, 116: 247-257. DOI:10.2307/2996814 |
Rambau, A., Suchard, M.A., Xie, D., et al., 2014. Tracer v1.6, available from http://beast.bio.ed.ac.uk/tracer.
|
Rana, S.K., Luo, D., Rana, H.K., et al, 2021. Molecular phylogeny, biogeography and character evolution of the montane genus Incarvillea Juss. (Bignoniaceae). Plant Divers., 43: 1-14. DOI:10.1016/j.pld.2020.09.002 |
F.Ronquist, J.P.Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
P.J.Rudall, J.G.Conran, M.W.Chase, 2000. Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Bot. J. Linn. Soc., 134: 73-92. DOI:10.1006/bojl.2000.0365 |
Shi, Y. -F., Li, J. -J., Li, B. -Y., 1998. Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the Late Cenozoic. Guangdong Science and Technology Press, Guangzhou, China.
|
R.A.Spicer, A.Farnsworth, T.Su, 2020. Cenozoic topography, monsoons and biodiversity conservation within the Tibetan Region: an evolving story. Plant Divers., 42: 229-254. DOI:10.1016/j.pld.2020.06.011 |
Spicer, R.A., Harris, N.B.W., Widdowson, M., et al, 2003. Constant elevation of southern Tibet over the past 15 million years. Nature, 421: 622-624. DOI:10.1038/nature01356 |
A.Stamatakis, 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
H.Sun, 2002. Evolution of Arctic-Tertiary flora in Himalayan-Hengduan mountains. Acta Bot. Yunnan., 24: 671-688. |
Sun, H., Zhang, J., Deng, T., et al, 2017. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers., 39: 161-166. DOI:10.1016/j.pld.2017.09.004 |
E.Therman, 1956. Cytotaxonomy of the tribe Polygonatae. Am. J. Bot., 43: 134-142. DOI:10.1002/j.1537-2197.1956.tb10474.x |
B.H.Tiffney, 1985. Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arnold Arbor., 66: 73-94. DOI:10.5962/bhl.part.13179 |
G.Y.Wang, Y.P.Yang, 2018. Hypothesizing the origin, migration routes, and distribution patterns of Ophiopogon (Asparagaceae) in East and Southeast Asia. J. Syst. Evol., 56: 194-201. DOI:10.1111/jse.12304 |
Wang, J.-J., Yang, Y.-P., Sun, H., et al, 2016. The biogeographic south-north divide of Polygonatum (Asparagaceae tribe Polygonateae) within eastern Asia and its recent dispersals in the Northern Hemisphere. PLoS One, 11: e0166134. DOI:10.1371/journal.pone.0166134 |
Wang, Y.-J., Susanna, A., Raab-Straube, E.V., et al, 2009. Island-like radiation of Saussurea (Asteraceae: Cardueae) triggered by uplifts of the Qinghai-Tibetan Plateau. Biol. J. Linn. Soc., 97: 893-903. DOI:10.1111/j.1095-8312.2009.01225.x |
J.Wen, 1999. Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Syst., 30: 421-455. DOI:10.1146/annurev.ecolsys.30.1.421 |
Wen, J., Ickert-Bond, S., Nie, Z. -L., et al., 2010. Timing and modes of evolution of eastern Asian - North American biogeographic disjunctions in seed plants In: Long, M., Gu, H., Zhou, Z. (Eds. ), Darwin's Heritage Today: Proceedings of the Darwin 2010 Beijing International Conference. Higher Education Press, Beijing, pp. 252-269.
|
Wen, J., Zhang, J.-Q., Nie, Z.-L., et al, 2014. Evolutionary diversifications of plants on the Qinghai-Tibetan plateau. Front. Genet., 5: 4. |
C.Y.Wu, 1988. Hengduan Mountains flora and her significance. J. Jap. Bot., 63: 297-311. |
Xiang, Q.-Y., Zhang, W.H., Ricklefs, R.E., et al, 2004. Regional differences in rates of plant speciation and molecular evolution: a comparison between eastern Asia and eastern North America. Evolution, 58: 2175-2184. |
Yue, J.-P., Sun, H., Baum, D.A., et al, 2009. Molecular phylogeny of Solms-laubachia (Brassicaceae) s.l., based on multiple nuclear and plastid DNA sequences, and its biogeographic implications. J. Syst. Evol., 47: 402-415. DOI:10.1111/j.1759-6831.2009.00041.x |
Zhang, J.-W., Nie, Z.-L., Wen, J., et al, 2011. Molecular phylogeny and biogeography of three closely related genera, Soroseris, Stebbinsia, and Syncalathium (Asteraceae, Cichorieae), endemic to the Tibetan Plateau, SW China. Taxon, 60: 15-26. DOI:10.1002/tax.601003 |
Zhou, Z., Hong, D.-Y., Niu, Y., et al, 2013. Phylogenetic and biogeographic analyses of the Sino-Himalayan endemic genus Cyananthus (Campanulaceae) and implications for the evolution of its sexual system. Mol. Phylogenet. Evol., 68: 482-497. DOI:10.1016/j.ympev.2013.04.027 |



